Glyceroglycolipid Metabolism Regulations under Phosphate Starvation Revealed by Transcriptome Analysis in Synechococcus elongatus PCC 7942
Abstract
:1. Introduction
2. Results and Discussion
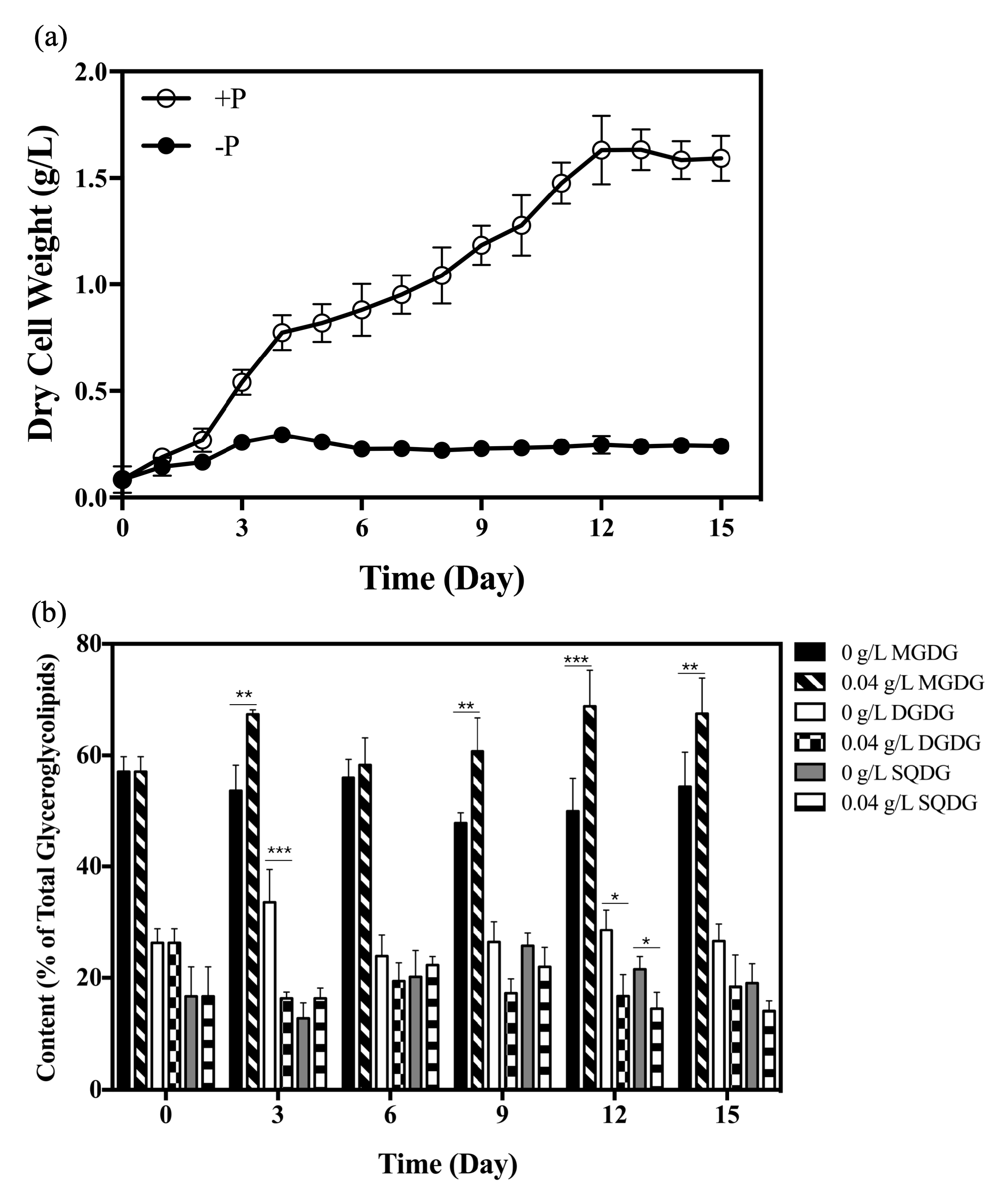
2.1. Changes in Glyceroglycolipid Composition in Synechococcus elongatus PCC 7942 under Phosphate Starvation
2.2. Expressions of Glyceroglycolipid Synthase Genes in Synechococcus elongatus PCC 7942 under Different Phosphate Concentrations
2.3. Global Transcriptomic Analysis under Different Phosphate Concentrations
2.4. Differential Expressions of Genes Involved in Glyceroglycolipid Synthesis
2.5. Glyceroglycolipid Homeostasis in Glyceroglycolipid Synthesis under Phosphate Starvation
2.6. Regulatory Networks Involved in Glyceroglycolipid Synthesis
2.7. qRT-PCR Confirmations of Differentially Expressed Transcripts
2.8. Reconstruction of Putative Glyceroglycolipid Regulatory Networks Based on Transcriptomic Evidence
3. Materials and Methods
3.1. Cyanobacteria Species and Treatments
3.2. Glyceroglycolipid Analysis
3.3. RNA Extraction, Library Preparation and Sequencing
3.4. Transcript Quantification and Differential Expression Analysis
3.5. Experimental Validation of Gene Expression with qRT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Boudiere, L.; Michaud, M.; Petroutsos, D.; Rebeille, F.; Falconet, D.; Bastien, O.; Roy, S.; Finazzi, G.; Rolland, N.; Jouhet, J.; et al. Glycerolipids in photosynthesis: Composition, synthesis and trafficking. Biochim. Biophys. Acta 2014, 1837, 470–480. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Z.; Miao, X. Nitrogen and hydrophosphate affects glycolipids composition in microalgae. Sci. Rep. 2016, 6, 30145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, C.; Yu, G.; Guan, H. Total synthesis and structure-activity relationship of glycoglycerolipids from marine organisms. Mar. Drugs 2014, 12, 3634–3659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrianasolo, E.H.; Haramaty, L.; Vardi, A.; White, E.; Lutz, R.; Falkowski, P. Apoptosis-Inducing Galactolipids from a Cultured Marine Diatom, Phaeodactylum tricornutum. J. Nat. Prod. 2008, 71, 1197–1201. [Google Scholar] [CrossRef] [Green Version]
- Riccio, G.; De Luca, D.; Lauritano, C. Monogalactosyldiacylglycerol and Sulfolipid Synthesis in Microalgae. Mar. Drugs 2020, 18, 237. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, T.; Nagatsu, A.; Murakami, N.; Sakakibara, J.; Tokuda, H.; Nishino, H.; Iwashima, A. Anti-tumour-promoting glyceroglycolipids from the green alga, chlorella vulgaris. Phytochemistry 1995, 40, 1433–1437. [Google Scholar] [CrossRef]
- Shirahashi, H.; Murakami, N.; Watanabe, M.; Nagatsu, A.; Sakakibara, J.; Tokuda, H.; Nishino, H.; Iwashima, A. Isolation and identification of anti-tumor-promoting principles from the fresh-water cyanobacterium Phormidium tenue. Chem. Pharm. Bull. 1993, 41, 1664–1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushina, Y.; Watanabe, I.; Ohtaa, K.; Takemura, M.; Sahara, H.; Takahashi, N.; Gasa, S.; Sugawara, F.; Matsukage, A.; Yoshida, S.; et al. Studies on inhibitors of mammalian DNA polymerase α and β: Sulfolipids from a pteridophyte, Athyrium niponicum. Biochem. Pharmacol. 1998, 55, 537–541. [Google Scholar] [CrossRef]
- Gustafson, K.R.; Cardellina, J.H.; Fuller, R.W.; Weislow, O.S.; Kiser, R.F.; Snader, K.M.; Patterson, G.M.L.; Boyd, M.R. AIDS-Antiviral Sulfolipids From Cyanobacteria (Blue-Green Algae). J. Natl. Cancer Inst. 1989, 81, 1254–1258. [Google Scholar] [CrossRef]
- Loya, S.; Reshef, V.; Mizrachi, E.; Silberstein, C.; Rachamim, Y.; Carmeli, S.; Hizi, A. The Inhibition of the Reverse Transcriptase of HIV-1 by the Natural Sulfoglycolipids from Cyanobacteria: Contribution of Different Moieties to Their High Potency. J. Nat. Prod. 1998, 61, 891–895. [Google Scholar] [CrossRef]
- Alipanah, L.; Winge, P.; Rohloff, J.; Najafi, J.; Brembu, T.; Bones, A.M. Molecular adaptations to phosphorus deprivation and comparison with nitrogen deprivation responses in the diatom Phaeodactylum tricornutum. PLoS ONE 2018, 13, e0193335. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Nobusawa, T.; Hori, K.; Shimojima, M.; Ohta, H. Betaine Lipid Is Crucial for Adapting to Low Temperature and Phosphate Deficiency in Nannochloropsis. Plant Physiol. 2018, 177, 181–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 2013, 52, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, M. Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 2011, 50, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Wei, D.; Chen, F.; Yang, S.T. Lipidomic profiling reveals lipid regulation in the snow alga Chlamydomonas nivalis in response to nitrate or phosphate deprivation. Process. Biochem. 2013, 48, 605–613. [Google Scholar] [CrossRef]
- Liang, K.; Zhang, Q.; Gu, M.; Cong, W. Effect of phosphorus on lipid accumulation in freshwater microalga Chlorella sp. J. Appl. Phycol. 2012, 25, 311–318. [Google Scholar] [CrossRef]
- Muhlroth, A.; Winge, P.; El Assimi, A.; Jouhet, J.; Marechal, E.; Hohmann-Marriott, M.F.; Vadstein, O.; Bones, A.M. Mechanisms of Phosphorus Acquisition and Lipid Class Remodeling under P Limitation in a Marine Microalga. Plant Physiol. 2017, 175, 1543–1559. [Google Scholar] [CrossRef] [Green Version]
- Gasparovic, B.; Godrijan, J.; Frka, S.; Tomazic, I.; Penezic, A.; Maric, D.; Djakovac, T.; Ivancic, I.; Paliaga, P.; Lyons, D.; et al. Adaptation of marine plankton to environmental stress by glycolipid accumulation. Mar. Environ. Res. 2013, 92, 120–132. [Google Scholar] [CrossRef]
- Yu, B.; Xu, C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737. [Google Scholar] [CrossRef] [Green Version]
- Li-Beisson, Y.; Thelen, J.J.; Fedosejevs, E.; Harwood, J.L. The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 2019, 74, 31–68. [Google Scholar] [CrossRef]
- Frentzen, M. Phosphatidylglycerol and sulfoquinovosyldiacylglycerol: Anionic membrane lipids and phosphate regulation. Curr. Opin. Plant Biol. 2004, 7, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, L.; Bale, N.; Hopmans, E.C.; Schouten, S.; Damste, J.S. Diversity and distribution of a key sulpholipid biosynthetic gene in marine microbial assemblages. Environ. Microbiol. 2014, 16, 774–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hölzl, G.; Dörmann, P. Chloroplast Lipids and Their Biosynthesis. Annu. Rev. Plant Biol. 2019, 70, 51–81. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, H.; Schottler, M.A.; Kelly, A.A.; Sundqvist, C.; Dormann, P.; Karim, S.; Jarvis, P. Monogalactosyldiacylglycerol deficiency in Arabidopsis affects pigment composition in the prolamellar body and impairs thylakoid membrane energization and photoprotection in leaves. Plant Physiol. 2008, 148, 580–592. [Google Scholar] [CrossRef] [Green Version]
- Murakawa, M.; Shimojima, M.; Shimomura, Y.; Kobayashi, K.; Awai, K.; Ohta, H. Monogalactosyldiacylglycerol synthesis in the outer envelope membrane of chloroplasts is required for enhanced growth under sucrose supplementation. Front. Plant Sci. 2014, 5, 280. [Google Scholar] [CrossRef] [Green Version]
- Miège, C.; Maréchal, E.; Shimojima, M.; Awai, K.; Block, M.A.; Ohta, H.; Takamiya, K.; Douce, R.; Joyard, J. Biochemical and topological properties of type A MGDG synthase, a spinach chloroplast envelope enzyme catalyzing the synthesis of both prokaryotic and eukaryotic MGDG. Eur. J. Biochem. 1999, 265, 990–1001. [Google Scholar] [CrossRef] [Green Version]
- Froehlich, J.E.; Benning, C.; Dormann, P. The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J. Biol. Chem. 2001, 276, 31806–31812. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.A.; Dormann, P. DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 2002, 277, 1166–1173. [Google Scholar] [CrossRef] [Green Version]
- Awai, K.; Marechal, E.; Block, M.A.; Brun, D.; Masuda, T.; Shimada, H.; Takamiya, K.; Ohta, H.; Joyard, J. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2001, 98, 10960–10965. [Google Scholar] [CrossRef] [Green Version]
- Riekhof, W.R.; Sears, B.B.; Benning, C. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: Discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 2005, 4, 242–252. [Google Scholar] [CrossRef] [Green Version]
- Awai, K.; Watanabe, H.; Benning, C.; Nishida, I. Digalactosyldiacylglycerol is required for better photosynthetic growth of Synechocystis sp. PCC6803 under phosphate limitation. Plant Cell Physiol. 2007, 48, 1517–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maida, E.; Awai, K. Digalactosyldiacylglycerol is essential in Synechococcus elongatus PCC 7942, but its function does not depend on its biosynthetic pathway. Biochim. Biophys. Acta 2016, 1861, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Awai, K. Diversity in Biosynthetic Pathways of Galactolipids in the Light of Endosymbiotic Origin of Chloroplasts. Front. Plant Sci. 2016, 7, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuzawa, Y.; Shimojima, M.; Sato, R.; Mizusawa, N.; Ikeda, K.; Suzuki, M.; Iwai, M.; Hori, K.; Wada, H.; Masuda, S.; et al. Cyanobacterial monogalactosyldiacylglycerol-synthesis pathway is involved in normal unsaturation of galactolipids and low-temperature adaptation of Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 2014, 1841, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awai, K.; Kakimoto, T.; Awai, C.; Kaneko, T.; Nakamura, Y.; Takamiya, K.; Wada, H.; Ohta, H. Comparative genomic analysis revealed a gene for monoglucosyldiacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol. 2006, 141, 1120–1127. [Google Scholar] [CrossRef] [Green Version]
- Awai, K.; Ohta, H.; Sato, N. Oxygenic photosynthesis without galactolipids. Proc. Natl. Acad. Sci. USA 2014, 111, 13571–13575. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Awai, K.; Nakamura, M.; Nagatani, A.; Masuda, T.; Ohta, H. Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 2009, 57, 322–331. [Google Scholar] [CrossRef]
- Kelly, A.A.; Froehlich, J.E.; Dormann, P. Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 2003, 15, 2694–2706. [Google Scholar] [CrossRef] [Green Version]
- Okazaki, Y.; Shimojima, M.; Sawada, Y.; Toyooka, K.; Narisawa, T.; Mochida, K.; Tanaka, H.; Matsuda, F.; Hirai, A.; Hirai, M.Y.; et al. A Chloroplastic UDP-Glucose Pyrophosphorylase from Arabidopsis Is the Committed Enzyme for the First Step of Sulfolipid Biosynthesis. Plant Cell 2009, 21, 892–909. [Google Scholar] [CrossRef] [Green Version]
- Kang, N.K.; Kim, E.K.; Sung, M.G.; Kim, Y.U.; Jeong, B.R.; Chang, Y.K. Increased biomass and lipid production by continuous cultivation of Nannochloropsis salina transformant overexpressing a bHLH transcription factor. Biotechnol. Bioeng. 2019, 116, 555–568. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.; Kang, N.K.; Koh, H.G.; Shin, S.E.; Lee, B.; Jeong, B.R.; Chang, Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018, 115, 331–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Hao, Q.; Bai, L.; Xu, J.; Yin, W.; Song, L.; Xu, L.; Guo, X.; Fan, C.; Chen, Y.; et al. Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. Biofuels 2014, 7, 128. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Feng, L.; Wang, X.; Miao, X. Adaptation of Synechococcus sp. PCC 7942 to phosphate starvation by glycolipid accumulation and membrane lipid remodeling. Biochim. Biophys. Acta 2019, 1864, 158522. [Google Scholar] [CrossRef]
- Abida, H.; Dolch, L.J.; Mei, C.; Villanova, V.; Conte, M.; Block, M.A.; Finazzi, G.; Bastien, O.; Tirichine, L.; Bowler, C.; et al. Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol. 2015, 167, 118–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selao, T.T.; Zhang, L.; Arioz, C.; Wieslander, A.; Norling, B. Subcellular localization of monoglucosyldiacylglycerol synthase in Synechocystis sp. PCC6803 and its unique regulation by lipid environment. PLoS ONE 2014, 9, e88153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimojima, M.; Tsuchiya, M.; Ohta, H. Temperature-dependent hyper-activation of monoglucosyldiacylglycerol synthase is post-translationally regulated in Synechocystis sp. PCC 6803. FEBS Lett. 2009, 583, 2372–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Masuda, T.; Takamiya, K.; Ohta, H. Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J. 2006, 47, 238–248. [Google Scholar] [CrossRef]
- Sato, N.; Kamimura, R.; Kaneta, K.; Yoshikawa, M.; Tsuzuki, M. Species-specific roles of sulfolipid metabolism in acclimation of photosynthetic microbes to sulfur-starvation stress. PLoS ONE 2017, 12, e0186154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voshol, G.P.; Meyer, V.; van den Hondel, C.A. GTP-binding protein Era: A novel gene target for biofuel production. BMC Biotechnol. 2015, 15, 21. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, B.H.; Handler, A.A.; Chao, D.K.; Nguyen, V.; Smith, M.; Ryu, S.Y.; Simons, E.L.; Anderson, P.E.; Simons, R.W. The widely conserved Era G-protein contains an RNA-binding domain required for Era function in vivo. Mol. Microbiol. 1999, 33, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- Cheng, P.; Li, H.; Yuan, L.; Li, H.; Xi, L.; Zhang, J.; Liu, J.; Wang, Y.; Zhao, H.; Zhao, H.; et al. The ERA-Related GTPase AtERG2 Associated with Mitochondria 18S RNA Is Essential for Early Embryo Development in Arabidopsis. Front. Plant Sci. 2018, 9, 182. [Google Scholar] [CrossRef] [Green Version]
- Suwastika, I.N.; Denawa, M.; Yomogihara, S.; Im, C.H.; Bang, W.Y.; Ohniwa, R.L.; Bahk, J.D.; Takeyasu, K.; Shiina, T. Evidence for lateral gene transfer (LGT) in the evolution of eubacteria-derived small GTPases in plant organelles. Front. Plant Sci. 2014, 5, 678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, Y.; Ahn, C.S.; Jung, H.J.; Kang, H.; Park, G.T.; Choi, Y.; Hwang, J.; Pai, H.S. DER containing two consecutive GTP-binding domains plays an essential role in chloroplast ribosomal RNA processing and ribosome biogenesis in higher plants. J. Exp. Bot. 2014, 65, 117–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Tian, Y.; Cheng, S.; Wang, Y.; Hao, Y.; Zhu, J.; Zhu, X.; Zhang, Y.; Yu, M.; Lei, J.; et al. WSL6 encoding an Era-type GTP-binding protein is essential for chloroplast development in rice. Plant Mol. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zheng, X.; Apaliya, M.T.; Yang, H.; Zhang, H. Transcriptome characterization and expression profile of defense-related genes in pear induced by Meyerozyma guilliermondii. Postharvest Biol. Technol. 2018, 141, 63–70. [Google Scholar] [CrossRef]
- Choi, S.Y.; Park, B.; Choi, I.G.; Sim, S.J.; Lee, S.M.; Um, Y.; Woo, H.M. Transcriptome landscape of Synechococcus elongatus PCC 7942 for nitrogen starvation responses using RNA-seq. Sci. Rep. 2016, 6, 30584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstraeten, N.; Fauvart, M.; Versees, W.; Michiels, J. The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 2011, 75, 507–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, W.; Liang, F.; Duan, Y.; Tan, X.; Lu, X. Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab. Eng. 2013, 19, 17–25. [Google Scholar] [CrossRef]
- Maeda, K.; Narikawa, R.; Ikeuchi, M. CugP is a novel ubiquitous non-GalU-type bacterial UDP-glucose pyrophosphorylase found in cyanobacteria. J. Bacteriol. 2014, 196, 2348–2354. [Google Scholar] [CrossRef] [Green Version]
- Zavaleta-Pastor, M.; Sohlenkamp, C.; Gao, J.L.; Guan, Z.; Zaheer, R.; Finan, T.M.; Raetz, C.R.; Lopez-Lara, I.M.; Geiger, O. Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc. Natl. Acad. Sci. USA 2010, 107, 302–307. [Google Scholar] [CrossRef] [Green Version]
- Boudiere, L.; Botte, C.Y.; Saidani, N.; Lajoie, M.; Marion, J.; Brehelin, L.; Yamaryo-Botte, Y.; Satiat-Jeunemaitre, B.; Breton, C.; Girard-Egrot, A.; et al. Galvestine-1, a novel chemical probe for the study of the glycerolipid homeostasis system in plant cells. Mol. Biosyst. 2012, 8, 2023–2035. [Google Scholar] [CrossRef]
- Wada, H.; Murata, N. Membrane Lipids in Cyanobacteria. In Lipids in Photosynthesis: Structure, Function and Genetics; Springer: Dordrecht, The Netherlands, 1998. [Google Scholar]
- Gu, Y.; He, L.; Zhao, C.; Wang, F.; Yan, B.; Gao, Y.; Li, Z.; Yang, K.; Xu, J. Biochemical and Transcriptional Regulation of Membrane Lipid Metabolism in Maize Leaves under Low Temperature. Front. Plant Sci. 2017, 8, 2053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laczko-Dobos, H.; Frycak, P.; Ughy, B.; Domonkos, I.; Wada, H.; Prokai, L.; Gombos, Z. Remodeling of phosphatidylglycerol in Synechocystis PCC6803. Biochim. Biophys. Acta 2010, 1801, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Fujii, S.; Sasaki, D.; Baba, S.; Ohta, H.; Masuda, T.; Wada, H. Transcriptional regulation of thylakoid galactolipid biosynthesis coordinated with chlorophyll biosynthesis during the development of chloroplasts in Arabidopsis. Front. Plant Sci. 2014, 5, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K. Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J. Plant Res. 2016, 129, 565–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, N.; Ebiya, Y.; Kobayashi, R.; Nishiyama, Y.; Tsuzuki, M. Disturbance of cell-size determination by forced overproduction of sulfoquinovosyl diacylglycerol in the cyanobacterium Synechococcus elongatus PCC 7942. Biochem. Biophys. Res. Commun. 2017, 487, 734–739. [Google Scholar] [CrossRef]
- Khozin-Goldberg, I.; Cohen, Z. The effect of phosphate starvation on the lipid and fatty acid composition of the fresh water eustigmatophyte Monodus subterraneus. Phytochemistry 2006, 67, 696–701. [Google Scholar] [CrossRef]
- Masuda, S.; Harada, J.; Yokono, M.; Yuzawa, Y.; Shimojima, M.; Murofushi, K.; Tanaka, H.; Masuda, H.; Murakawa, M.; Haraguchi, T.; et al. A Monogalactosyldiacylglycerol Synthase Found in the Green Sulfur Bacterium Chlorobaculum tepidum Reveals Important Roles for Galactolipids in Photosynthesis. Plant Cell 2011, 23, 2644–2658. [Google Scholar] [CrossRef] [Green Version]
- Dubots, E.; Botte, C.; Boudiere, L.; Yamaryo-Botte, Y.; Jouhet, J.; Marechal, E.; Block, M.A. Role of phosphatidic acid in plant galactolipid synthesis. Biochimie 2012, 94, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, I.; Mizusawa, N.; Wada, H.; Sato, N. Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol. 2007, 145, 1361–1370. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Narise, T.; Sonoike, K.; Hashimoto, H.; Sato, N.; Kondo, M.; Nishimura, M.; Sato, M.; Toyooka, K.; Sugimoto, K.; et al. Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J. 2013, 73, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Manan, S.; Chen, B.; She, G.; Wan, X.; Zhao, J. Transport and transcriptional regulation of oil production in plants. Crit. Rev. Biotechnol. 2016, 37, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.; Singh, S.; Kaushik, M.S.; Mishra, A.K. Regulation of organophosphate metabolism in cyanobacteria. A review. Microbiology 2015, 84, 291–302. [Google Scholar] [CrossRef]
- Salinas, P.; Ruiz, D.; Cantos, R.; Lopez-Redondo, M.L.; Marina, A.; Contreras, A. The regulatory factor SipA provides a link between NblS and NblR signal transduction pathways in the cyanobacterium Synechococcus sp. PCC 7942. Mol. Microbiol. 2007, 66, 1607–1619. [Google Scholar] [CrossRef]
- Yamaryo, Y.; Kanai, D.; Awai, K.; Shimojima, M.; Masuda, T.; Shimada, H.; Takamiya, K.-I.; Ohta, H. Light and Cytokinin Play a Co-operative Role in MGDG Synthesis in Greening Cucumber Cotyledons. Plant Cell Physiol. 2003, 44, 844–855. [Google Scholar] [CrossRef] [Green Version]
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef]
- Wang, Y.; Pang, C.; Li, X.; Hu, Z.; Lv, Z.; Zheng, B.; Chen, P. Identification of tRNA nucleoside modification genes critical for stress response and development in rice and Arabidopsis. BMC Plant Biol. 2017, 17, 261. [Google Scholar] [CrossRef] [PubMed]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hartel, H.; Dormann, P.; Benning, C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 10649–10654. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Miao, X. Biodiesel quality and biochemical changes of microalgae Chlorella pyrenoidosa and Scenedesmus obliquus in response to nitrate levels. Bioresour. Technol. 2014, 170, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Miao, X. Monoglucosyldiacylglycerol participates in phosphate stress adaptation in Synechococcus sp. PCC 7942. Biochem. Biophys. Res. Commun. 2020, 522, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.G.; Kim, K.H.; Lee, B.M.; Moon, J.C. Transcriptome analysis for identifying possible gene regulations during maize root emergence and formation at the initial growth stage. Genes Genom. 2018, 40, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Zhou, L.; Cheng, D.; Wang, L.; Gao, J.; Zhao, Q.; Wei, W.; Sun, Y. Comparative transcriptomic analysis reveals phenol tolerance mechanism of evolved Chlorella strain. Bioresour. Technol. 2017, 227, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Shen, J.; Bai, F.; Xu, N. Transcriptome profiling of the microalga Chlorella pyrenoidosa in response to different carbon dioxide concentrations. Mar. Genom. 2016, 29, 81–87. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.; Blake, J.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.; Dolinski, K.; Dwight, S.; Eppig, J.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Scherer, P.I.; Raeder, U.; Geist, J.; Zwirglmaier, K. Influence of temperature, mixing, and addition of microcystin-LR on microcystin gene expression in Microcystis aeruginosa. Microbiologyopen 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Woodger, F.J.; Badger, M.R.; Price, G.D. Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol. 2003, 133, 2069–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene ID | Annotation | FC | p-Value |
|---|---|---|---|
| Synpcc7942_0938 | transcriptional regulator, ArsR family | 8.79281803 | 5.0074E-13 |
| Synpcc7942_2585 | transcriptional regulator, BadM/Rrf2 family | 2.43751415 | 0.00232243 |
| Synpcc7942_2416 | two component transcriptional regulator, winged helix family | 2.15739652 | 0.04603509 |
| Synpcc7942_0110 | transcriptional regulator, XRE family | 2.13386852 | 0.02139503 |
| Synpcc7942_1897 | putative transcription factor DevT-like | 2.12534589 | 0.00718275 |
| Synpcc7942_1725 | transcriptional regulator, GntR family | 1.94907168 | 0.04567919 |
| Synpcc7942_2305 | two component transcriptional regulator, winged helix family, nblR | 0.72020702 | 0.04515754 |
| Synpcc7942_1739 | transcriptional regulator, MerR family | 0.61129814 | 0.02357565 |
| Synpcc7942_0556 | two component transcriptional regulator, winged helix family | 0.61086684 | 0.01166383 |
| Synpcc7942_2466 | two component transcriptional regulator, winged helix family | 0.58625772 | 0.03952819 |
| Synpcc7942_1159 | transcriptional regulator, MarR family | 0.57296615 | 0.0038297 |
| Synpcc7942_0764 | transcriptional regulator, XRE family | 0.47965835 | 0.00047173 |
| Gene ID | Annotation | FC | qRT-PCR |
|---|---|---|---|
| Synpcc7942_0938 | transcriptional regulator, ArsR family | 8.792818 | 23.18 |
| Synpcc7942_2585 | transcriptional regulator, BadM/Rrf2 family | 2.437514 | 23.82 |
| Synpcc7942_2416 | two component transcriptional regulator, winged helix family | 2.157397 | 2−0.84 |
| Synpcc7942_0110 | transcriptional regulator, XRE family | 2.133869 | 22.78 |
| Synpcc7942_1897 | putative transcription factor DevT-like | 2.125346 | 2−1.01 |
| Synpcc7942_1725 | transcriptional regulator, GntR family | 1.949072 | 2−1.58 |
| Synpcc7942_1083 | a probable glycosyltransferase, mgdA | 1.33778 | 2−0.14 |
| Synpcc7942_0861 | a conserved hypothetical protein, mgdE | 1.205566 | 2−0.43 |
| Synpcc7942_0986 | a probable glycosyltransferase, dgdA | 0.90492 | 2−0.32 |
| Synpcc7942_2305 | two component transcriptional regulator, winged helix family | 0.720207 | 2−2.79 |
| Synpcc7942_0578 | UDP-sulfoquinovose synthase, sqdB | 0.634708 | 2−3.54 |
| Synpcc7942_1739 | transcriptional regulator, MerR family | 0.611298 | 2−3.70 |
| Synpcc7942_0556 | two component transcriptional regulator, winged helix family | 0.610867 | 2−3.49 |
| Synpcc7942_2466 | two component transcriptional regulator, winged helix family | 0.586258 | 2−4.39 |
| Synpcc7942_1159 | transcriptional regulator, MarR family | 0.572966 | 2−4.38 |
| Synpcc7942_0764 | transcriptional regulator, XRE family | 0.479658 | 2−1.02 |
| Synpcc7942_0579 | sulfolipid sulfoquinovosyl diacylglycerol biosynthesis protein, sqdX | 0.425591 | 2−3.34 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Miao, X. Glyceroglycolipid Metabolism Regulations under Phosphate Starvation Revealed by Transcriptome Analysis in Synechococcus elongatus PCC 7942. Mar. Drugs 2020, 18, 360. https://doi.org/10.3390/md18070360
Xu X, Miao X. Glyceroglycolipid Metabolism Regulations under Phosphate Starvation Revealed by Transcriptome Analysis in Synechococcus elongatus PCC 7942. Marine Drugs. 2020; 18(7):360. https://doi.org/10.3390/md18070360
Chicago/Turabian StyleXu, Xinrui, and Xiaoling Miao. 2020. "Glyceroglycolipid Metabolism Regulations under Phosphate Starvation Revealed by Transcriptome Analysis in Synechococcus elongatus PCC 7942" Marine Drugs 18, no. 7: 360. https://doi.org/10.3390/md18070360
APA StyleXu, X., & Miao, X. (2020). Glyceroglycolipid Metabolism Regulations under Phosphate Starvation Revealed by Transcriptome Analysis in Synechococcus elongatus PCC 7942. Marine Drugs, 18(7), 360. https://doi.org/10.3390/md18070360




