Octominin: A Novel Synthetic Anticandidal Peptide Derived from Defense Protein of Octopus minor
Abstract
:1. Introduction
2. Results
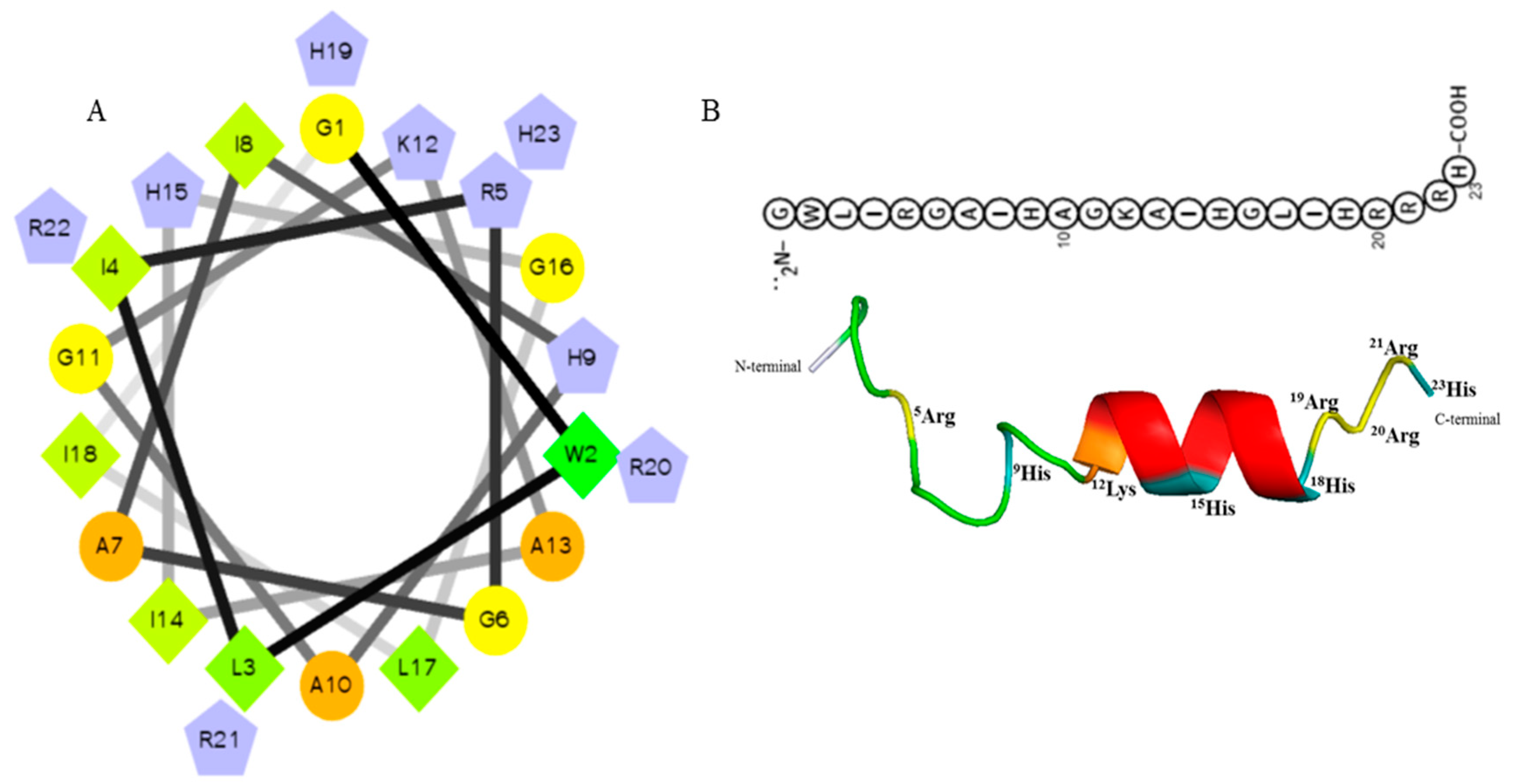
2.1. Designing, Synthesis, and Characteristics of Octominin
2.2. MIC, MFC, and Growth Inhibition Profile of C. albicans Exposed to Octominin
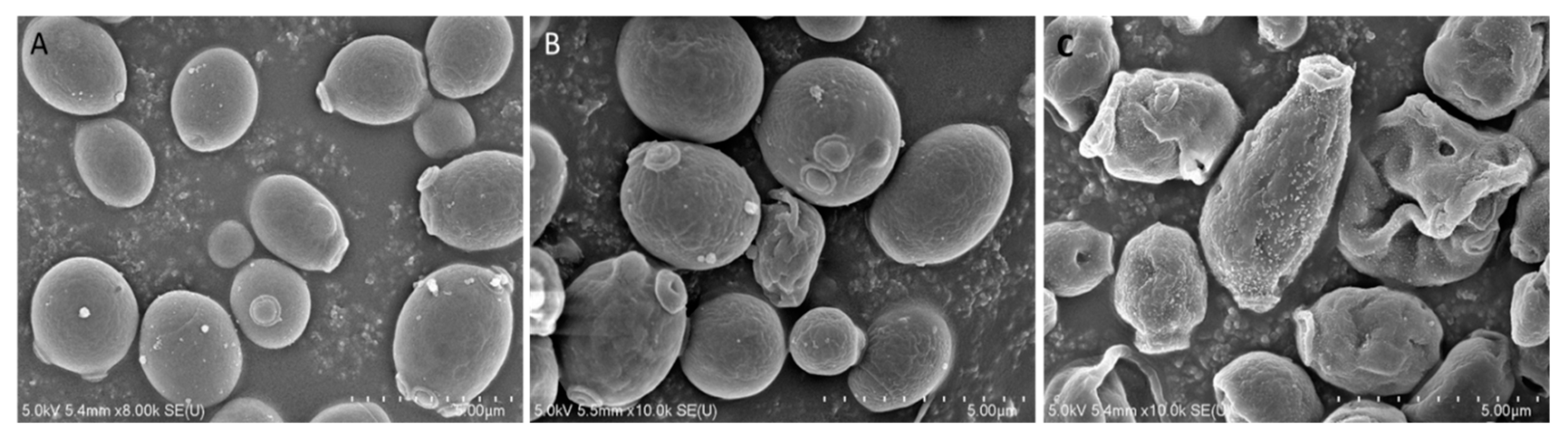
2.3. Effects of Octominin on Morphological Changes of C. albicans
2.4. Effects of Octominin on the Viability of C. albicans
2.5. Effect of Octominin on the Membrane Permeability of C. albicans
2.6. Effects of Octominin on ROS Production in C. albicans
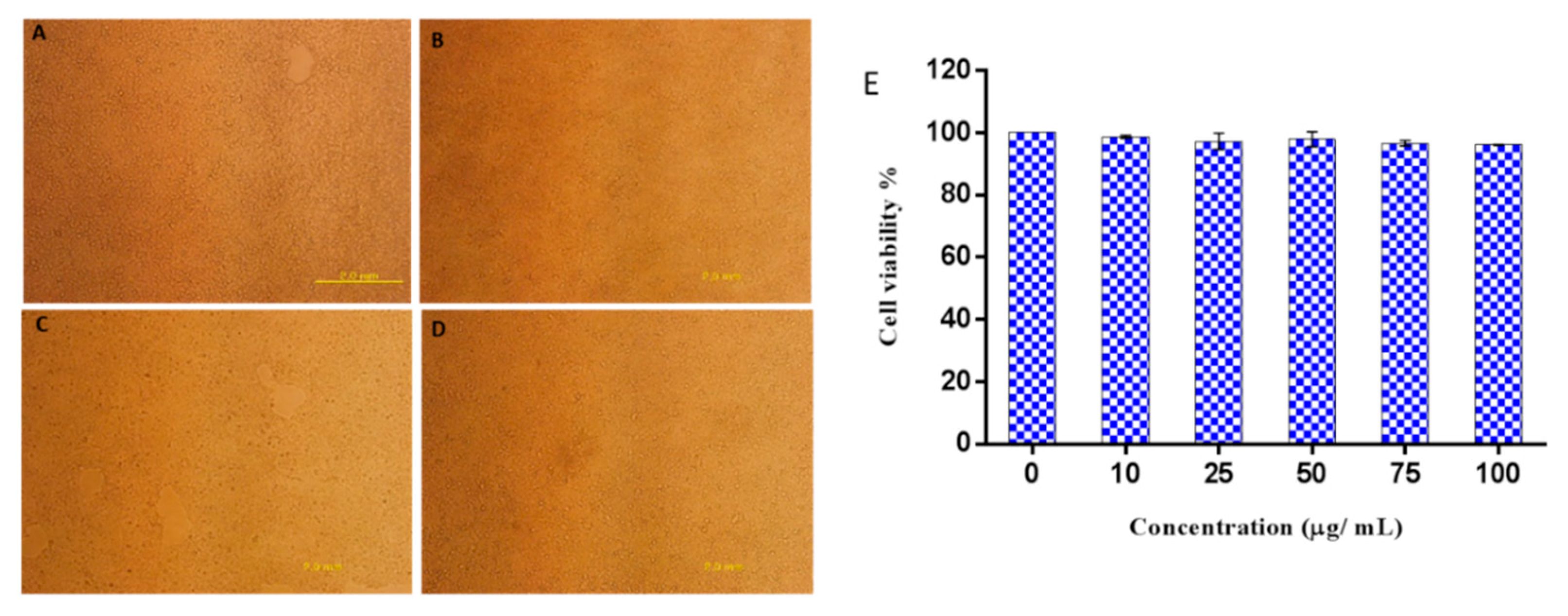
2.7. Cytotoxicity of Octominin to Mammalian Cells
2.8. Efficacy of Octominin Treatment on C. albicans Infection in a Zebrafish Model
3. Discussion
4. Materials and Methods
4.1. Design and Synthesis of the Screened AMP in O. minor
4.2. Culture of C. albicans
4.3. Determination of MIC and MFC of Octominin for C. albicans
4.4. Determination of the Effect of Octominin on C. albicans Cell Viability
4.5. Analysis of Morphological Changes of C. albicans upon Octominin Treatment
4.6. Determination of the Effect of Octominin on the Membrane Permeability of C. albicans
4.7. Effect of Octominin on ROS Production in C. albicans
4.8. Analysis of Octominin Cytotoxicity on Mammalian Cells
4.9. In Vivo Efficacy of Octominin upon C. albicans Infection in a Zebrafish Model
4.9.1. Zebrafish Husbandry
4.9.2. Determination of In Vivo Efficacy of Octominin upon C. albicans Infection
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- De Alteriis, E.; Maselli, V.; Falanga, A.; Galdiero, S.; Di Lella, F.M.; Gesuele, R.; Guida, M.; Galdiero, E. Efficiency of gold nanoparticles coated with the antimicrobial peptide indolicidin against biofilm formation and development of Candida spp. clinical isolates. Infect. Drug Resist. 2018, 11, 915–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manohar, V.; Ingram, C.; Gray, J.; Talpur, N.A.; Echard, B.W.; Bagchi, D.; Preuss, H.G. Antifungal activities of origanum oil against Candida albicans. Mol. Cell Biochem. 2001, 228, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Amigo-Benavent, M.; Wang, S.; Mateos, R.; Sarriá, B.; Bravo, L. Antiproliferative and cytotoxic effects of green coffee and yerba mate extracts, their main hydroxycinnamic acids, methylxanthine and metabolites in different human cell lines. Food Chem. Toxicol. 2017, 106, 125–138. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Lee, D.G. A novel mechanism of fluconazole: Fungicidal activity through dose-dependent apoptotic responses in Candida albicans. Microbiology 2018, 164, 194–204. [Google Scholar] [CrossRef]
- Samaranayake, Y.H.; Cheung, B.P.K.; Wang, Y.; Yau, J.Y.Y.; Yeung, K.W.S.; Samaranayake, L.P. Fluconazole resistance in Candida glabrata is associated with modification of its virulence attributes. J. Med. Microbiol. 2012, 62, 303–318. [Google Scholar] [CrossRef] [Green Version]
- Pfaller, M.A. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012, 125, 3–13. [Google Scholar] [CrossRef]
- Zhang, L.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R1–R21. [Google Scholar] [CrossRef]
- Giuliani, A.; Pirri, G.; Nicoletto, S.F. Antimicrobial peptides: An overview of a promising class of therapeutics. Cent. Eur. J. Biol. 2007, 2, 1–33. [Google Scholar] [CrossRef]
- Lai, Y.; Gallo, R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Abarrategui, C.; Alba, A.; Silva, O.N.; Reyes-Acosta, O.; Vasconcelos, I.M.; Oliveira, J.T.; Migliolo, L.; Costa, M.P.; Costa, C.R.; Silva, M.R.; et al. Functional characterization of a synthetic hydrophilic antifungal peptide derived from the marine snail Cenchritis muricatus. Biochimie 2012, 94, 968–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silphaduang, U.; Noga, E.J. Peptide antibiotics in mast cells of fish. Nature 2001, 414, 268. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.G.; Yan, H.; Hancock, R.E. Biological properties of structurally related α-helical cationic antimicrobial peptides. Infect. Immun. 1999, 67, 2005–2009. [Google Scholar] [PubMed]
- Gallo, R.L.; Ono, M.; Povsic, T.; Page, C.; Eriksson, E.; Klagsbrun, M.; Bernfield, M. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proc. Natl. Acad. Sci. USA 1994, 91, 11035–11039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Zoysa, M.; Nikapitiya, C.; Whang, I.; Lee, J.S.; Lee, J. Abhisin: A potential antibacterial peptide derived from histone H2A of disk abalone. Fish Shellfish Immun. 2009, 27, 639–646. [Google Scholar] [CrossRef]
- Ribeiro, S.F.; Carvalho, A.O.; Da Cunha, M.; Rodrigues, R.; Cruz, L.P.; Melo, V.M.; Vasconcelos, I.M.; Melo, E.J.; Gomes, V.M. Isolation and characterization of novel peptides from chilli pepper seeds: Antimicrobial activities against pathogenic yeasts. Toxicon 2007, 50, 600–611. [Google Scholar] [CrossRef]
- Ye, X.Y.; Ng, T.B. Isolation of a new cyclophilin-like protein from chickpeas with mitogenic, antifungal and anti-HIV-1 reverse transcriptase activities. Life Sci. 2002, 70, 1129–1138. [Google Scholar] [CrossRef]
- Chia, T.J.; Wu, Y.C.; Chen, J.Y.; Chi, S.C. Antimicrobial peptides (AMP) with antiviral activity against fish nodavirus. Fish Shellfish Immunol. 2010, 28, 434–439. [Google Scholar] [CrossRef]
- Park, Y.; Jang, S.H.; Lee, D.G.; Hahm, K.S. Antinematodal effect of antimicrobial peptide, PMAP-23, isolated from porcine myeloid against Caenorhabditis elegans. J. Pept. Sci. 2004, 10, 304–311. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Park, S.C.; Kim, J.Y.; Jeong, C.; Yoo, S.; Hahm, K.S.; Park, Y. A plausible mode of action of pseudin-2, an antimicrobial peptide from Pseudis paradoxa. Biochim. Biophys. Acta. 2011, 1808, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diehnelt, C.W. Peptide array based discovery of synthetic antimicrobial peptides. Front. Microbiol. 2013, 4, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Wang, G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004, 32, D590–D592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Zoysa, M. Chapter 35: Antimicrobial Peptides in Marine Mollusks and their Potential Applications. In Marine Proteins and Peptides: Biological Activities and Applications, 1st ed.; Wiley-Blackwell: Chichester, UK, 2013; pp. 695–707. [Google Scholar]
- Iijima, N.; Tanimoto, N.; Emoto, Y.; Morita, Y.; Uematsu, K.; Murakami, T.; Nakai, T. Purification and characterization of three isoforms of chrysophsin, a novel antimicrobial peptide in the gills of the red sea bream, Chrysophrys major. Eur. J. Biochem. 2003, 270, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Kim, D.H.; Park, Y.; Kim, H.K.; Kim, H.N.; Shin, Y.K.; Choi, C.H.; Hahm, K.S. Fungicidal Effect of Antimicrobial Peptide, PMAP-23, Isolated from Porcine Myeloid against Candida albicans. Biochem. Biophys. Res. Commun. 2001, 282, 570–574. [Google Scholar] [CrossRef]
- Nikawa, H.; Fukushima, H.; Makihira, S.; Hamada, T.; Samaranayake, L.P. Fungicidal effect of three new synthetic cationic peptides against Candida albicans. Oral Dis. 2004, 10, 221–228. [Google Scholar] [CrossRef] [Green Version]
- Bellamy, W.; Wakabayashi, H.; Takase, M.; Kawase, K.; Shimamura, S.; Tomita, M. Killing of Candida albicans by lactoferricin B, a potent antimicrobial peptide derived from the N-terminal region of bovine lactoferrin. Med. Microbiol. Immunol. 1993, 182, 97–105. [Google Scholar] [CrossRef]
- Lee, D.G.; Park, Y.; Kim, H.N.; Kim, H.K.; Kim, P.I.; Choi, B.H.; Hahm, K.S. Antifungal mechanism of an antimicrobial peptide, HP (2–20), derived from N-terminus of Helicobacter pylori ribosomal protein L1 against Candida albicans. Biochim. Biophys. Research Commun. 2002, 291, 1006–1013. [Google Scholar] [CrossRef]
- Dananjaya, S.H.S.; Udayangani, R.M.C.; Oh, C.; Nikapitiya, C.; Lee, J.; De Zoysa, M. Green synthesis, physio-chemical characterization and anti-candidal function of a biocompatible chitosan gold nanocomposite as a promising antifungal therapeutic agent. RSC Adv. 2017, 7, 9182–9193. [Google Scholar] [CrossRef] [Green Version]
- Maurya, I.K.; Pathak, S.; Sharma, M.; Sanwal, H.; Chaudhary, P.; Tupe, S.; Deshpande, M.; Chauhan, V.S.; Prasad, R. Antifungal activity of novel synthetic peptides by accumulation of reactive oxygen species (ROS) and disruption of cell wall against Candida albicans. Peptides 2011, 32, 1732–1740. [Google Scholar] [CrossRef]
- Veerman, E.C.; Nazmi, K.; Van’t Hof, W.; Bolscher, J.G.; Den Hertog, A.L.; Nieuw Amerongen, A.V. Reactive oxygen species play no role in the candidacidal activity of the salivary antimicrobial peptide histatin 5. Biochem. J. 2004, 381, 447–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavanagh, K.; Dowd, S. Histatins: Antimicrobial peptides with therapeutic potential. J. Pharm. Pharmacol. 2004, 56, 285–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peixoto, L.R.; Rosalen, P.L.; Ferreira, G.L.; Freires, I.A.; de Carvalho, F.G.; Castellano, L.R.; de Castro, R.D. Antifungal activity, mode of action and anti-biofilm effects of Laurus nobilis Linnaeus essential oil against Candida spp. Arch. Oral Biol. 2017, 73, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Kulatunga, D.C.M.; Dananjaya, S.H.S.; Godahewa, G.I.; Lee, J.; De Zoysa, M. Chitosan silver nanocomposite (CAgNC) as an antifungal agent against Candida albicans. Med. Mycol. 2017, 55, 213–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikapitiya, C.; Dananjaya, S.H.S.; Chandrarathna, H.P.S.U.; De Zoysa, M.; Whang, I. Octominin: A Novel Synthetic Anticandidal Peptide Derived from Defense Protein of Octopus minor. Mar. Drugs 2020, 18, 56. https://doi.org/10.3390/md18010056
Nikapitiya C, Dananjaya SHS, Chandrarathna HPSU, De Zoysa M, Whang I. Octominin: A Novel Synthetic Anticandidal Peptide Derived from Defense Protein of Octopus minor. Marine Drugs. 2020; 18(1):56. https://doi.org/10.3390/md18010056
Chicago/Turabian StyleNikapitiya, Chamilani, S.H.S. Dananjaya, H.P.S.U. Chandrarathna, Mahanama De Zoysa, and Ilson Whang. 2020. "Octominin: A Novel Synthetic Anticandidal Peptide Derived from Defense Protein of Octopus minor" Marine Drugs 18, no. 1: 56. https://doi.org/10.3390/md18010056
APA StyleNikapitiya, C., Dananjaya, S. H. S., Chandrarathna, H. P. S. U., De Zoysa, M., & Whang, I. (2020). Octominin: A Novel Synthetic Anticandidal Peptide Derived from Defense Protein of Octopus minor. Marine Drugs, 18(1), 56. https://doi.org/10.3390/md18010056




