Verrucosamide, a Cytotoxic 1,4-Thiazepane-Containing Thiodepsipeptide from a Marine-Derived Actinomycete
Abstract
1. Introduction
2. Results
2.1. Isolation and Structure Elucidation
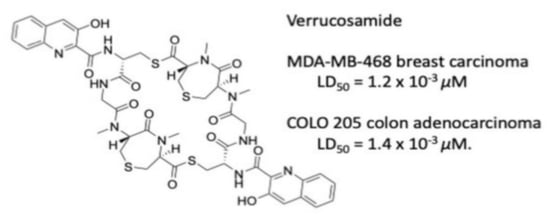
2.2. Bioactivity
2.3. Structures of 4–6
3. Conclusions
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Strain Isolation and Identification
4.3. Strain Cultivation
4.4. Extraction and Purification
4.5. X-ray Analysis of Verrucosamide (1)
4.6. Bioactivity Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981/ to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Jahromi, S.T.; Poorsaheli, H.B.; Vianello, F. Metabolites from marine microorganisms, Micro, and Macroalgae: Immense Scope for Pharmacology. Mar. Drugs 2019, 17, 464. [Google Scholar] [CrossRef] [PubMed]
- Laatsch, H. Marine bacterial metabolites. In Frontiers in Marine Biotechnology; Proksch, P., Müller, W.E.G., Eds.; Horizon Bioscience: Norfolk, UK, 2006; pp. 225–288. ISBN 1-904933-18-1. [Google Scholar]
- Okada, H.; Suzuki, H.; Yoshinari, T.; Arakawa, H.; Okura, A.; Suda, H. A new topoisomerase II inhibitor, BE-22179, produced by a Streptomycete. I. producing strain, fermentation, isolation and biological activity. J. Antibiot. 1994, 47, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Baz, J.P.; Canedo, L.M.; Fernandez-Puentes, J.L.; Silva Elipe, M.V. Thiocoraline, a novel depsipeptide with antitumor activity produced by a marine Micromonospora. II. Physico-chemical properties and structure determination. J. Antibiot. 1997, 50, 738–741. [Google Scholar] [CrossRef]
- Romero, F.; Espliego, F.; Baz, J.P.; de Quesada, T.G.; Gravalos, D.; de la Calle, F.; Fernandez-Puentes, J.L. Thiocoraline, a new depsipeptide with antitumor activity produced by a marine Micromospora. J. Antibiot. 1997, 50, 734–737. [Google Scholar] [CrossRef]
- Wyche, T.P.; Hou, Y.; Braun, D.; Cohen, H.C.; Xiong, M.P.; Bugni, T.S. First natural analogs of the cytotoxic thiodepsipeptide thiocoraline A from a marine Verrucosispora sp. J. Org. Chem. 2011, 76, 6542–6547. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, H.; Shoji, J.; Kawano, K.; Kyogoku, Y. Structure confirmation of triostin A by 1H and 13C magnetic resonance. J. Antibiot. 1976, 29, 107–110. [Google Scholar] [CrossRef]
- Waring, M.J. Echinomycin and related quinoxaline antibiotics. In Molecular Aspects of Anticancer Drug-DNA Interactions; Neidle, S., Waring, M.J., Eds.; Macmillan & Co.: London, UK, 1993; Volume 1, pp. 213–242. [Google Scholar]
- Waring, M.J.; Wakelin, L.P.G. Echinomycin: A bifunctional intercalating antibiotic. Nature 1974, 252, 653–657. [Google Scholar] [CrossRef]
- Ughetto, G.; Wang, A.H.; Quigley, G.J.; van der Marel, G.A.; van Boom, J.H.; Rich, A. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985, 13, 2305–2323. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Machado, H.; Jang, K.H.; Trzoss, L.; Jensen, P.R.; Fenical, W. Integration of genomc data with NMR analysis enables assignment of the full stereostructure of neaumycin B, a potent inhibitor of glioblastoma from a marine-derived Micromonospora. J. Am. Chem. Soc. 2018, 140, 10775–10784. [Google Scholar] [CrossRef]
- Kim, M.C.; Cullum, R.; Machado, H.; Smith, A.J.; Yang, I.; Rodvold, J.J.; Fenical, W. Photopiperazines A-D, photosensitive interconverting diketopiperazines with significant and selective activity against U87 glioblastoma cells, from a rare, marine-derived actinomycete of the family Streptomycetaceae. J. Nat. Prod. 2019, 82, 2262–2267. [Google Scholar] [CrossRef]
- Hammons, J.C.; Trzoss, L.; Jimenez, P.C.; Hirata, A.S.; Lotufo, L.C.; La Clair, J.J.; Fenical, W. Advance of Seriniquinone Analogs as Melanoma Agents. ACS Med. Chem. Lett. 2019, 10, 186–190. [Google Scholar] [CrossRef]
- Flack, H.D. On enantiomorph-polarity estimation. Acta Crystallogr. 1983, A39, 876–881. [Google Scholar] [CrossRef]
- Shaaban, K.A.; Shepherd, M.D.; Ahmed, T.A.; Nybo, S.E.; Leggas, M.; Rohr, J. Pyramidamycins A-D and 4-hydroxyquinoline-2-carboxamide; cytotoxic benzamides from Streptomyces sp. DGC1. J. Antibiot. 2012, 65, 615–622. [Google Scholar] [CrossRef]
- Ortiz-Lopez, F.J.; Alcalde, E.; Sarmiento-Vizcaino, A.; Diaz, C.; Cautain, B.; Garcia, L.A.; Blanco, G.; Reyes, F. New 3-Hydroxyquinaldic Acid Derivatives from Cultures of the Marine Derived Actinomycete Streptomyces cyaneofuscatus M-157. Mar. Drugs 2018, 16, 371. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Koshino, H.; Esumi, Y.; Tsuda, E.; Kurosawa, K. SW-163C and E, novel antitumor depsipeptide produced by Streptomyces sp. II. Structure elucidation. J. Antibiot. 2001, 54, 622–627. [Google Scholar] [CrossRef]
- Matson, J.A.; Bush, J.A. Sandramycin, a novel antitumor antibiotic produced by a Nocardioides sp. Production, isolation, characterization and biological properties. J. Antibiot. 1989, 42, 1763–1767. [Google Scholar] [CrossRef]
- Mellor, A.L.; Munn, D.; Chandler, P.; Keskin, D.; Johnson, T.; Marshall, B.; Jhaver, K.; Baban, B. Tryptophan catabolism and T cell responses. Adv. Exp. Med. Biol. 2003, 527, 27–35. [Google Scholar] [CrossRef]
- Stone, T.W.; Darlington, L.G. Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug. Discov. 2002, 1, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Kurnasov, O.; Jablonski, L.; Polanuyer, B.; Dorrestein, P.; Begley, T.; Osterman, A. Aerobic tryptophan degradation pathway in bacteria: Novel kynurenine formamidase. FEMS Microbiol. Lett. 2003, 227, 219–227. [Google Scholar] [CrossRef]



| No. | 1 | |||
|---|---|---|---|---|
| δC, mult a | δH, mult (J in Hz) b | COSY | HMBC | |
| OH | - | 11.89, br s | - | - |
| 1 | 196.8, C | - | - | - |
| 2 | 69.8, CH | 4.71, t (5.5) | H-6 | C-1, C-3, C-6, C-14 |
| 3 | 172.0, C | - | - | - |
| 4 | 59.6, CH | 5.33, dd (10.6, 2.9) | H-5 | C3, C-5, C-7, C-15 |
| 5 | 27.8, CH2 | 2.81, m c 2.37, m c | H-4 | C-3, C-4, C-6 |
| 6 | 29.0, CH2 | 2.90, m c | H-2 | C-1, C-2, C-5 |
| 7 | 170.0, C | - | - | - |
| 8 | 40.8, CH2 | 4.47, dd (10.6, 2.9), 4.32, dd (10.6, 2.9) | H-9 | C-7 |
| 9 | - | 8.03, br s | H-8 | - |
| 10 | 169.2, C | - | - | - |
| 11 | 55.1, CH | 4.61, m | H-16 | C-10, C-16, C-13 |
| 12 | - | 9.87, br d (0.6) | H-11 | - |
| 13 | 168.8, C | - | - | - |
| 14 | 38.6, CH3 | 3.14, s (N-CH3) | - | C-2, C-3 |
| 15 | 31.4, CH3 | 3.23, s (N-CH3) | - | C-4, C-7 |
| 16 | 27.3, CH2 | 4.11, d (5.4) 3.32, d (3.4) | H-11 | C-1, C-10, C-11 |
| 17 | 135.4, C | - | - | - |
| 18 | 154.2, C | - | - | - |
| 19 | 119.8, CH | 7.52, m c | C-17, C-18, C-23, C-23a | |
| 19a | 132.2, C | - | - | - |
| 20 | 126.7, CH | 7.75, m c | H-21, H-22 | C-19, C-22, C-23a |
| 21 | 129.3, CH | 7.68, m c | H-20, H-22, H-23 | C-19a, C-23 |
| 22 | 128.7, CH | 7.53, m c | H-20, H-21, H-23 | C-20, C-23, C-23a |
| 23 | 127.4, CH | 7.53, m c | H-21, H-22 | C-21, C-23, C-23a |
| 23a | 141.2, C | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nair, V.; Kim, M.C.; Golen, J.A.; Rheingold, A.L.; Castro, G.A.; Jensen, P.R.; Fenical, W. Verrucosamide, a Cytotoxic 1,4-Thiazepane-Containing Thiodepsipeptide from a Marine-Derived Actinomycete. Mar. Drugs 2020, 18, 549. https://doi.org/10.3390/md18110549
Nair V, Kim MC, Golen JA, Rheingold AL, Castro GA, Jensen PR, Fenical W. Verrucosamide, a Cytotoxic 1,4-Thiazepane-Containing Thiodepsipeptide from a Marine-Derived Actinomycete. Marine Drugs. 2020; 18(11):549. https://doi.org/10.3390/md18110549
Chicago/Turabian StyleNair, Vimal, Min Cheol Kim, James A. Golen, Arnold L. Rheingold, Gabriel A. Castro, Paul R. Jensen, and William Fenical. 2020. "Verrucosamide, a Cytotoxic 1,4-Thiazepane-Containing Thiodepsipeptide from a Marine-Derived Actinomycete" Marine Drugs 18, no. 11: 549. https://doi.org/10.3390/md18110549
APA StyleNair, V., Kim, M. C., Golen, J. A., Rheingold, A. L., Castro, G. A., Jensen, P. R., & Fenical, W. (2020). Verrucosamide, a Cytotoxic 1,4-Thiazepane-Containing Thiodepsipeptide from a Marine-Derived Actinomycete. Marine Drugs, 18(11), 549. https://doi.org/10.3390/md18110549