A Green Enzymatic Extraction Optimization and Oxidative Stability of Krill Oil from Euphausia Superba
Abstract
:1. Introduction
2. Result and Discussion
2.1. Optimization of Extraction Parameters
2.1.1. Predicted Model and Statistical Analysis
2.1.2. Response Surface Plot
2.1.3. Verification of Predictive Model
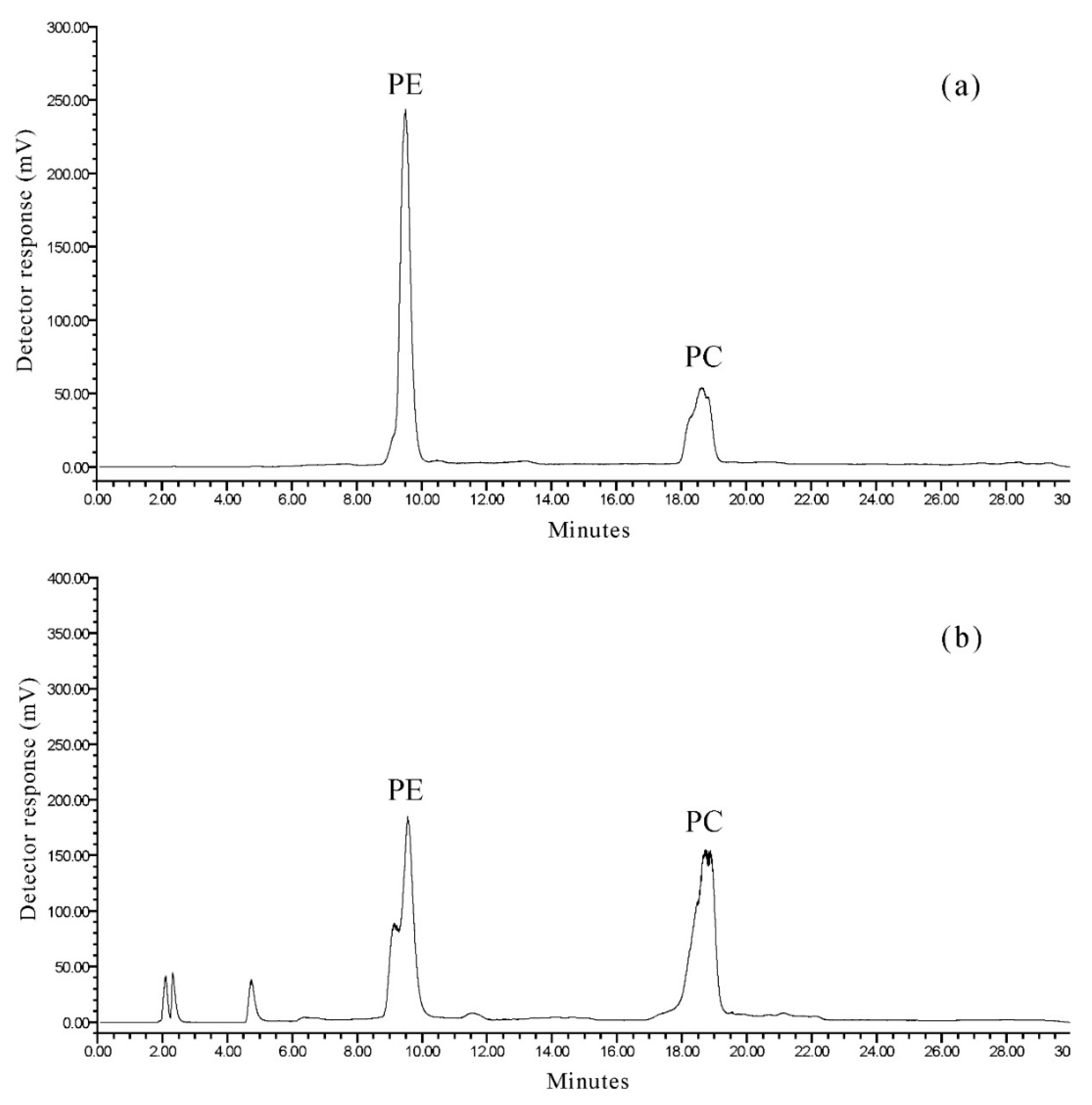
2.2. Separation of Phospholipid Classes
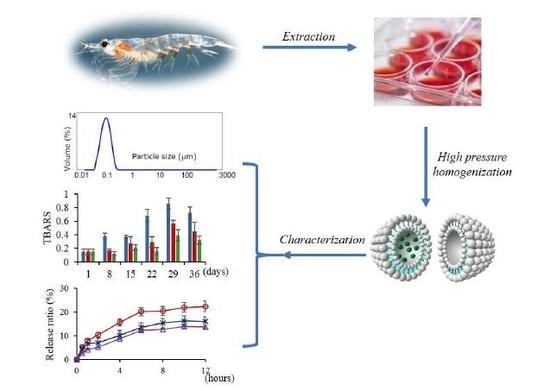
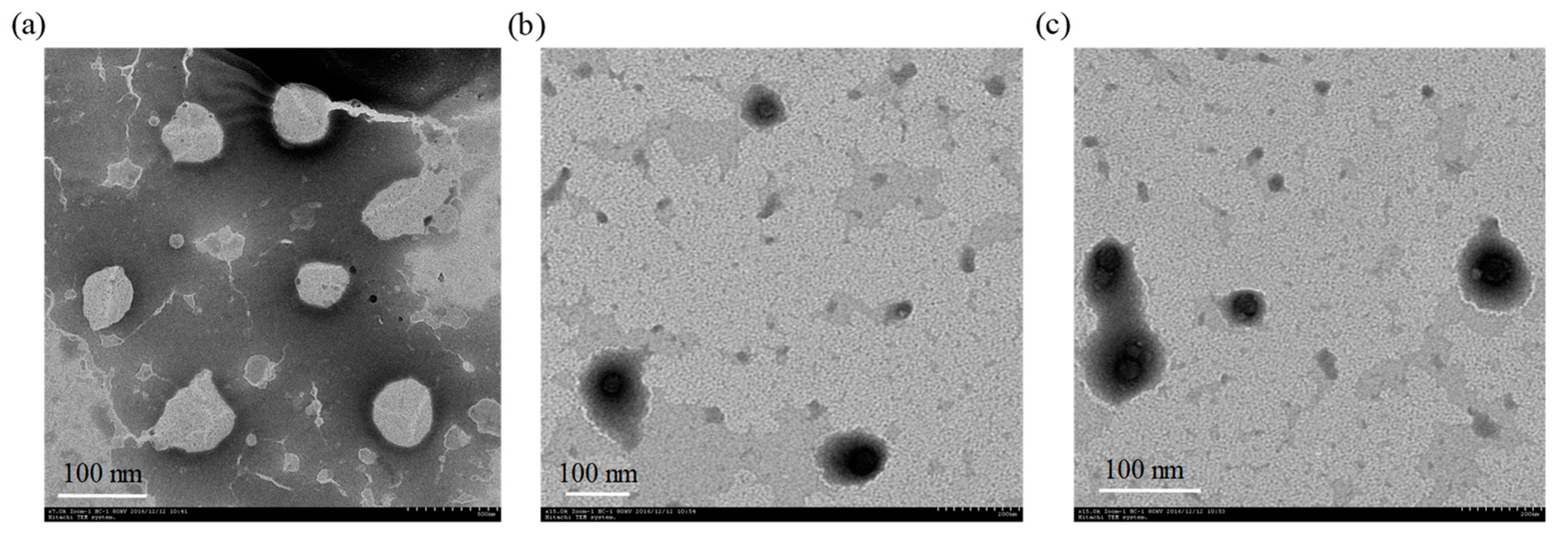
2.3. Characterization of Nanoliposomes
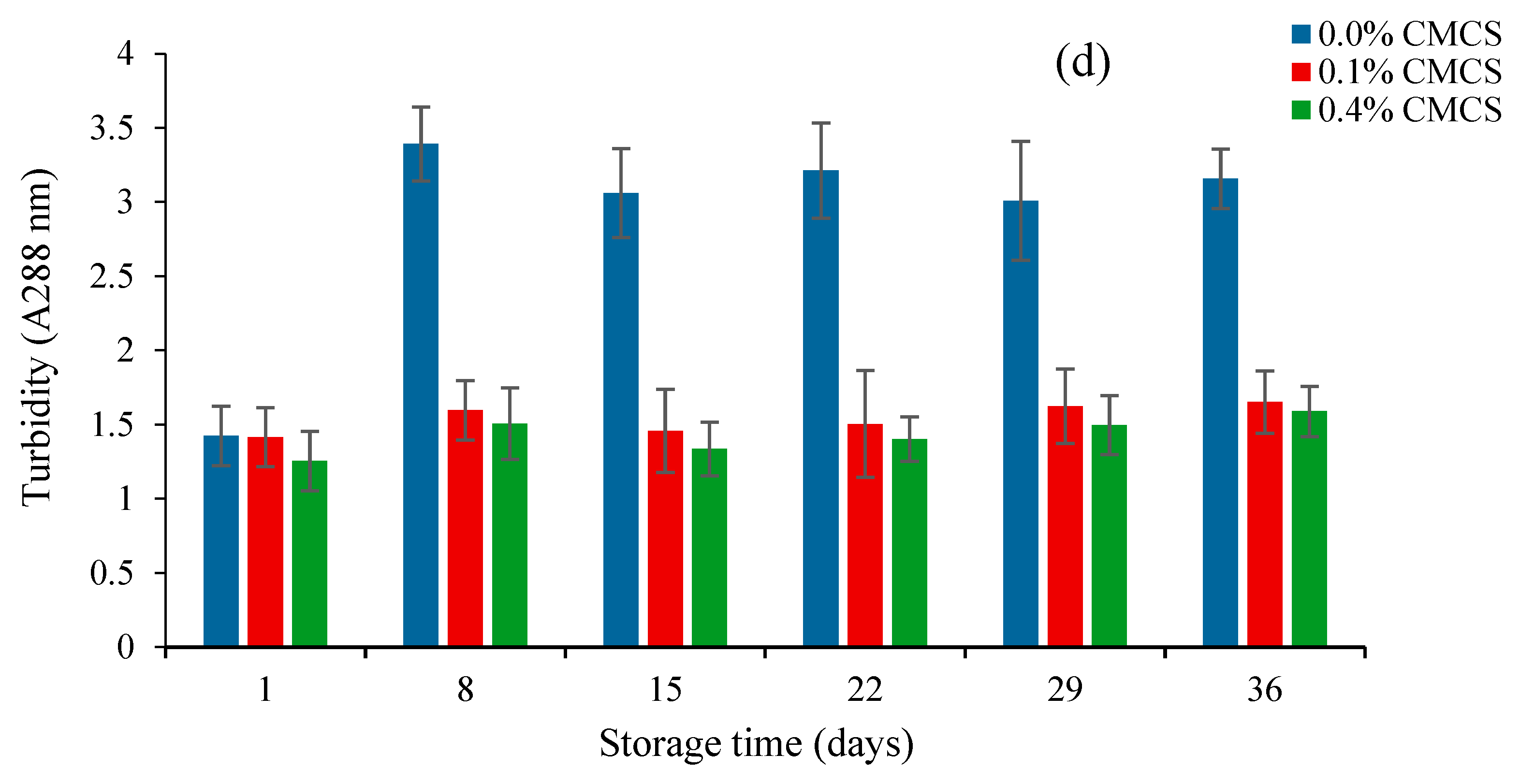
2.4. Oxidative Stability of Nanoliposomes
2.5. In Vitro Release of Krill Oil from Nanoliposomes
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Krill Oil Extraction
3.3. Experimental Design of RSM
3.4. Separation of Phospholipid Classes of Krill Oil
3.5. Preparation of Nanoliposome
3.6. Characterization of Nanoliposomes
3.7. Encapsulated Ratio of Krill Oil
3.8. Measurements of Lipid Oxidation
3.8.1. Peroxide Value (POV)
3.8.2. Thiobarbituric Acid-Reactive Substances (TBARS)
3.8.3. pH Value
3.8.4. Turbidity
3.9. In Vitro Release of Krill Oil from Nanoliposomes
3.10. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lu, F.S.H.; Nielsen, N.S.; Timm-Heinrich, M.; Jacobsen, C. Oxidative stability of marine phospholipids in the liposomal form and their applications. Lipids 2011, 46, 3–23. [Google Scholar] [PubMed]
- Zhou, L.; Li, P.X.; Zhao, Y.L.; Hou, S.; Cong, B.L.; Huang, J.; Ding, Y.; Zeng, X.X. Optimization of lipid extraction and determination of fatty acid compositions in edible meats of freshwater and marine shrimps. J. Aquat. Food Prod. Technol. 2017, 26, 824–834. [Google Scholar] [CrossRef]
- Burri, L.; Hoem, N.; Banni, S.; Berge, K. Marine omega-3 phospholipids: Metabolism and biological activities. Int. J. Mol. Sci. 2012, 13, 15401–15419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijendran, V.; Huang, M.C.; Diau, G.Y.; Boehm, G.; Nathanielsz, P.W.; Brenna, J.T. Efficacy of dietary arachidonic acid provided as triglyceride or phospholipid as substrates for brain arachidonic acid accretion in baboon neonates. Pediatr. Res. 2002, 51, 265–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Zhao, M.J.; Ennahar, S.; Bindler, F.; Marchioni, E. Identification of oxidation compounds of 1-stearoyl-2-linoleoyl-sn-glycero-3-phosphoethanolamine during thermal oxidation. J. Agric. Food Chem. 2015, 63, 9615–9620. [Google Scholar] [CrossRef]
- Zhou, L.; Le Grandois, J.; Marchioni, E.; Zhao, M.J.; Ennahar, S.; Bindler, F. Improvement of total lipid and glycerophospholipid recoveries from various food matrices using pressurized liquid extraction. J. Agric. Food Chem. 2010, 58, 9912–9917. [Google Scholar] [CrossRef]
- Gigliotti, J.C.; Davenport, M.P.; Beamer, S.K.; Tou, J.C.; Jaczynski, J. Extraction and characterization of lipids from Antarctic krill (Euphausia superba). Food Chem. 2011, 125, 1028–1036. [Google Scholar] [CrossRef]
- Zhang, Y.; Kong, X.Y.; Wang, Z.M.; Sun, Y.M.; Zhu, S.; Li, L.H.; Lv, P.M. Optimization of enzymatic hydrolysis for effective lipid extraction from microalgae Scenedesmus sp. Renew Energ. 2018, 125, 1049–1057. [Google Scholar] [CrossRef]
- Araujo, P.; Zhu, H.; Breivik, J.F.; Hjelle, J.I.; Zeng, Y. Determination and structural elucidation of triacylglycerols in krill oil by chromatographic techniques. Lipids 2014, 49, 163–172. [Google Scholar] [CrossRef]
- Wu, Q.; Uluata, S.; Cui, L.; Wang, C.; Li, D.S.; Mcclements, J.; Decker, E.A. Physical and oxidation stability of self-emulsifying krill oil-in-water emulsions. Food Funct. 2016, 7, 3590–3598. [Google Scholar] [CrossRef]
- Zamora-Mora, V.; Fernández-Gutiérrez, M.; González-Gómez, Á.; Sanz, B.; Román, J.; Goya, G.F.; Hernández, R.; Mijangos, C. Chitosan nanoparticles for combined drug delivery and magnetic hyperthermia: From preparation to in vitro studies. Carbohyd. Polym. 2017, 157, 361–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekar, V.; Rajendran, K.; Vallinayagam, S.; Deepak, V.; Mahadevan, S. Synthesis and characterization of chitosan ascorbate nanoparticles for therapeutic inhibition for cervical cancer and their in silico modeling. J. Ind. Eng Chem. 2018, 62, 239–249. [Google Scholar] [CrossRef]
- Bowman, K.; Leong, K.W. Chitosan nanoparticles for oral drug and gene delivery. Int. J. Nanomed. 2016, 1, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wang, Y.J.; Yip, X.P.; Glynn, F.; Shepherd, R.K.; Caruso, F. Nanoporous peptide particles for encapsulating and releasing neurotrophic factors in an animal model of neurodegeneration. Adv. Mater. 2012, 24, 3362–3366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumirska, J.; Weinhold, M.X.; Thoming, J.; Stepnowski, P. Biomedical activity of chitin/chitosan based materials-influence of physicochemical properties apart from molecular weight and degree of N-Acetylation. Polymers 2011, 3, 1875–1901. [Google Scholar] [CrossRef]
- Banik, N.; Ramteke, A.; Maji, T.K. Carboxymethyl chitosan-montmorillonite nanoparticles for controlled delivery of isoniazid: Evaluation of the effect of the glutaraldehyde and montmorillonite. Polym. Adv Technol. 2014, 25, 1580–1589. [Google Scholar] [CrossRef]
- Sheng, Z.P.; Bhail, S.; Sanguansri, L. Improving the oxidative stability of krill oil-in-water emulsions. J. Am. Oil Chem. Soc. 2014, 91, 1347–1354. [Google Scholar] [CrossRef]
- Haider, H.; Majeed, H.; Williams, P.A.; Safdar, W.; Zhong, F. Formation of chitosan nanoparticles to encapsulate krill oil (Euphausia superba) for application as a dietary supplement. Food Hydrocolloid. 2017, 63, 27–34. [Google Scholar] [CrossRef]
- Firatligil-Durmus, E.; Evranuz, O. Response surface methodology for protein extraction optimization of red pepper seed (Capsicum frutescens). LWT-Food Sci. Technol. 2010, 43, 226–231. [Google Scholar] [CrossRef]
- Wei, Z.J.; Liao, A.M.; Zhang, H.X.; Liu, J.; Jiang, S.T. Optimization of supercritical carbon dioxide extraction of silkworm pupal oil applying the response surface methodology. Bioresource. Technol. 2009, 110, 232–238. [Google Scholar] [CrossRef]
- Héron, S.; Dreux, M.; Tchapla, A. Post-column addition as a method of controlling triacylglycerol response coefficient of an evaporative light scattering detector in liquid chromatography-evaporative light-scattering detection. J. Chromatogr. A 2004, 1035, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Mertins, O.; Dimova, R. Binding of chitosan to phospholipid vesicles studied with isothermal titration calorimetry. Langmuir 2011, 27, 5506–5515. [Google Scholar] [CrossRef] [PubMed]
- Roohinejad, S.; Oey, I.; Wen, J.; Lee, S.J.; Everett, D.W.; Burritt, D.J. Formation of oil-in-water β-carotene microemulsions: Effect of oil type and fatty acid chain length. Food Chem. 2015, 174, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Wilson, K.; Walle, C.F.V.D.; Sattar, N.; Petrie, J.R.; Kumar, M.N.V.R. Microemulsions for oral delivery of insulin: Design, development and evaluation in streptozotocin induced diabetic rats. Eur. J. Pharm. Biopharm. 2010, 76, 159–169. [Google Scholar] [CrossRef]
- Takeungwongtrakul, S.; Benjakul, S. Oxidative stability of shrimp oil-in-water emulsions as affected by antioxidant incorporation. Int. Aquatic. Res. 2013, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Chaijan, M.; Benjakul, S.; Visessanguan, W.; Faustman, C. Changes of lipids in sardine (Sardinella gibbosa) muscle during iced storage. Food Chem. 2006, 99, 83–91. [Google Scholar] [CrossRef]
- Sahoo, S.; Sasmal, A.; Nanda, R.; Phani, A.R.; Nayak, P.L. Synthesis of chitosan-polycaprolactone blend for control delivery of ofloxacin drug. Carbohyd. Polym. 2010, 79, 106–113. [Google Scholar] [CrossRef]
- McDonald, R.E.; Hultin, H.O. Some characteristics of the enzymic lipid peroxidation system in the microsomal fraction of flounder skeletal muscle. J. Food Sci. 1987, 52, 15–21. [Google Scholar] [CrossRef]
- Hu, M.; McClements, D.J.; Decker, E.A. Impact of whey protein emulsifiers on the oxidative stability of salmon oil-in-water emulsions. J. Agric Food Chem. 2003, 51, 1435–1439. [Google Scholar] [CrossRef]
- Xia, S.Q.; Xu, S.Y. Ferrous sulfate liposomes: Preparation, stability and application in fluid milk. Food Res. Int. 2005, 38, 289–296. [Google Scholar] [CrossRef]



| Source | Df d | Mean Square | F-Value | p-Value |
|---|---|---|---|---|
| Model | 9 | 102.34 | 54.46 | <0.0001 |
| X1 a | 1 | 4.71 | 2.51 | 0.1573 |
| X2 b | 1 | 3.21 | 1.71 | 0.2323 |
| X3 c | 1 | 94.60 | 50.34 | 0.0002 |
| X1X2 | 1 | 1.13 | 0.60 | 0.4627 |
| X1X3 | 1 | 0.14 | 0.075 | 0.7923 |
| X2X3 | 1 | 4.12 | 2.19 | 0.1822 |
| X12 | 1 | 39.75 | 21.15 | 0.0025 |
| X22 | 1 | 15.93 | 8.48 | 0.0226 |
| X32 | 1 | 719.26 | 382.74 | <0.0001 |
| Lack of fit | 3 | 13.15 | 4.38 | |
| Pure error | 4 | 0.000 | 0.000 | |
| R2 | 0.9859 | |||
| CV% | 1.76% |
| PL Class | Equation: ax2 + bx + c | R2 | ||
|---|---|---|---|---|
| a | b | c | ||
| PE | 6.0 × 107 | 1.0 × 107 | 155,617 | 0.9991 |
| PC | 5.0 × 107 | 1.0 × 107 | 40,616 | 0.9998 |
| CMCS (w/v) | Particle Size (nm) | PDI | Zeta Potential | Encapsulated Ratio (%) |
|---|---|---|---|---|
| 0.0% | 185.07 ± 2.15 | 0.14 ± 0.03 | −38.43 ± 0.11 | 85.4 ± 1.4 |
| 0.1% | 181.56 ± 3.16 | 0.16 ± 0.02 | −38.84 ± 1.36 | 87.33 ± 0.92 |
| 0.2% | 194.3 ± 2.2 | 0.19 ± 0.02 | −40.26 ± 0.82 | 84.6 ± 0.2 |
| 0.3% | 202.73 ± 3.14 | 0.15 ± 0.00 | −41.67 ± 0.47 | 84.79 ± 1.58 |
| 0.4% | 185.67 ± 2.45 | 0.15 ± 0.03 | −42.18 ± 0.92 | 88.23 ± 1.27 |
| 0.5% | 228.16 ± 4.19 | 0.19 ± 0.01 | −42.09 ± 1.39 | 86.45 ± 2.04 |
| Run | Uncoded Variables | Coded Variables a | Lipid Yield (Y), % | |||||
|---|---|---|---|---|---|---|---|---|
| χ1 | χ2 | χ3 | X1 | X2 | X3 | Experimental | Predicted | |
| 1 | 0.15 | 3 | 45 | 0 | 0 | 0 | 86.33 | 86.33 |
| 2 | 0.10 | 2 | 45 | −1 | −1 | 0 | 81.33 | 81.71 |
| 3 | 0.15 | 4 | 40 | 0 | 1 | −1 | 76.46 | 75.13 |
| 4 | 0.20 | 2 | 45 | 1 | −1 | 0 | 82.96 | 82.18 |
| 5 | 0.15 | 3 | 45 | 0 | 0 | 0 | 86.33 | 86.33 |
| 6 | 0.15 | 2 | 50 | 0 | −1 | 1 | 68.2 | 69.52 |
| 7 | 0.10 | 4 | 45 | −1 | 1 | 0 | 78.6 | 79.38 |
| 8 | 0.10 | 3 | 50 | −1 | 0 | 1 | 67.5 | 65.79 |
| 9 | 0.10 | 3 | 40 | −1 | 0 | −1 | 72.5 | 73.05 |
| 10 | 0.15 | 3 | 45 | 0 | 0 | 0 | 86.33 | 86.33 |
| 11 | 0.15 | 3 | 45 | 0 | 0 | 0 | 86.33 | 86.33 |
| 12 | 0.20 | 4 | 45 | 1 | 1 | 0 | 82.36 | 81.98 |
| 13 | 0.15 | 2 | 40 | 0 | −1 | −1 | 75.3 | 74.37 |
| 14 | 0.20 | 3 | 40 | 1 | 0 | −1 | 72.5 | 74.21 |
| 15 | 0.15 | 3 | 45 | 0 | 0 | 0 | 86.33 | 86.33 |
| 16 | 0.15 | 4 | 50 | 0 | 1 | 1 | 65.3 | 66.23 |
| 17 | 0.20 | 3 | 50 | 1 | 0 | 1 | 68.25 | 67.70 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Yang, F.; Zhang, M.; Liu, J. A Green Enzymatic Extraction Optimization and Oxidative Stability of Krill Oil from Euphausia Superba. Mar. Drugs 2020, 18, 82. https://doi.org/10.3390/md18020082
Zhou L, Yang F, Zhang M, Liu J. A Green Enzymatic Extraction Optimization and Oxidative Stability of Krill Oil from Euphausia Superba. Marine Drugs. 2020; 18(2):82. https://doi.org/10.3390/md18020082
Chicago/Turabian StyleZhou, Li, Fu Yang, Minghao Zhang, and Jikai Liu. 2020. "A Green Enzymatic Extraction Optimization and Oxidative Stability of Krill Oil from Euphausia Superba" Marine Drugs 18, no. 2: 82. https://doi.org/10.3390/md18020082
APA StyleZhou, L., Yang, F., Zhang, M., & Liu, J. (2020). A Green Enzymatic Extraction Optimization and Oxidative Stability of Krill Oil from Euphausia Superba. Marine Drugs, 18(2), 82. https://doi.org/10.3390/md18020082