Modulation of the Liver Protein Carbonylome by the Combined Effect of Marine Omega-3 PUFAs and Grape Polyphenols Supplementation in Rats Fed an Obesogenic High Fat and High Sucrose Diet
Abstract
1. Introduction
2. Results
2.1. Biochemical and Biometrics
2.1.1. Effects of Fish Oil Mixture (FOM) and Grape Seed Polyphenols Extract (GSE) Supplements on HFHS-Fed Rats
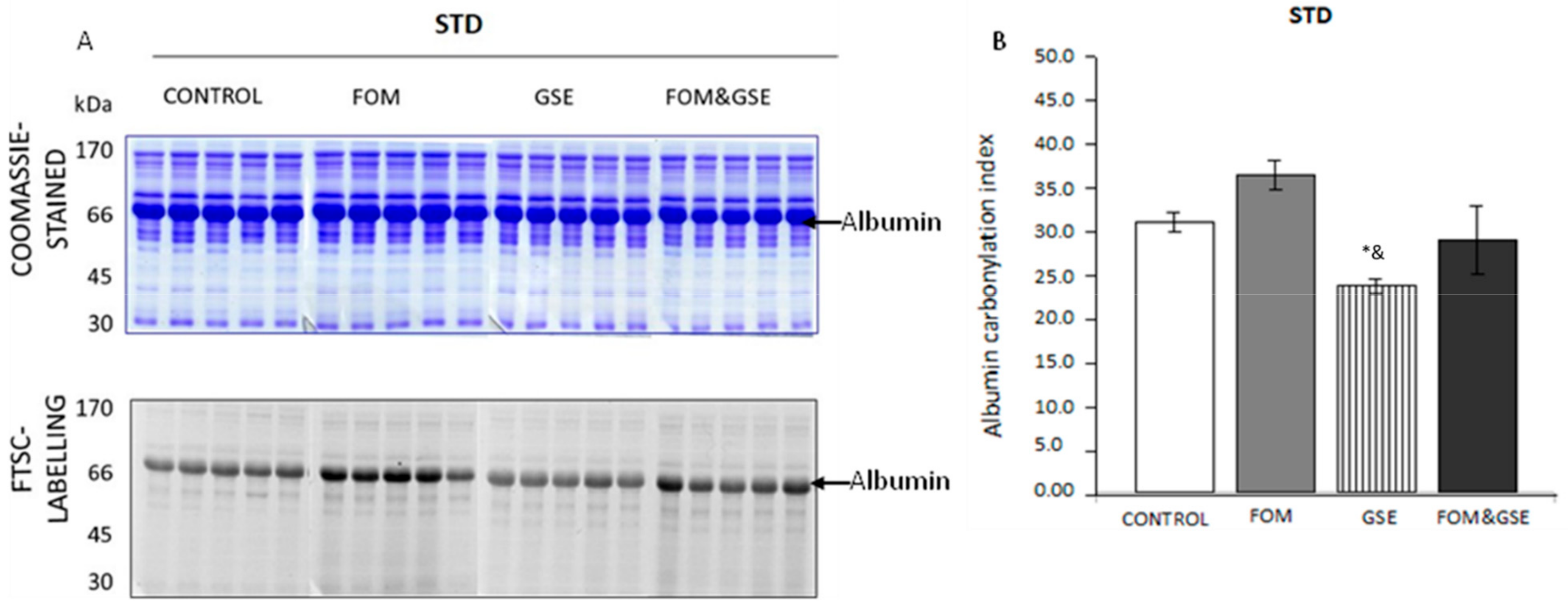
2.1.2. Effects of Fish Oil Mixture (FOM) and Grape Seed Polyphenols Extract (GSE) Supplements on STD-Fed Rats
2.2. Modulation of Total Level of Carbonylation in Plasma by Fish Oil and Grape Polyphenol Supplementations
2.3. Modulation of Total Level of Carbonylation in Liver by Fish Oil and Grape Polyphenol Supplementations
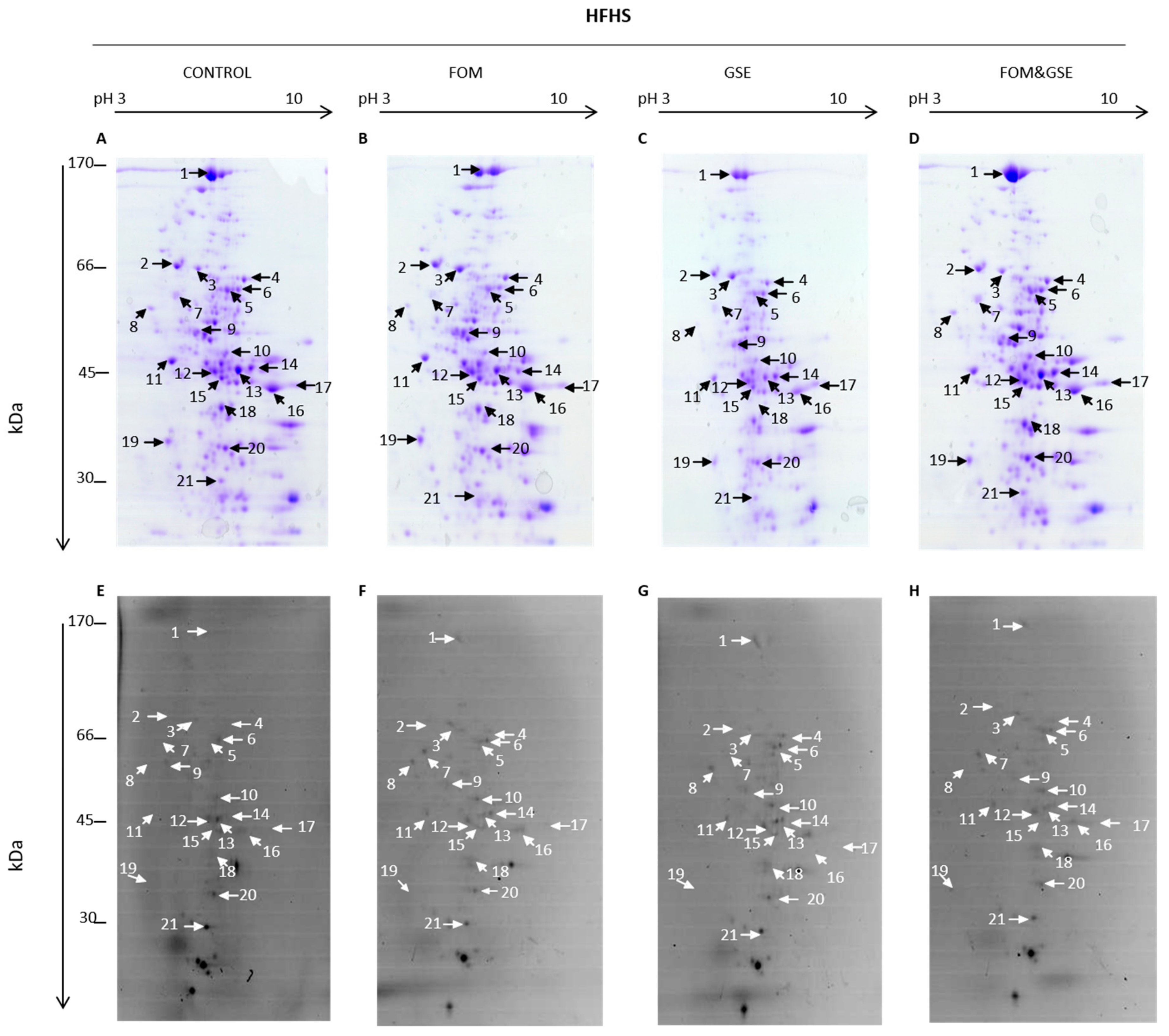
2.4. Focusing on the Protein Liver Carbonylome: Modulation of Specific Protein Carbonylation
- (a)
- Proteins which were mainly sensitive to the oxidative changes induced by one but not both supplements. Proteins belonging to this first subset were albumin, actin, Otc, and catalase, because FOM supplementation explained the behavior of the three first and GSE seemed to be the main responsible for the decrease of catalase carbonylation.
- (b)
- Proteins whose changes in carbonylation index can be explained by the “direct sum” of individual effect of the supplements (additive effect). We can include in this subset: Aldh2, because FOM and GSE showed opposite individual behavior and the combination did not show any effect compared to HFHS control; the mixture of proteins corresponding to spot 12, because both supplements exerted the same individual effects that were exacerbated after their combination. Got2 and Akr1c9 could be also included in this group. GSE increased their carbonylation index but the addition of FOM, which did not show any individual effect, made the proteins reached the carbonylation level of controls. Therefore, the addition of FOM seemed to counteract the GSE effect on these proteins.
- (c)
- Proteins that showed an “unexpected” response to the combination of supplements (synergistic effect). Serotransferrin was one of the proteins belonging to this group because the combination FOM&GSE increased its carbonylation, while individual supplementation with FOM did not have any response and GSE significantly decreased its carbonylation level. The second protein in this group was the mitochondrial 60 kDa heat shock protein (Hspd1) (spot 7), which significantly reduced its carbonylation in the FOM&GSE group but individual FOM and GSE supplements did not exert any effect. Finally, Pgd and Nit2 significantly decreased its carbonylation level in the FOM&GSE-HFHS diet, but FOM supplement exerted the opposite effect increasing protein carbonylation and GSE did not show any measurable effect.
- (a)
- Proteins which were mainly sensitive to one but not both supplements: Actb, which carbonylation index increased only through the GSE supplementation, and Nit2, whose carbonylation was reduced by FOM supplements.
- (b)
- Proteins whose changes can be explained by the sum of the individual effects of supplements (additive effect). This group included P4hb, Akr1c9, albumin, Hsp1, transketolase (Tkt) (spot 4), Pgd, alcohol dehydrogenase 1 (Adh1) (spot 16), and catalase, which were significantly influenced by one supplement but not the combination.
- (c)
- Proteins that showed an “unexpected” response to the combination of supplements (synergistic effect). The proteins included in this group were: Aldh2 and argininosuccinate synthase (Ass1) (spot 13), which presented higher carbonylation index after the combination of FOM and GSE; Got2 and fumarylacetoacetase (Fah) (spot 15), which reached the highest carbonylation level in FOM&GSE group even if FOM reduced its carbonylation and GSE increased it. Additionally, the mixture of proteins identified in the spot 12 (Acadl, Hpd, and Upb1) showed the lowest carbonylation value with the combination of both supplements and finally, Otc, which significantly increased its carbonylation whereas GSE produced the opposite effect and FOM did not affect.
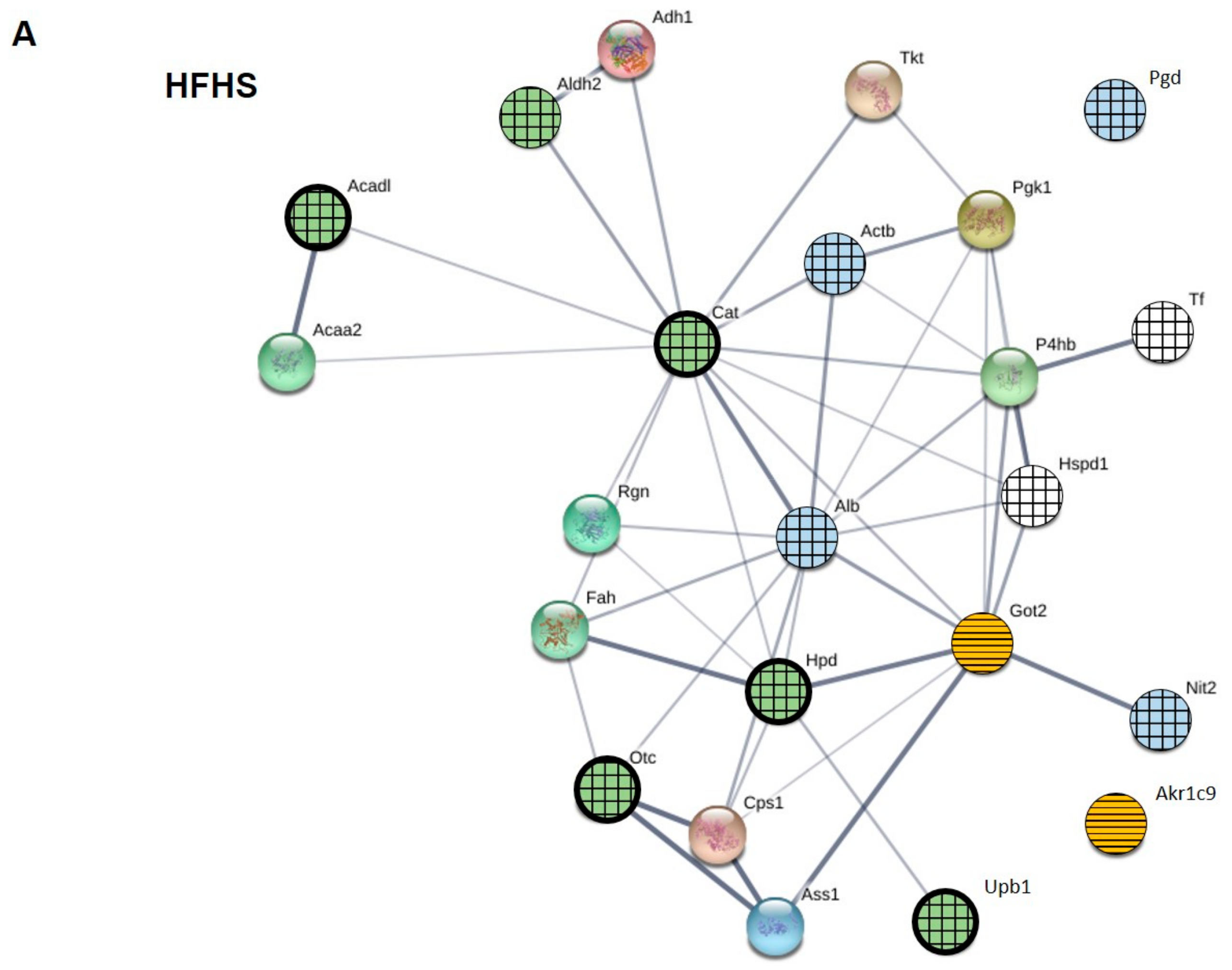
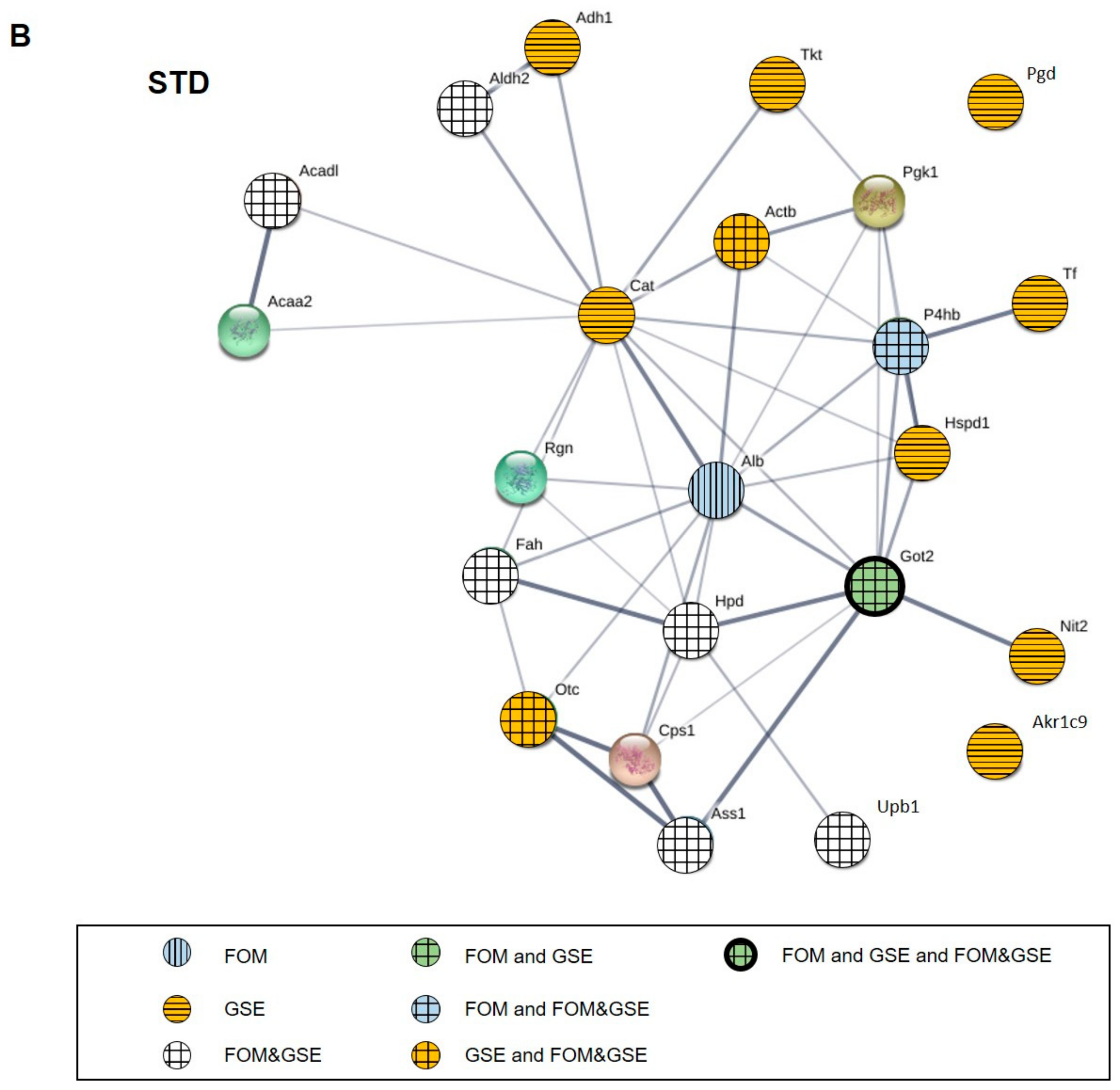
2.5. Protein–Protein Interaction (PPI) Network of Carbonylated Liver Proteins and Functional Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Animals and Diets
4.3. Biochemical Measurements
4.4. Extraction of Liver Proteins and FTSC Labelling of In Vivo-Generated Protein Carbonyls in Both Liver and Plasma
4.5. Relative Quantification of Total and Specific Protein Carbonylation
4.6. Image Analysis, Carbonylation Index Calculation, and Statistics
4.7. In-Gel Digestion and Carbonylated Protein Identification by nanoLC–ESI–IT–MS/MS
4.8. Protein–Protein Interaction (PPI) Network and Pathway and Process Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rasool, S.; Geetha, T.; Broderick, T.L.; Babu, J.R. High fat with high sucrose diet leads to obesity and induces myodegeneration. Front. Physiol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Free radicals, antioxidants, and human disease: curiosity, cause, or consequence? Lancet 1994, 344, 721–724. [Google Scholar] [CrossRef]
- Jones, D.P. Clinical Measures of the Balance. Antioxid. Redox Signal. 2006, 8, 1865–1879. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.P.; Jung, T.; Grune, T.; Siems, W. 4-Hydroxynonenal (HNE) modified proteins in metabolic diseases. Free Radic. Biol. Med. 2017, 111, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Giustarini, D.; Colombo, R.; Rossi, R.; Milzani, A. Protein carbonylation in human diseases. Trends Mol. Med. 2003, 9, 169–176. [Google Scholar] [CrossRef]
- Fedorova, M.; Bollineni, R.C.; Hoffmann, R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom. Rev. 2014, 33, 79–97. [Google Scholar] [CrossRef] [PubMed]
- Hauck, A.K.; Zhou, T.; Hahn, W.; Petegrosso, R.; Kuang, R.; Chen, Y.; Bernlohr, D.A. Obesity-induced protein carbonylation in murine adipose tissue regulates the DNA-binding domain of nuclear zinc finger proteins. J. Biol. Chem. 2018, 293, 13464–13476. [Google Scholar] [CrossRef]
- Hauck, A.K.; Huang, Y.; Hertzel, A.-V.; Bernlohr, D.A. Adipose oxidative stress and protein carbonylation. J. Biol. Chem. 2019, 294, 1083–1088. [Google Scholar] [CrossRef]
- Hecker, M.; Wagner, A.H. Role of protein carbonylation in diabetes. J. Inherit. Metab. Dis. 2018, 41, 29–38. [Google Scholar] [CrossRef]
- Boden, G.; Homko, C.; Barrero, C.A.; Stein, T.P.; Chen, X.; Cheung, P.; Fecchio, C.; Koller, S.; Merali, S. Excessive caloric intake acutely causes oxidative stress, GLUT4 carbonylation, and insulin resistance in healthy men. Sci. Transl. Med. 2015, 7. [Google Scholar] [CrossRef]
- Méndez, L.; Pazos, M.; Molinar-Toribio, E.; Sánchez-Martos, V.; Gallardo, J.M.; Nogués, M.R.; Torres, J.L.; Medina, I. Protein carbonylation associated to high-fat, high-sucrose diet and its metabolic effects. J. Nutr. Biochem. 2014, 25, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Mutch, D.M.; Wahli, W.; Williamson, G. Nutrigenomics and nutrigenetics: the emerging faces of nutrition. FASEB J. 2005, 19, 1602–1616. [Google Scholar] [CrossRef] [PubMed]
- Taltavull, N.; Ras, R.; Mariné, S.; Romeu, M.; Giralt, M.; Méndez, L.; Medina, I.; Ramos-Romero, S.; Torres, J.L.; Nogués, M.R. Protective effects of fish oil on pre-diabetes: A lipidomic analysis of liver ceramides in rats. Food Funct. 2016, 7, 3981–3988. [Google Scholar] [CrossRef] [PubMed]
- Albracht-Schulte, K.; Kalupahana, N.S.; Ramalingam, L.; Wang, S.; Rahman, S.M.; Robert-McComb, J.; Moustaid-Moussa, N. Omega-3 fatty acids in obesity and metabolic syndrome: a mechanistic update. J. Nutr. Biochem. 2018, 58, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dasilva, G.; Pazos, M.; García-Egido, E.; Gallardo, J.M.; Ramos-Romero, S.; Torres, J.L.; Romeu, M.; Nogués, M.R.; Medina, I. A lipidomic study on the regulation of inflammation and oxidative stress targeted by marine ω-3 PUFA and polyphenols in high-fat high-sucrose diets. J. Nutr. Biochem. 2017, 43, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, S.; Méndez, L.; Dasilva, G.; Torres, J.L.; Ramos-Romero, S.; Romeu, M.; Nogués, M.R.; Medina, I. Targeting hepatic protein carbonylation and oxidative stress occurring on diet-induced metabolic diseases through the supplementation with fish oils. Mar. Drugs 2018, 16, 353. [Google Scholar] [CrossRef]
- Sakai, C.; Ishida, M.; Ohba, H.; Yamashita, H.; Uchida, H.; Yoshizumi, M.; Ishida, T. Fish oil omega-3 polyunsaturated fatty acids attenuate oxidative stress-induced DNA damage in vascular endothelial cells. PLoS ONE 2017, 12, 1–13. [Google Scholar] [CrossRef]
- Giovinazzo, G.; Grieco, F. Functional Properties of Grape and Wine Polyphenols. Plant Foods Hum. Nutr. 2015, 70, 454–462. [Google Scholar] [CrossRef]
- Ramos-Romero, S.; Hereu, M.; Molinar-Toribio, E.; Almajano, M.P.M.P.; Méndez, L.; Medina, I.; Taltavull, N.; Romeu, M.; Nogués, M.R.; Torres, J.L.J.L. Effects of the combination of ω-3 PUFAs and proanthocyanidins on the gut microbiota of healthy rats. Food Res. Int. 2017, 97, 364–371. [Google Scholar] [CrossRef]
- Molinar-Toribio, E.; Fuguet, E.; Ramos-Romero, S.; Taltavull, N.; Méndez, L.; Nogués, M.R.R.; Medina, I.; Torres, J.L.J.L.; Pérez-Jiménez, J. A high-fat high-sucrose diet affects the long-term metabolic fate of grape proanthocyanidins in rats. Eur. J. Nutr. 2018, 57, 339–349. [Google Scholar] [CrossRef]
- Maestre, R.; Douglass, J.D.; Kodukula, S.; Medina, I.; Storch, J. Alterations in the Intestinal Assimilation of Oxidized PUFAs Are Ameliorated by a Polyphenol-Rich Grape Seed Extract in an In Vitro Model and Caco-2 Cells. J. Nutr. 2013, 143, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Romero, S.; Molinar-Toribio, E.; Pérez-Jiménez, J.; Taltavull, N.; Dasilva, G.; Romeu, M.; Medina, I.; Torres, J.L. The combined action of omega-3 polyunsaturated fatty acids and grape proanthocyanidins on a rat model of diet-induced metabolic alterations. Food Funct. 2016, 7, 3516–3523. [Google Scholar] [CrossRef] [PubMed]
- Méndez, L.; Ciordia, S.; Fernández, M.S.; Juárez, S.; Ramos, A.; Pazos, M.; Gallardo, J.M.; Torres, J.L.; Nogués, M.R.; Medina, I. Changes in liver proteins of rats fed standard and high-fat and sucrose diets induced by fish omega-3 PUFAs and their combination with grape polyphenols according to quantitative proteomics. J. Nutr. Biochem. 2017, 41, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Méndez, L.; Pazos, M.; Gallardo, J.M.; Torres, J.L.; Pérez-Jiménez, J.; Nogués, M.R.; Romeu, M.; Medina, I. Reduced protein oxidation in Wistar rats supplemented with marine ω3 PUFAs. Free Radic. Biol. Med. 2013, 55, 8–20. [Google Scholar] [CrossRef]
- Razack, S.; Kumar, K.; Nallamuthu, I.; Naika, M.; Khanum, F. Antioxidant, Biomolecule Oxidation Protective Activities of Nardostachys jatamansi DC and Its Phytochemical Analysis by RP-HPLC and GC-MS. Antioxidants 2015, 4, 185–203. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Aldini, G.; Domingues, M.R.; Spickett, C.M.; Domingues, P.; Altomare, A.; Sánchez-Gómez, F.J.; Oeste, C.L.; Pérez-Sala, D. Protein lipoxidation: Detection strategies and challenges. Redox Biol. 2015, 5, 253–266. [Google Scholar] [CrossRef]
- Chaudiere, J.; Gerard, D.; Clement, M.; Bourre, J.M. Induction of selenium-glutathione peroxidase by stimulation of metabolic hydrogen peroxide production in vivo. Bioelectrochem. Bioenerg. 1987, 18, 247–256. [Google Scholar] [CrossRef]
- Hu, M. Dietary Polyphenols as Antioxidants and Anticancer Agents: More Questions than Answers. Chang Gung Med. J. 2011, 34, 449–459. [Google Scholar]
- Méndez, L.; Pazos, M.; Giralt, M.; Nogués, M.R.; Pérez-Jiménez, J.; Torres, J.L.; Gallardo, J.M.; Medina, I. Targets of protein carbonylation in spontaneously hypertensive obese Koletsky rats and healthy Wistar counterparts: A potential role on metabolic disorders. J. Proteomics 2014, 106, 246–259. [Google Scholar] [CrossRef]
- Madian, A.G.; Myracle, A.D.; Diaz-Maldonado, N.; Rochelle, N.S.; Janle, E.M.; Regnier, F.E. Differential carbonylation of proteins as a function of in vivo oxidative stress. J. Proteome Res. 2011, 10, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Song, B.J.; Abdelmegeed, M.A.; Yoo, S.H.; Kim, B.J.; Jo, S.A.; Jo, I.; Moon, K.H. Post-translational modifications of mitochondrial aldehyde dehydrogenase and biomedical implications. J. Proteomics 2011, 74, 2691–2702. [Google Scholar] [CrossRef] [PubMed]
- Aldini, G.; Granata, P.; Orioli, M.; Santaniello, E.; Carini, M. Detoxification of 4-hydroxynonenal (HNE) in keratinocytes: Characterization of conjugated metabolites by liquid chromatography/electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2003, 38, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Lawless, M.; O’Byrne, K.; Gray, S. Targeting oxidative stress in cancer. Expert Opin. Ther. Targets 2010, 14, 1225–1245. [Google Scholar] [CrossRef]
- Nordgren, M.; Fransen, M. Peroxisomal metabolism and oxidative stress. Biochimie 2014, 98, 56–62. [Google Scholar] [CrossRef]
- Ottaviani, J.I.; Borges, G.; Momma, T.Y.; Spencer, J.P.E.; Keen, C.L.; Crozier, A.; Schroeter, H. The metabolome of [2-14C](-)-epicatechin in humans: Implications for the assessment of efficacy, safety, and mechanisms of action of polyphenolic bioactives. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Breitzig, M.; Bhimineni, C.; Lockey, R.; Kolliputi, N. 4-Hydroxy-2-nonenal: a critical target in oxidative stress? Am. J. Physiol. Physiol. 2016, 311, C537–C543. [Google Scholar] [CrossRef]
- Young, J.D.; Egnatchik, R.A.; Shiota, M.; Leamy, A.K.; Sacco, S.A.; Cheah, Y.E. Glutamate-oxaloacetate transaminase activity promotes palmitate lipotoxicity in rat hepatocytes by enhancing anaplerosis and citric acid cycle flux. J. Biol. Chem. 2018, 294, 3081–3090. [Google Scholar]
- Taverna, M.; Marie, A.L.; Mira, J.P.; Guidet, B. Specific antioxidant properties of human serum albumin. Ann. Intensive Care 2013, 3, 1–7. [Google Scholar] [CrossRef]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. 2008, 22, 659–661. [Google Scholar] [CrossRef]
- Knaze, V.; Zamora-Ros, R.; Luján-Barroso, L.; Romieu, I.; Scalbert, A.; Slimani, N.; Riboli, E.; Van Rossum, C.T.M.; Bueno-De-Mesquita, H.B.; Trichopoulou, A.; et al. Intake estimation of total and individual flavan-3-ols, proanthocyanidins and theaflavins, their food sources and determinants in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Br. J. Nutr. 2012, 108, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Sacks, H. Notes on methodology. Struct. Soc. Action 2010, 26, 21–27. [Google Scholar]
- Puttmann, M.; Krug, H.; Von Ochsenstein, E.; Kattermann, R. Fast HPLC determination of serum free fatty acids in the picomole range. Clin. Chem. 1993, 39, 825–832. [Google Scholar] [PubMed]
- Sharp, P.; Rainbow, S. Continuous glucose monitoring and haemoglobin A 1c. Ann. Clin. Biochem. 2002, 39, 516–517. [Google Scholar] [CrossRef] [PubMed]
- Bucolo, G.; David, H. Quantitative determination of serum triglycerides by the use of enzymes. Clin. Chem. 1973, 19, 476–482. [Google Scholar]
- Young, D.; Friedman, R. Effects of Disease on Clinical Laboratory Tests; AACC Press: Washington, DC, USA, 2001. [Google Scholar]
- Fukunaga, K.; Suzuki, T.; Takama, K. Highly sensitive high-performance liquid chromatography for the measurement of malondialdehyde in biological samples. J. Chromatogr. B Biomed. Sci. Appl. 1993, 621, 77–81. [Google Scholar] [CrossRef]
- Mendes, R.; Cardoso, C.; Pestana, C. Measurement of malondialdehyde in fish: A comparison study between HPLC methods and the traditional spectrophotometric test. Food Chem. 2009, 112, 1038–1045. [Google Scholar] [CrossRef]
- Mateos, R.; Lecumberri, E.; Ramos, S.; Goya, L.; Bravo, L. Determination of malondialdehyde (MDA) by high-performance liquid chromatography in serum and liver as a biomarker for oxidative stress: Application to a rat model for hypercholesterolemia and evaluation of the effect of diets rich in phenolic antioxidant. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005, 827, 76–82. [Google Scholar] [CrossRef]
- Cao, G.; Booth, S.L.; Sadowski, J.A.; Prior, R.L. Increased plasma antioxidant capacity with consumption of diets high in fruits and vegetables. FASEB J. 1998, 12, 3–9. [Google Scholar]
- Wheeler, C.R.; Salzman, J.A.; Elsayed, N.M.; Omaye, S.T.; Korte, D.W. Automated assays for superoxide dismutase, catalase, glutathione peroxidase, and glutathione reductase activity. Anal. Biochem. 1990, 184, 193–199. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; M, G.N.; Olson, B.J.; Klenk, D.C. Measurement of Protein Using Bicinchoninic Acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Zar, J. Biostatistical Analysis, 4th ed.; Hall, N.P., Ed.; Pearson Education: Bengaluru, India, 1994. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, XX, 37–46. [Google Scholar] [CrossRef]




| HFHS-C | HFHS-FOM | HFHS-GSE | HFHS-FOM&GSE | |
|---|---|---|---|---|
| Weight Increase (%) | 101.8 (16.9) | 114.9 (20.3) | 93.6 (14.4) | 117.4 (20.6) |
| Adiposity index (%) × | 6.62 (3.95) | 6.12 (2.62) | 4.98 (1.31) | 4.32 (1.54) |
| Hepatosomatic index (%) ∆ | 2.4 (0.1) | 2.4 (0.1) | 2.3 (0.1) | 2.7 (0.4) |
| Total cholesterol (mmol/L) | 3.5 (0.3) | 2.7 (0.5) *# | 3.7 (0.3) | 2.8 (0.3) *# |
| Triglycerides (mmol/L) | 1.3 (0.2) | 1.6 (0.3) | 2.2 (0.2) *&$ | 1.5 (0.3) |
| FFA (μg/mL) | 180.7 (24.2) | 140.2 (15.9) | 187.8 (4.5) &$ | 136.6 (25.2) *# |
| TFA (μg/mL) | 2442.2 (245.0) | 2233.0 (425.4) | 3764.9 (374.1) *&$ | 2101.4 (231.1) |
| HbA1c (%) | 3.25 (0.14) | 3.14 (0.12) | 3.11 (0.12) | 2.70 (0.70) |
| Fasting Glucose (mg/dL) | 65.86 (3.63) | 64.14 (6.87) | 65.43 (4.69) | 67.00 (3.92) |
| Plasma insulin (ng/mL) | 2.0 (0.6) | 1.6 (0.4) | 2.6 (1.2) | 1.2 (0.5) *# |
| ORAC (µmol Trolox/mL plasma) | 17.7 (7.9) | 10.7 (5.7) | 19.3 (5.0) | 24.0 (3.5) & |
| Plasma GPX(U/g Hb) | 0.4 (0.04) | 3.8 (0.9) *#$ | 13.4 (1.9) *&$ | 36.7 (8.5) *&# |
| TBARS (mg MDA/kg liver) | 3.6 (0.7) | 10.7 (2.0) *#$ | 2.7 (0.3) | 6.6 (1.4) *&# |
| Liver CRP (mg/mL) | 93.9 (53.7) | 64.3 (18.9) | 76.7 (41.7) | 60.3 (46.9) |
| Liver TNFα (mg/mL) | 91.5 (47.8) | 55.2 (49.3) * | 85.2 (59.5) | 54.5 (65.4) * |
| STD-C | STD-FOM | STD-GSE | STD-FOM&GSE | |
|---|---|---|---|---|
| Weight Increase (%) | 83.8 (22.1) | 92.9 (21.3) | 81.4 (11.6) | 88.8 (23.2) |
| Adiposity index (%) × | 1.92 (0.48) | 3.99 (1.20) * | 6.17 (3.02) * | 4.84 (1.19) * |
| Hepatosomatic index (%) ∆ | 2.6 (0.5) | 2.7 (0.2) | 2.7 (0.2) | 2.7 (0.2) |
| Total cholesterol (mmol/L) | 4.3 (1.0) | 3.8 (0.2) | 4.4 (0.3) | 3.5 (0.4) |
| Triglycerides (mmol/L) | 1.5 (0.4) | 1.7 (0.4) | 1.9 (0.3) | 1.6 (0.3) |
| FFA (μg/mL) | 212.0 (22.1) | 164.2 (32.0) | 166.6 (24.2) | 147.5 (30.0) * |
| TFA (μg/mL) | 2982.0 (772.8) | 2873.8 (343.7) | 3426.7 (338.5) | 2770.9 (438.6) |
| HbA1c (%) | 3.24 (0.15) | 3.15 (0.10) | 3.25 (0.22) | 3.35 (0.20) |
| Fasting Glucose (mg/dL) | 64.57 (2.70) | 71.00 (6.53) | 61.00 (3.37) | 64.86 (5.87) |
| Plasma insulin (ng/mL) | 0.9 (0.3) | 1.2 (0.6) | 1.2 (0.5) | 1.5 (0.5) |
| ORAC (µmol Trolox/mL plasma) | 17.6 (8.3) | 18.8 (4.4) | 16.6 (5.1) | 22.1 (2.9) |
| Plasma GPX(U/g Hb) | 0.4 (0.1) | 1.2 (0.7) *#$ | 6.1 (0.5) *&$ | 19.2 (3.01) *&# |
| TBARS (mg MDA/kg liver) | 4.5 (1.4) | 10.0 (2.4) *# | 3.2 (0.2 7) &$ | 7.6 (1.7) # |
| Liver CRP (mg/mL) | 91.4 (21.6) | 45.4 (15.7) * | 49.7 (7.9) * | 56.1 (26.5) * |
| Liver TNFα (mg/mL) | 116.0 (72.5) | 63.8 (26.3) * | 73.1 (66.7) * | 62.6 (47.6) * |
| Spot N° | UniprotKB Accession | Protein Description | Gene Name | Subcellular Localization | Avg. Mass (Da) | Coverage (%) | #Peptides (#Unique) |
|---|---|---|---|---|---|---|---|
| 1 | P07756 | Carbamoyl-phosphate synthase [ammonia] mitochondrial | Cps1 | Mitochondria | 164,579 | 70 | 73 (69) |
| 2 | P12346 | Serotransferrin | Tf | Extracellular region or secreted | 76,346 | 38 | 26 (26) |
| 3 | P02770 | Serum albumin | Alb | Extracellular region or secreted | 68,731 | 42 | 45 (45) |
| 4 | P50137 | Transketolase | Tkt | Endoplasmic reticulum/Peroxisome | 67,644 | 49 | 43 (43) |
| 5,6 | P04762 | Catalase | Cat | Peroxisome | 59,757 | 27 | 18 (18) |
| 7 | P63039 | 60 kDa heat shock protein mitochondrial | Hspd1 | Mitochondria | 60,956 | 31 | 21 (21) |
| 8 | P04785 | Protein disulfide-isomerase | P4hb | Endoplasmic reticulum | 56,951 | 38 | 25 (25) |
| 9 | P11884 | Aldehyde dehydrogenase mitochondrial | Aldh2 | Mitochondria | 56,488 | 31 | 24 (24) |
| 10 | P85968 | 6-phosphogluconate dehydrogenase decarboxylating | Pgd | Mitochondria | 53,236 | 20 | 10 (10) |
| 11 | P60711 | Actin cytoplasmic 1 | Actb | Cytoskeleton | 41,737 | 56 | 49 (24) |
| 12 | P15650 | Long-chain specific acyl-CoA dehydrogenase mitochondrial | Acadl | Mitochondria | 47,873 | 36 | 24 (24) |
| P32755 | 4-hydroxyphenylpyruvate dioxygenase | Hpd | Endoplasmic reticulum | 45,112 | 45 | 24 (24) | |
| Q03248 | Beta-ureidopropionase | Upb1 | Cytoplasm | 44,042 | 28 | 12 (12) | |
| 13 | P09034 | Argininosuccinate synthase | Ass1 | Cytosol | 46,496 | 54 | 48 (48) |
| 14 | P16617 | Phosphoglycerate kinase 1 | Pgk1 | Cytoplasm | 44,538 | 20 | 7 (7) |
| P13437 | 3-ketoacyl-CoA thiolase mitochondrial | Acaa2 | Mitochondria | 41,871 | 41 | 19 (19) | |
| 15 | P25093 | Fumarylacetoacetase | Fah | Cytosol/Extracellular region or secreted | 45,976 | 10 | 4 (4) |
| 16 | P06757 | Alcohol dehydrogenase 1 | Adh1 | Cytoplasm | 39,645 | 46 | 28 (24) |
| 17 | P00507 | Aspartate aminotransferase mitochondrial | Got2 | Mitochondria | 47,314 | 43 | 31 (30) |
| 18 | P00481 | Ornithine carbamoyltransferase mitochondrial | Otc | Mitochondria | 39,886 | 27 | 9 (9) |
| 19 | Q03336 | Regucalcin | Rgn | Cytoplasm | 33,390 | 53 | 27 (27) |
| 20 | P23457 | 3-alpha-hydroxysteroid dehydrogenase | Akr1c9 | Cytoplasm | 37,028 | 40 | 20 (12) |
| 21 | Q497B0 | Omega-amidase NIT2 | Nit2 | Cytoplasm | 30,701 | 28 | 10 (10) |
| CARBONYLATION INDEX 1 | FOLD CHANGE2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Spot N° | Protein Description | Gene Name | HFHS-C | HFHS-FOM | HFHS-GSE | HFHS-FOM&GSE | HFHS-FOM/HFHS-C | HFHS-GSE/HFHS-C | HFHS-FOM&GSE/HFHS-C |
| 1 | Carbamoyl-phosphate synthase [ammonia] mitochondrial | Cps1 | 0.27 (0.01) | 0.32 (0.14) | 0.22 (0.11) | 0.31 (0.06) | 1.19 | 0.81 | 1.15 |
| 2 | Serotransferrin | Tf | 0.24 (0.05) | 0.53 (0.25) | 0.19 (0.10) | 0.47 (0.04) *# | 2.21 | 0.79 | 1.96 |
| 3 | Serum albumin | Alb | 1.18 (0.04) | 0.50 (0.18) *# | 1.33 (0.52) & | 0.66 (0.12) * | 0.42 | 1.13 | 0.56 |
| 4 | Transketolase | Tkt | 0.64 (0.13) | 0.67 (0.01) | 0.52 (0.22) | 0.53 (0.52) | 1.05 | 0.81 | 0.83 |
| 5,6 | Catalase | Cat | 1.33 (0.43) | 0.93 (0.50) | 0.77 (0.14) * | 0.57 (0.30) * | 0.70 | 0.58 | 0.43 |
| 7 | 60 kDa heat shock protein mitochondrial | Hspd1 | 0.55 (0.07) | 0.60 (0.12) | 1.45 (0.82) | 0.31 (0.03) *& | 1.09 | 2.64 | 0.56 |
| 8 | Protein disulfide-isomerase | P4hb | 0.76 (0.15) | 0.85 (0.08) | 0.98 (3.42) | 0.93 (0.62) | 1.12 | 1.29 | 1.22 |
| 9 | Aldehyde dehydrogenase mitochondrial | Aldh2 | 0.61 (0.12) | 0.17 (0.08) *# | 2.38 (0.26) *&$ | 0.54 (0.23) # | 0.28 | 3.90 | 0.89 |
| 10 | 6-phosphogluconate dehydrogenase decarboxylating | Pgd | 1.01 (0.02) | 3.98 (1.20) *$ | 1.31 (1.20) | 0.35 (0.01) *& | 3.94 | 1.30 | 0.35 |
| 11 | Actin cytoplasmic 1 | Actb | 0.11 (0.05) | 0.43 (0.14) *# | 0.15 (0.09) | 0.69 (0.27) *# | 3.91 | 1.36 | 6.27 |
| 12 | Long-chain specific acyl-CoA dehydrogenase mitochondrial | Acadl | 1.89 (0.35) | 0.99 (0.18) *$ | 0.82 (0.13) *$ | 0.37 (0.01) *&# | 0.52 | 0.43 | 0.20 |
| 4-hydroxyphenylpyruvate dioxygenase | Hpd | ||||||||
| Beta-ureidopropionase | Upb1 | ||||||||
| 13 | Argininosuccinate synthase | Ass1 | 0.75 (0.42) | 0.42 (0.13) | 0.35 (0.17) | 0.25 (0.26) | 0.56 | 0.47 | 0.33 |
| 14 | Phosphoglycerate kinase 1 3-ketoacyl-CoA thiolase mitochondrial | Pgk1 Acaa2 | 0.22 (0.10) | 0.33 (0.09) | 0.13 (0.12) | 0.36 (0.01) | 1.50 | 0.59 | 1.64 |
| 15 | Fumarylacetoacetase | Fah | 0.80 (0.76) | 0.55 (0.37) | 0.55 (0.25) | 0.39 (0.01) | 0.69 | 0.69 | 0.49 |
| 16 | Alcohol dehydrogenase 1 | Adh1 | 1.21 (0.14) | 1.69 (0.98) | 1.55 (0.55) | 1.23 (0.21) | 1.40 | 1.28 | 1.02 |
| 17 | Aspartate aminotransferase mitochondrial | Got2 | 1.41 (0.61) | 1.01 (0.89) | 4.46 (0.94) *& | 2.63 (1.02) | 0.72 | 3.16 | 1.87 |
| 18 | Ornithine carbamoyltransferase mitochondrial | Otc | 0.86 (0.18) | 0.61 (0.06) *$ | 0.26 (0.22) * | 0.32 (0.01) *& | 0.71 | 0.30 | 0.37 |
| 19 | Regucalcin | Rgn | 0.18 (0.25) | 0.29 (0.13) | 0.51 (0.23) | 0.29 (0.15) | 1.61 | 2.83 | 1.61 |
| 20 | 3-alpha-hydroxysteroid dehydrogenase | Akr1c9 | 0.20 (0.07) | 0.19 (0.14) | 0.55 (0.15) *& | 0.30 (0.23) | 0.95 | 2.75 | 1.50 |
| 21 | Omega-amidase NIT2 | Nit2 | 1.88 (0.29) | 4.76 (1.20) *#$ | 1.76 (0.18) &$ | 0.79 (0.01) *&# | 2.53 | 0.94 | 0.42 |
| CARBONYLATION INDEX 1 | FOLD CHANGE 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Spot N° | Protein Description | Gene Name | STD-C | STD-FOM | STD-GSE | STD-FOM&GSE | STD-FOM/STD-C | STD-GSE/STD-C | STD-FOM&GSE/STD-C |
| 1 | Carbamoyl-phosphate synthase [ammonia] mitochondrial | Cps1 | 0.15 (0.14) | 0.11 (0.01) | 0.20 (0.11) | 0.19 (0.07) | 0.73 | 1.33 | 1.27 |
| 2 | Serotransferrin | Tf | 0.13 (0.06) | 0.08 (0.07) | 0.09 (0.08) | 0.19 (0.09) | 0.62 | 0.69 | 1.46 |
| 3 | Serum albumin | Alb | 0.50 (0.03) | 0.20 (0.08) *# | 0.62 (0.22) | 0.40 (0.16) | 0.40 | 1.24 | 0.80 |
| 4 | Transketolase | Tkt | 0.61 (0.32) | 0.39 (0.09) | 0.91 (0.21) &$ | 0.43 (0.10) | 0.64 | 1.49 | 0.70 |
| 5,6 | Catalase | Cat | 0.72 (0.16) | 0.96 (0.08) | 1.63 (0.46) *&$ | 1.05 (0.14) | 1.33 | 2.26 | 1.46 |
| 7 | 60 kDa heat shock protein mitochondrial | Hspd1 | 0.45 (0.25) | 0.55 (0.22) | 0.17 (0.08) &$ | 0.88 (0.27) | 1.22 | 0.38 | 1.96 |
| 8 | Protein disulfide-isomerase | P4hb | 0.60 (0.16) | 2.68 (0.79) *#$ | 0.77 (0.07) &$ | 1.37 (0.45) *&# | 4.47 | 1.28 | 2.28 |
| 9 | Aldehyde dehydrogenase mitochondrial | Aldh2 | 3.84 (0.54) | 5.43 (1.15) | 4.73 (0.97) | 6.51 (0.95) * | 1.41 | 1.23 | 1.70 |
| 10 | 6-phosphogluconate dehydrogenase decarboxylating | Pgd | 1.84 (0.16) | 1.55 (0.73) | 1.01 (0.27) * | 1.41 (0.14) | 0.84 | 0.55 | 0.77 |
| 11 | Actin cytoplasmic 1 | Actb | 0.19 (0.10) | 0.28 (0.09) | 0.42 (0.06) *& | 0.49 (0.04) *& | 1.47 | 2.21 | 2.58 |
| 12 | Long-chain specific acyl-CoA dehydrogenase mitochondrial | Acadl | 0.90 (0.54) | 2.02 (0.87) | 1.25 (0.27) | 0.46 (0.07) &# | 2.24 | 1.39 | 0.51 |
| 4-hydroxyphenylpyruvate dioxygenase | Hpd | ||||||||
| Beta-ureidopropionase | Upb1 | ||||||||
| 13 | Argininosuccinate synthase | Ass1 | 0.23 (0.06) | 0.21 (0.04) $ | 0.36 (0.03) *$ | 0.50 (0.04) *&# | 0.91 | 1.57 | 2.17 |
| 14 | Phosphoglycerate kinase 1 3-ketoacyl-CoA thiolase mitochondrial | Pgk1 Acaa2 | 0.31 (0.07) | 0.24 (0.11) | 0.27 (0.10) | 0.42 (0.38) | 0.77 | 0.87 | 1.35 |
| 15 | Fumarylacetoacetase | Fah | 0.90 (0.80) | 0.89 (0.23) #$ | 1.47 (0.21) &$ | 2.23 (0.13) *&# | 0.99 | 1.63 | 2.48 |
| 16 | Alcohol dehydrogenase 1 | Adh1 | 0.79 (0.11) | 0.62 (0.12) | 1.03 (0.22) &$ | 0.58 (0.21) | 0.78 | 1.30 | 0.73 |
| 17 | Aspartate aminotransferase mitochondrial | Got2 | 0.23 (0.06) | 0.13 (0.01) *#$ | 0.47 (0.00) *& | 0.54 (0.12) *& | 0.57 | 2.04 | 2.35 |
| 18 | Ornithine carbamoyltransferase mitochondrial | Otc | 0.69 (1.16) | 0.70 (0.13) #$ | 0.38 (0.11) &$ | 1.56 (0.15) &# | 1.01 | 0.55 | 2.26 |
| 19 | Regucalcin | Rgn | 0.28 (0.09) | 0.60 (0.27) | 0.19 (0.21) | 0.38 (0.19) | 2.14 | 0.68 | 1.36 |
| 20 | 3-alpha-hydroxysteroid dehydrogenase | Akr1c9 | 0.85 (0.14) | 0.97 (0.22) # | 0.65 (0.09) *& | 0.76 (0.19) | 1.14 | 0.76 | 0.89 |
| 21 | Omega-amidase NIT2 | Nit2 | 4.84 (2.42) | 2.60 (0.34) *# | 3.82 (0.44) * | 2.68 (0.32) *# | 0.54 | 0.79 | 0.55 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méndez, L.; Muñoz, S.; Miralles-Pérez, B.; Nogués, M.R.; Ramos-Romero, S.; Torres, J.L.; Medina, I. Modulation of the Liver Protein Carbonylome by the Combined Effect of Marine Omega-3 PUFAs and Grape Polyphenols Supplementation in Rats Fed an Obesogenic High Fat and High Sucrose Diet. Mar. Drugs 2020, 18, 34. https://doi.org/10.3390/md18010034
Méndez L, Muñoz S, Miralles-Pérez B, Nogués MR, Ramos-Romero S, Torres JL, Medina I. Modulation of the Liver Protein Carbonylome by the Combined Effect of Marine Omega-3 PUFAs and Grape Polyphenols Supplementation in Rats Fed an Obesogenic High Fat and High Sucrose Diet. Marine Drugs. 2020; 18(1):34. https://doi.org/10.3390/md18010034
Chicago/Turabian StyleMéndez, Lucía, Silvia Muñoz, Bernat Miralles-Pérez, Maria Rosa Nogués, Sara Ramos-Romero, Josep Lluis Torres, and Isabel Medina. 2020. "Modulation of the Liver Protein Carbonylome by the Combined Effect of Marine Omega-3 PUFAs and Grape Polyphenols Supplementation in Rats Fed an Obesogenic High Fat and High Sucrose Diet" Marine Drugs 18, no. 1: 34. https://doi.org/10.3390/md18010034
APA StyleMéndez, L., Muñoz, S., Miralles-Pérez, B., Nogués, M. R., Ramos-Romero, S., Torres, J. L., & Medina, I. (2020). Modulation of the Liver Protein Carbonylome by the Combined Effect of Marine Omega-3 PUFAs and Grape Polyphenols Supplementation in Rats Fed an Obesogenic High Fat and High Sucrose Diet. Marine Drugs, 18(1), 34. https://doi.org/10.3390/md18010034






