Mass Spectrometry-Based Metabolomics Combined with Quantitative Analysis of the Microalgal Diatom (Chaetoceros calcitrans)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Metabolites and Profiling of C. calcitrans Extract by UHPLC-ESI-Orbitrap MS/MS
2.1.1. Identification of Carotenoids and Chlorophylls
2.1.2. Identification of Fatty Acids
2.1.3. Identification of Glycerolipids, Glycerolphospholipids and Sterol Lipid
2.1.4. Identification of Other Possible Compounds
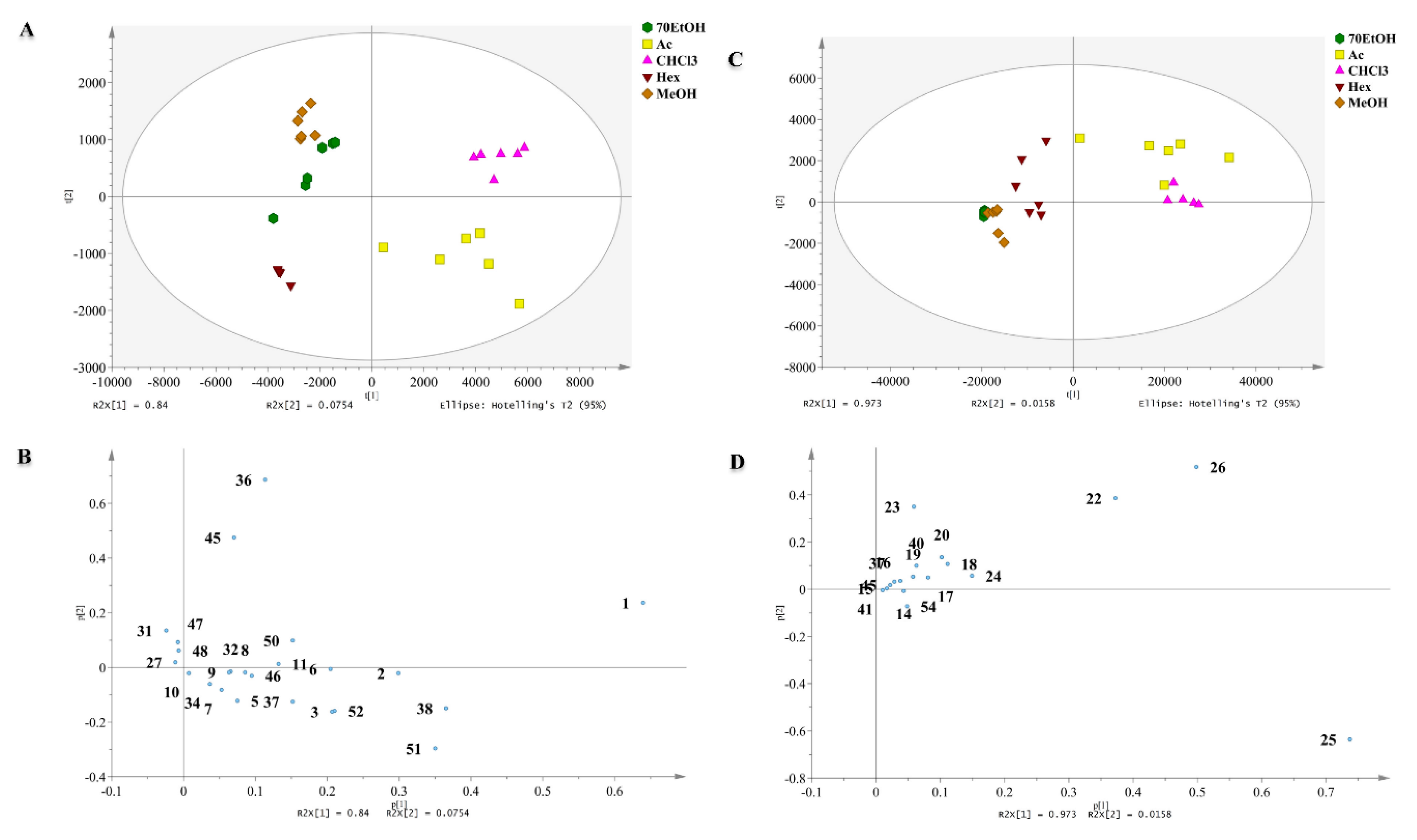
2.2. Chemometric Analysis of MS Data
2.2.1. Discrimination through Unsupervised Principal Component Analysis (PCA)
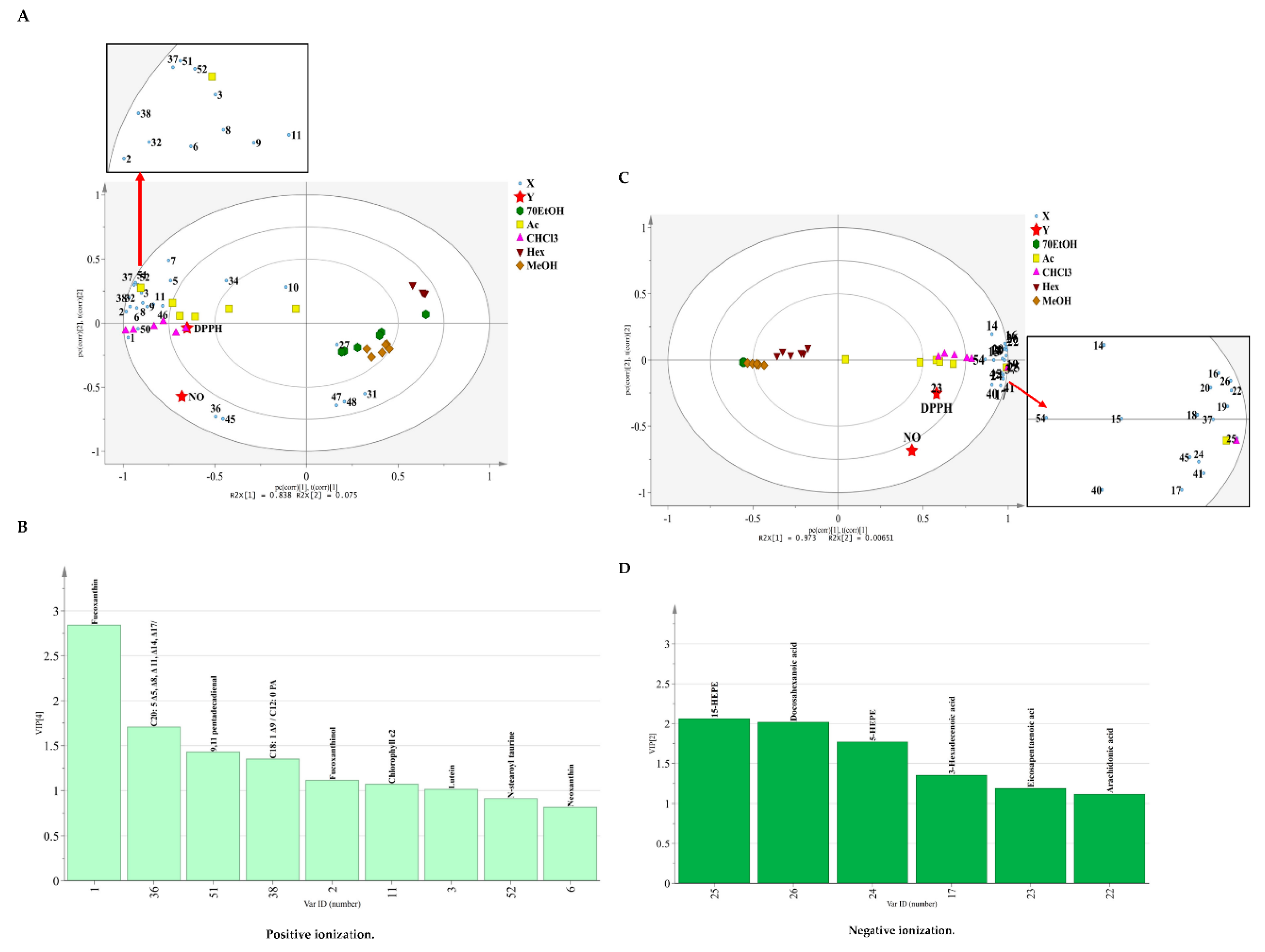
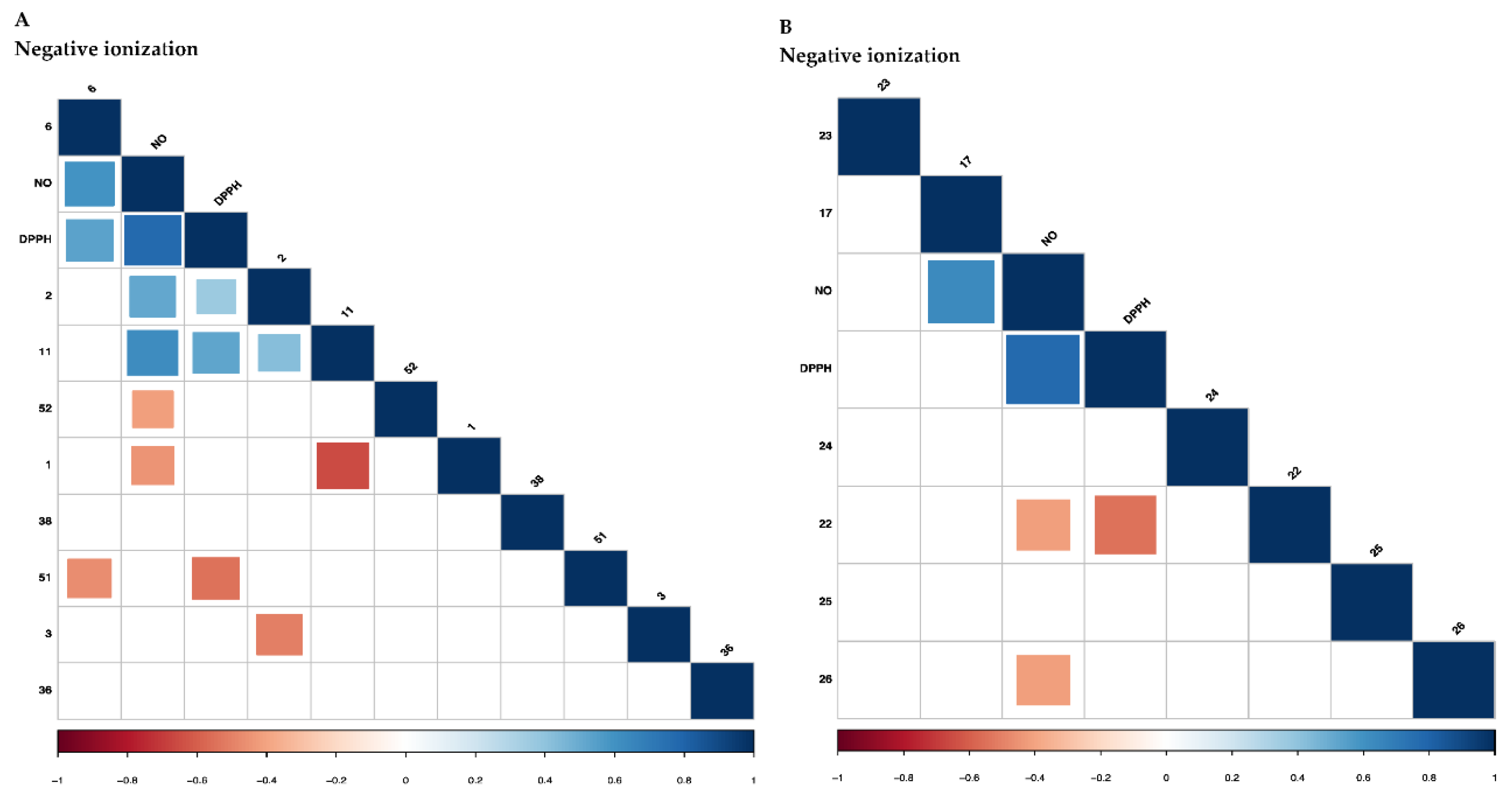
2.2.2. Correlation of the Metabolites with Antioxidant and Anti-Inflammatory Activities Using Partial Least Square (PLS) Regression Analysis
2.3. Quantification of the Metabolites in Microalgal Diatom C. calcitrans Extracts through UHPLC-ESI-Orbitrap MS
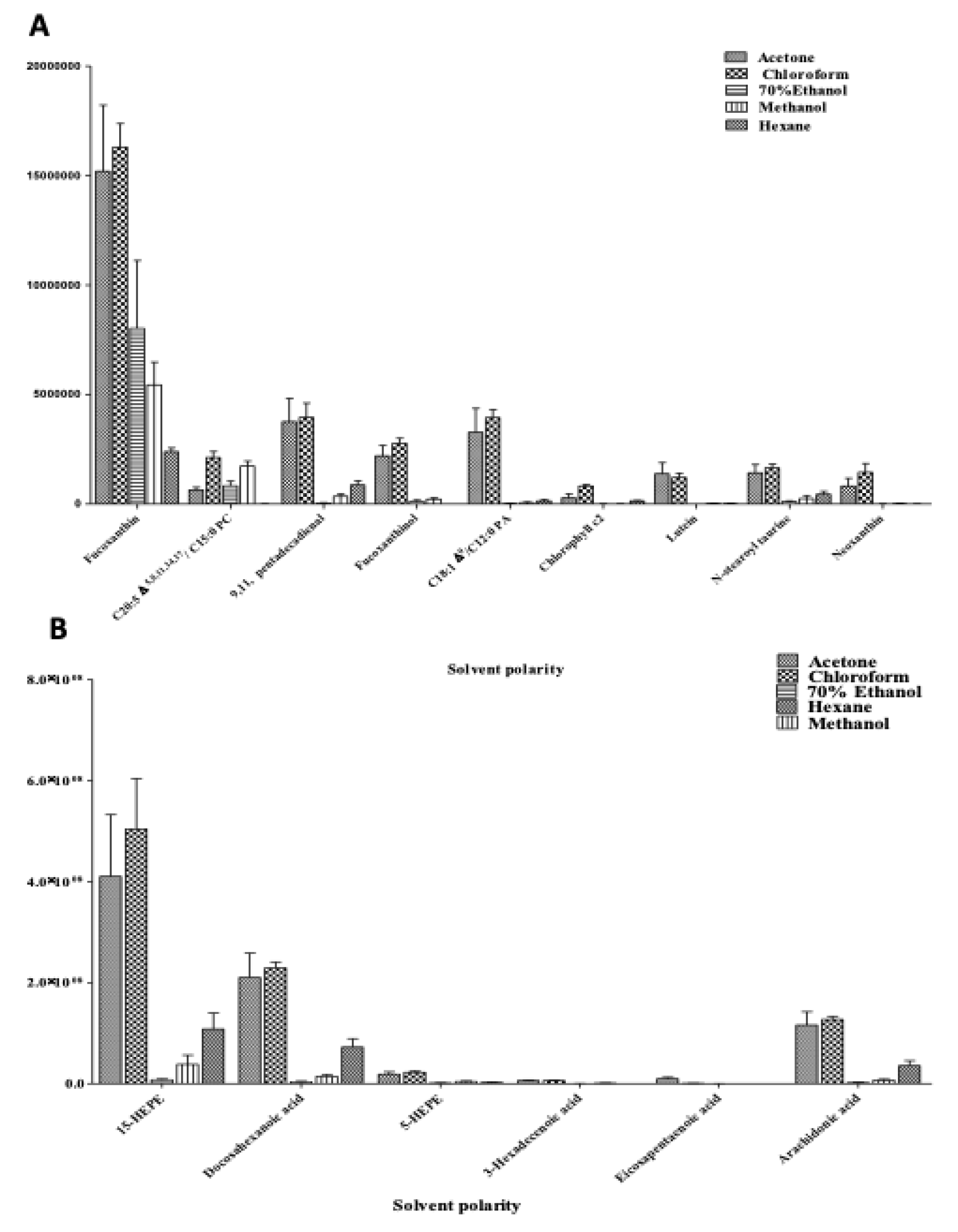
2.3.1. Relative Quantification of Metabolites in C. calcitrans
2.3.2. Validation of the Proposed Method
Linearity
LOD and LOQ
Application of Method: Quantification of the Targeted Metabolites in Active Extracts of C. calcitrans
3. Materials and Methods
3.1. Chemicals and Samples
3.2. In Vitro Preliminary Assays
3.2.1. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) Radical Scavenging Assay
3.2.2. Nitric Oxide (NO) Inhibitory Assay
3.3. Targeted Metabolite Profiling Using UHPLC-ESI-Orbitrap MS
3.3.1. Sample Preparation of UHPLC-ESI-Orbitrap MS Approach
3.3.2. UHPLC-ESI-Orbitrap MS Analysis
3.3.3. UHPLC-ESI-Orbitrap MS Data Preprocessing
3.4. Targeted Metabolites Quantification Using UHPLC-ESI-MS/MS Orbitrap
3.4.1. Relative Quantification of the Identified Metabolites Using UHPLC-ESI-MS/MS Orbitrap Approach
3.4.2. Targeted Metabolites Quantification Using UHPLC-ESI-MS/MS Orbitrap Analysis
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Méndez-Martínez, Y.; García-Guerrero, M.U.; Lora-Vilchis, M.C.; Martínez-Córdova, L.R.; Arcos-Ortega, F.G.; Alpuche, J.J.; Cortés-Jacinto, E. Nutritional effect of Artemia nauplii enriched with Tetraselmis suecica and Chaetoceros calcitrans microalgae on growth and survival on the river prawn Macrobrachium americanum larvae. Aquacult. Int. 2018, 26, 1001–1015. [Google Scholar] [CrossRef]
- Zanella, L.; Pertile, P.; Massironi, M.; Caviola, E. Extracts of Microalgae and Their Application. U.S. Patent 9,974,819, 22 May 2018. [Google Scholar]
- Azizan, A.; Bustamam, M.S.A.; Maulidiani, M.; Shaari, K.; Ismail, I.S.; Nagao, N.; Abas, F. Metabolite profiling of the microalgal diatom Chaetoceros calcitrans and correlation with antioxidant and nitric oxide inhibitory activities via 1H NMR-based metabolomics. Mar. Drugs 2018, 16, 154. [Google Scholar] [CrossRef] [Green Version]
- Dang, T.T.; Bowyer, M.C.; Van Altena, I.A.; Scarlett, C.J. Comparison of chemical profile and antioxidant properties of the brown algae. Int. J. Food Sci. Technol. 2018, 53, 174–181. [Google Scholar] [CrossRef]
- Yamano, N.; Mizoguchi, T.; Fujii, R. The pH-dependent photophysical properties of chlorophyll-c bound to the light-harvesting complex from a diatom, Chaetoceros calcitrans. J. Photochem. Photobiol. A Chem. 2018, 358, 379–385. [Google Scholar] [CrossRef]
- Juin, C.; Bonnet, A.; Nicolau, E.; Bérard, J.B.; Devillers, R.; Thiéry, V.; Cadoret, J.-P.; Picot, L. UPLC-MSE profiling of phytoplankton metabolites: Application to the identification of pigments and structural analysis of metabolites in Porphyridium purpureum. Mar. Drugs 2015, 13, 2541–2558. [Google Scholar] [CrossRef] [Green Version]
- Hano, T.; Tomaru, Y. Metabolomics-based approach to explore growth phase-dependent markers in cultured diatom Chaetoceros tenuissimus. J. Chrom. B 2019, 1128, 121779. [Google Scholar] [CrossRef]
- Kustka, A.B.; Allen, A.E.; Morel, F.M. Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms1. J. Phycol. 2007, 43, 715–729. [Google Scholar] [CrossRef]
- Allen, A.E.; LaRoche, J.; Maheswari, U.; Lommer, M.; Schauer, N.; Lopez, P.J.; Finazzi, G.; Fernie, A.R.; Bowler, C. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. USA 2008, 105, 10438–10443. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Hao, W.; Zou, Y.; Shi, M.; Jiang, Y.; Xiao, P.; Lei, A.; Hu, Z.; Zhang, W.; Zhao, L.; et al. Fatty acid and metabolomic profiling approaches differentiate heterotrophic and mixotrophic culture conditions in a microalgal food supplement ‘Euglena’. BMC Biotechnol. 2016, 16, 49. [Google Scholar] [CrossRef] [Green Version]
- Bianco, G.; Agerbirk, N.; Losito, I.; Cataldi, T.R. Acylated glucosinolates with diverse acyl groups investigated by high resolution mass spectrometry and infrared multiphoton dissociation. Phytochemistry 2014, 100, 92–102. [Google Scholar] [CrossRef]
- Cataldi, T.R.; Bianco, G.; Abate, S.; Losito, I. Identification of unsaturated N-acylhomoserine lactones in bacterial isolates of Rhodobacter sphaeroides by liquid chromatography coupled to electrospray ionization-hybrid linear ion trap-Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2011, 25, 1817–1826. [Google Scholar] [CrossRef]
- Porzel, A.; Farag, M.A.; Mülbradt, J.; Wessjohann, L.A. Metabolite profiling and fingerprinting of Hypericum species: A comparison of MS and NMR metabolomics. Metabolomics 2014, 10, 574–588. [Google Scholar] [CrossRef]
- Abdul-Hamid, N.A.; Abas, F.; Ismail, I.S.; Tham, C.L.; Maulidiani, M.; Mediani, A.; Swarup, S.; Umashankar, S.; Zolkeflee, N.K.Z. Metabolites and biological activities of Phoenix dactylifera L. pulp and seeds: A comparative MS and NMR based metabolomics approach. Phytochem. Lett. 2019, 31, 20–32. [Google Scholar] [CrossRef]
- Bird, S.S.; Marur, V.R.; Sniatynski, M.J.; Greenberg, H.K.; Kristal, B.S. Lipidomics profiling by high-resolution LC−MS and high-energy collisional dissociation fragmentation: Focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal. Chem. 2010, 83, 940–949. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.; Magnúsdóttir, M.; Brynjólfson, S.; Palsson, B.Ø.; Paglia, G. UPLC-UV-MS E analysis for quantification and identification of major carotenoid and chlorophyll species in algae. Anal. Bioanal. Chem. 2012, 404, 3145–3154. [Google Scholar] [CrossRef] [PubMed]
- Airs, R.L.; Llewellyn, C.A. Improved detection and characterisation of fucoxanthin-type carotenoids: Novel pigments in Emiliiana Huxleyi (prymnesiophyceae). J. Phycol. 2006, 42, 391–399. [Google Scholar] [CrossRef]
- Bijttebier, S.K.; D’Hondt, E.; Hermans, N.; Apers, S.; Voorspoels, S. Unravelling ionization and fragmentation pathways of carotenoids using orbitrap technology: A first step towards identification of unknowns. J. Mass Spectrom. 2013, 48, 740–754. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Dong, L.; Pajkovic, N.D. Atmospheric pressure chemical ionization tandem mass spectrometry of carotenoids. Int. J. Mass Spectrom. 2012, 312, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Ríos, J.J.; Pérez-Gálvez, A.; Roca, M. Comprehensive chlorophyll composition in the main edible seaweeds. Food Chem. 2017, 228, 625–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerwin, J.L.; Wiens, A.M.; Ericsson, L.H. Identification of fatty acids by electrospray mass spectrometry and tandem mass spectrometry. J. Mass Spectrom. 1996, 31, 184–192. [Google Scholar] [CrossRef]
- Mansour, M.P.; Frampton, D.M.; Nichols, P.D.; Volkman, J.K.; Blackburn, S.I. Lipid and fatty acid yield of nine stationary-phase microalgae: Applications and unusual C24–C28 polyunsaturated fatty acids. J. Appl. Phycol. 2005, 17, 287–300. [Google Scholar] [CrossRef]
- Lang, I.; Hodac, L.; Friedl, T.; Feussner, I. Fatty acid profiles and their distribution patterns in microalgae: A comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol. 2011, 11, 124–140. [Google Scholar] [CrossRef] [Green Version]
- Rettner, J.; Markus, W.; Meyer, N.; Werz, O.; Georg, P. Survey of the C20 and C22 Oxylipin Family in Marine Diatoms. Tetrahedron Lett. 2018, 59, 828–831. [Google Scholar] [CrossRef]
- Liu, B.; Vieler, A.; Li, C.; Jones, A.D.; Benning, C. Triacylglycerol profiling of microalgae Chlamydomonas reinhardtii and Nannochloropsis oceanica. Bioresour. Technol. 2013, 146, 310–316. [Google Scholar] [CrossRef]
- Hsu, F.F.; Turk, J. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4, 5-bisphosphate by electrospray ionization tandem mass spectrometry: A mechanistic study. J. Am. Soc. Mass Spectrom. 2000, 11, 986–999. [Google Scholar] [CrossRef] [Green Version]
- Murphy, R.C.; Axelsen, P.H. Mass spectrometric analysis of long-chain lipids. Mass Spectrom. Rev. 2011, 30, 579–599. [Google Scholar] [CrossRef] [Green Version]
- Danielewicz, M.A.; Anderson, L.A.; Franz, A.K. Triacylglycerol profiling of marine microalgae by mass spectrometry. J. Lipid Res. 2011, 52, 2101–2108. [Google Scholar] [CrossRef] [Green Version]
- Yao, L.; Gerde, J.A.; Lee, S.L.; Wang, T.; Harrata, K.A. Microalgae lipid characterization. J. Agric. Food Chem. 2015, 63, 1773–1787. [Google Scholar] [CrossRef] [Green Version]
- Kumari, P.; Kumar, M.; Reddy, C.R.K.; Jha, B. Algal lipids, fatty acids and sterols. In Functional Ingredients from Algae for Foods and Nutraceuticals; Dominguez, H., Ed.; Woodhead Publishing Limited: Cambridge, UK, 2013; pp. 87–134. [Google Scholar]
- Da Costa, E.; Silva, J.; Mendonça, S.H.; Abreu, M.H.; Domingues, M.R. Lipidomic approaches towards deciphering glycolipids from microalgae as a reservoir of bioactive lipids. Mar. Drugs 2016, 14, 101. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wang, J.; Ma, Q.; Yang, J.; Li, X.; Yuan, Y.J. Phospholipid metabolism in an industry microalga Chlorella sorokiniana: The impact of inoculum sizes. PLoS ONE 2013, 8, e70827. [Google Scholar] [CrossRef] [Green Version]
- Brembu, T.; Mühlroth, A.; Alipanah, L.; Bones, A.M. The effects of phosphorus limitation on carbon metabolism in diatoms. Philos. Trans. R. Soc. B 2017, 372, 20160406. [Google Scholar] [CrossRef] [PubMed]
- Barofsky, A.; Pohnert, G. Biosynthesis of polyunsaturated short chain aldehydes in the diatom Thalassiosira rotula. Org. Lett. 2007, 9, 1017–1020. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.H.; Zhu, J.; Jiang, J.G. Carotenoids biosynthesis and cleavage related genes from bacteria to plants. Crit. Rev. Food Sci. Nutr. 2018, 58, 2314–2333. [Google Scholar] [CrossRef]
- Alonso, D.L.; Belarbi, E.H.; Rodríguez-Ruiz, J.; Segura, C.I.; Giménez, A. Acyl lipids of three microalgae. Photochemistry 1998, 47, 1473–1481. [Google Scholar] [CrossRef]
- Kaiser, B.K.; Carleton, M.; Hickman, J.W.; Miller, C.; Lawson, D.; Budde, M.; Cross, F. Fatty aldehydes in cyanobacteria are a metabolically flexible precursor for a diversity of biofuel products. PLoS ONE 2013, 8, e58307. [Google Scholar] [CrossRef] [Green Version]
- Sachindra, N.M.; Sato, E.; Maeda, H.; Hosokawa, M.; Niwano, Y.; Kohno, M.; Miyashita, K. Radical scavenging and singlet oxygen quenching activity of marine carotenoid fucoxanthin and its metabolites. J. Agric. Food Chem. 2007, 55, 8516–8522. [Google Scholar] [CrossRef]
- Plaza, M.; Santoyo, S.; Jaime, L.; Reina, G.G.B.; Herrero, M.; Señoráns, F.J.; Ibáñez, E. Screening for bioactive compounds from algae. J. Pharm. Biomed. Anal. 2010, 51, 450–455. [Google Scholar] [CrossRef]
- Foo, S.C.; Yusoff, F.M.; Ismail, M.; Basri, M.; Chan, K.W.; Khong, N.M.; Yau, S.K. Production of fucoxanthin-rich fraction (FxRF) from a diatom, Chaetoceros calcitrans (Paulsen) Takano 1968. Algal Res. 2015, 12, 26–32. [Google Scholar] [CrossRef]
- Food and Drug Administration. Guidance for Industry: Bioanalytical Method Validation; US Department of Health and Human Services, FDA, Center for Drug Evaluation and Research: Rockville, MD, USA, 2001.
- Foo, S.C.; Yusoff, F.M.; Ismail, M.; Basri, M.; Khong, N.M.H.; Chan, K.W.; Yau, S.K. Efficient solvent extraction of antioxidant-rich extract from a tropical diatom, Chaetoceros calcitrans (Paulsen) Takano 1968. Asian Pac. J. Trop. Biomed. 2015, 5, 834–840. [Google Scholar] [CrossRef]
- Fu, W.; Guðmundsson, Ó.; Paglia, G.; Herjólfsson, G.; Andrésson, Ó.S.; Palsson, B.Ø.; Brynjólfsson, S. Enhancement of carotenoid biosynthesis in the green microalga Dunaliella salina with light-emitting diodes and adaptive laboratory evolution. Appl. Microbiol. Biotechnol. 2013, 97, 2395–2403. [Google Scholar] [CrossRef] [Green Version]
- Rivera, S.M.; Canela-Garayoa, R. Analytical tools for the analysis of carotenoids in diverse materials. J. Chromatogr. A 2012, 1224, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rivera, S.M.; Christou, P.; Canela-Garayoa, R. Identification of carotenoids using mass spectrometry. Mass Spectrom. Rev. 2014, 33, 353–372. [Google Scholar] [CrossRef] [Green Version]
- Jora, M.; Burns, A.P.; Ross, R.L.; Lobue, P.A.; Zhao, R.; Palumbo, C.M.; Jora, M.; Burns, A.P.; Ross, R.L.; Lobue, P.A.; et al. Differentiating positional isomers of nucleoside modifications by higher-energy collisional dissociation mass spectrometry (HCD MS). J. Am. Soc. Mass Spectrom. 2018, 29, 1745–1756. [Google Scholar] [CrossRef]
- Wei, X.; Shi, X.; Kim, S.; Zhang, L.; Patrick, J.S.; Binkley, J.; Zhang, X. Data preprocessing method for liquid chromatography–mass spectrometry-based metabolomics. Anal. Chem. 2012, 84, 7963–7971. [Google Scholar] [CrossRef] [Green Version]
- Glauser, G.; Veyrat, N.; Rochat, B.; Wolfender, J.L.; Turlings, T.C. Ultra-high pressure liquid chromatography–mass spectrometry for plant metabolomics: A systematic comparison of high-resolution quadrupole-time-of-flight and single stage Orbitrap mass spectrometers. J. Chromatogr. A 2013, 1292, 151–159. [Google Scholar] [CrossRef]
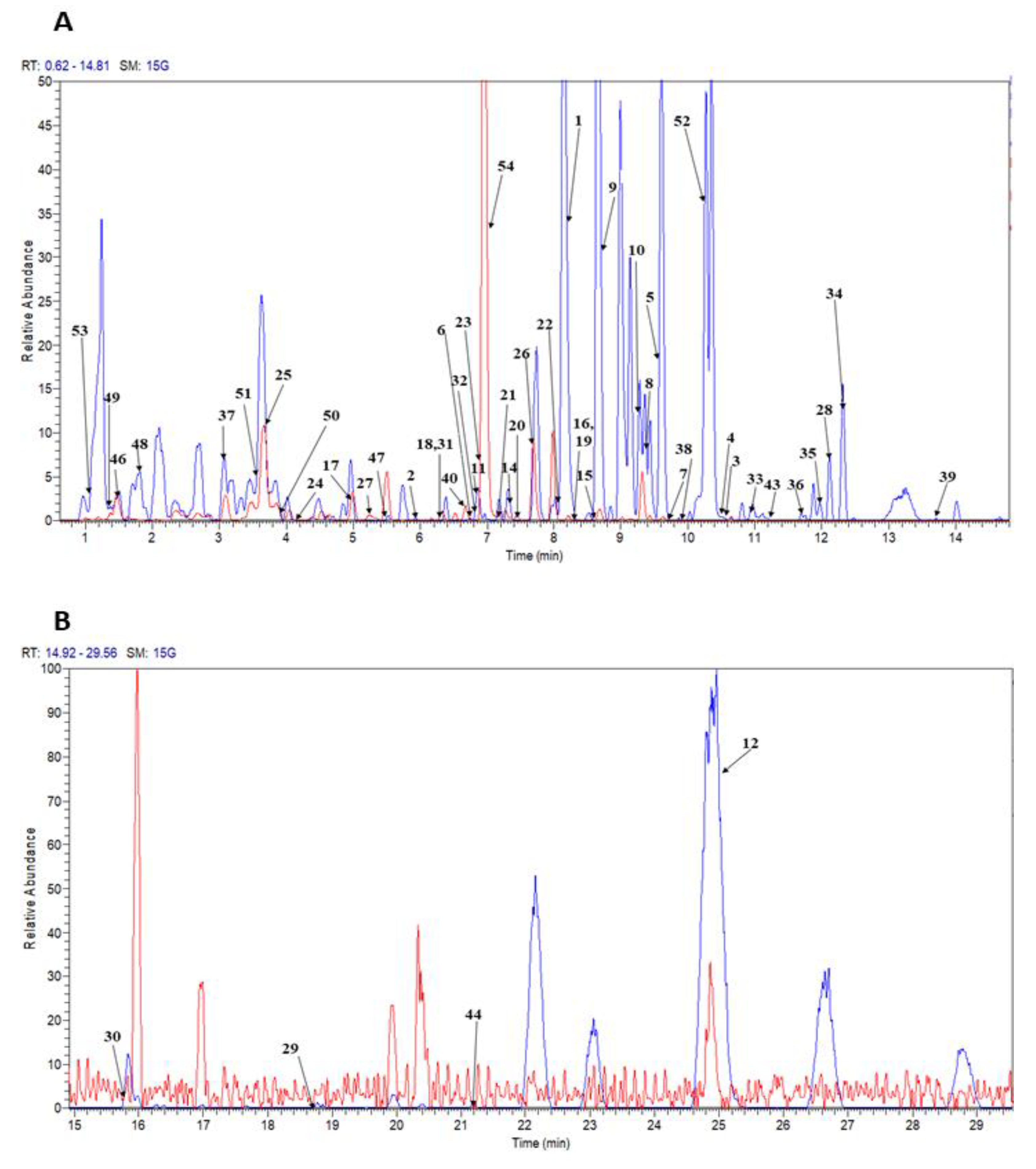
| Peak no. | tR (min) | Tentative Identification | Ion Mode | Molecular Formula | UV | [M + H]+/[M − H]− m/z | Fragment Ions of [M + H]+/[M − H]− | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Measured Mass (Da) | Theoretical Mass (Da) | Mass Error (ppm) | ||||||||
| 1 | 8.19 | Fucoxanthin | ESI+ | C42H58O6 | 412,442 | 659.4294 | 659.4306 | −1.82 | 641.4180, 581.3969, 489.2938 | [16,17] |
| 2 | 5.9 | Fucoxanthinol | ESI+ | C40H58O5 | 224,426 | 617.4172 | 617.4200 | 4.53 | 599.4075, 581.3972, | [17] |
| 3 | 10.54 | Lutein | ESI+ | C40H56O2 | 226,408 | 569.4210 | 569.4280 | −12.29 | 523.5612, 495.3164 | [18,19] |
| 4 | 10.52 | Zeaxanthin | ESI+ | C40H56O2 | 226, 408 | 569.4204 | 569.4280 | −13.34 | 523.5612. | [18,19] |
| 5 | 9.56 | (3′R,3′S)-Astaxanthin | ESI+ | C40H52O4 | 474 | 597.3903 | 597.3938 | −5.85 | 147.1163, 173.1319, 201.1262 | [19] |
| 6 | 6.73 | Neoxanthin | ESI+ | C40H56O4 | 400, 420 | 601.4238 | 601.4251 | −2.16 | 583.4016 | [6,18] |
| 7 | 9.75 | Diadinoxanthin | ESI+ | C42H54O3 | 404 | 583.4135 | 583.4145 | 1.71 | 565.4039, 547.3956 | [6,18] |
| 8 | 9.29 | (3,4,3′)-4-Hydroxy-alloxanthin | ESI+ | C40H52O3 | 408 | 581.3970 | 581.3989 | −0.69 | 563.3879 | [6] |
| 9 | 8.76 | Canthaxanthin | ESI+ | C40H52O2 | 408 | 565.4026 | 565.4040 | −2.48 | 547.3898 | [18] |
| 10 | 9.23 | 14′-apo-beta-carotenal | ESI+ | C22H31O | 414 | 311.2369 | 311.2369 | 0 | 293.2261 | [18] |
| 11 | 6.88 | Chlorophyll c2 | ESI+ | C35H28MgN4O5 | 424 | 609.1981 | 609.1983 | −0.33 | 591.1877, 549.1774 | [20] |
| 12 | 25.49 | Pheophytin a | ESI+ | C55H74MgN4 | 506,536 | 871.5694 | 871.5732 | 4.36 | 593.2754 | [20] |
| 13 | 19.26 | Pheophytin b | ESI+ | C55H74MgN6 | 536 | 885.5518 | 885.5525 | 0.79 | 593. 533 | [20] |
| 14 | 7.31 | Myristic acid | ESI− | C14 H28 O2 | N.D | 227.2104 | 227.2107 | −1.32 | 191.4482 | [21] |
| 15 | 8.58 | Palmitic acid | ESI− | C16 H32 O2 | N.D | 255.2330 | 255.2330 | 0 | 61.98716 | [22,23] |
| 16 | 8.38 | cis-Δ9-Palmitoleic acid | ESI− | C16 H30 O2 | N.D | 253.2175 | 253.2173 | 0.79 | 217.1967 | [22] |
| 17 | 4.96 | 3-Hexadecenoic acid | ESI− | C16 H30 O2 | N.D | 253.2171 | 253.2173 | 1.58 | 141.2954 | [22] |
| 18 | 6.27 | Stearidonic acid | ESI− | C18 H28 O2 | N.D | 275.2015 | 275.2017 | 0 | 231.2124, 217.8763 | [21] |
| 19 | 8.39 | Linoleic acid | ESI− | C18 H32 O2 | N.D | 279.2331 | 279.2330 | 0.36 | 111.3027 | [23] |
| 20 | 7.11 | α or γ -Linolenic acid | ESI− | C18 H30 O2 | N.D | 277.2173 | 277.2173 | 0 | 69.9031, 59.0124 | [21] |
| 21 | 7.43 | α or γ -Linolenic acid | ESI− | C18 H30 O2 | N.D | 277.2173 | 277.2173 | 0 | 69.9031, 59.0124 | [21] |
| 22 | 8.11 | Arachidonic acid | ESI− | C20 H32 O2 | N.D | 303.2329 | 303.233 | −0.33 | 287.0870, 259.2397 | [21,23] |
| 23 | 6.86 | Eicosapentaenoic acid | ESI− | C20 H30 O2 | N.D | 301.2177 | 301.2173 | 0.33 | 273.5861, 257.2269 | [21,24] |
| 24 | 4.19 | 5 or 15-HEPE | ESI− | C20H30O3 | N.D | 317.2124 | 317.2122 | 0.63 | 255.2231, 115.0391 | [24] |
| 25 | 3.72 | 5 or 15-HEPE | ESI− | C20H30O3 | N.D | 317.2132 | 317.2122 | 3.15 | 255.2231 | [24] |
| 26 | 7.63 | Docosahexaenoic acid | ESI− | C22 H32 O2 | N.D | 327.2331 | 327.233 | 0.31 | 283.2432, 269.2458 | [25] |
| 27 | 5.23 | 4,5-Epoxy-17R-HDHA | ESI+ | C25H34O4 | N.D | 399.2519 | 399.253 | −2.76 | 321.2201, 296.2936 | [25] |
| 28 | 12.14 | 6-bromo- Δ5-heptacosadienoic acid | ESI+ | C27H5079Br02- | N.D | 485.3008, 487.2993 | 485.2989, 487.2989 | −3.77, −0.82 | 405.3321, 112.5382, 57.0341 | [22] |
| 29 | 18.85 | C18:3 Δ6, Δ9, Δ12/C19:1 Δ9 phosphatidylinositol | ESI+ | C46H81O13P | N.D | 873.5482 | 873.5488 | −0.69 | 595.2531 | [26] |
| 30 | 15.73 | C16: 1 Δ9/C16:0 phosphatidylglycerol | ESI+ | C38H73O10P | N.D | 721.5011 | 721.5014 | −0.42 | 663.4510, 193.1956 | [27] |
| 31 | 6.28 | C18: 3 Δ9, Δ12, Δ15/C13:0 phosphatidylglycerol | ESI+ | C37H67O10P | N.D | 703.4542 | 703.4545 | 0.21 | 671.4276, 643.4357 | [27] |
| 32 | 6.8 | C20:1 Δ 11/C0:0 phosphatidic acid | ESI+ | C23H45O7P | N.D | 465.2977 | 465.2976 | 0.21 | 251.1775, 197.1311 | [27,28] |
| 33 | 10.89 | C22: 3 Δ10, Δ13, Δ16/C22: 5 Δ7, Δ10, Δ13, Δ16, Δ19/C0: 0 diacylglycerol | ESI+ | C47H76O5 | N.D | 721.5760 | 721.5765 | 0 | 316.5049, 313.1947, 173.1183 | [29] |
| 34 | 12.31 | C18: 3 Δ9, Δ12, Δ15/C19: 1 Δ9/C20: 5 Δ5, Δ8, Δ 11, Δ14, Δ17 triacylglycerol | ESI+ | C60H98O6 | N.D | 915.7436 | 915.7407 | 3.17 | 469.3639 | [6] |
| 35 | 12 | C18: 3 Δ6, Δ9, Δ12/C22:4 Δ7, Δ10, Δ13, Δ16 phosphatidic acid | ESI+ | C43H71O8P | N.D | 747.4981 | 747.4959 | 2.94 | 732.1207, 101.112 | [29] |
| 36 | 11.69 | C20: 5 Δ5, Δ8, Δ 11, Δ14, Δ17/C15: 0 phosphatidylcholine | ESI+ | C43H76NO8P | N.D | 766.5370 | 766.5381 | −1.43 | 740.5090, 631.427 | [29] |
| 37 | 3.06 | 5-Androstenetriol | ESI+ | C19H30O3 | N.D | 307.2272 | 307.2268 | 1.3 | 289.1795 | [29] |
| 38 | 9.88 | C18: 1 Δ9/C12: 0 phosphatidic acid | ESI− | C33H63O8P | N.D | 617.4211 | 617.4188 | 3.72 | 301.2803 | [30,31] |
| 39 | 13.62 | C17: 0/C22: 5 Δ7, Δ10, Δ13, Δ16, Δ19/C0: 0 diacylglycerol | ESI− | C42H72O5 | N.D | 655.5309 | 655.5307 | 0.31 | 356.3249, 256.2157, 115.0359 | [28] |
| 40 | 6.66 | C20: 3 Δ8, Δ11, Δ14/C20: 5 Δ5, Δ8, Δ11, Δ14, Δ17/C0: 0 diacylglycerol | ESI− | C43H68O5 | N.D | 663.5026 | 663.4994 | 4.82 | 375.2085, 112.9845 | [27,29] |
| 41 | 18.04 | C22: 2 Δ13, Δ16/C22: 6 Δ4, Δ7, Δ10, Δ13, Δ16, Δ19/C0: 0 diacylglycerol | ESI− | C47H76O5 | N.D | 719.5636 | 719.562 | 2.22 | 664.5043, 440.8061 | [29,30] |
| 42 | 25.9 | C16:0/C18: 2 Δ9, Δ12 diacylglyceryl-N,N,N-trimethyl homoserine (DGTS) | ESI− | C44H81NO7 | N.D | 734.5950 | 734.594 | 1.36 | 386.8379 | [31] |
| 43 | 11.22 | C16: 1 Δ9/C20: 5 Δ5, Δ8, Δ11, Δ14, Δ17 phosphatidylglycerol | ESI− | C42H71O10P | N.D | 765.4713 | 765.4712 | 0.13 | 529.2575, 253.2171 | [30,32] |
| 44 | 21.35 | O-C18:0/C19:0 phosphatidylserine | ESI− | C43H86NO9P | N.D | 790.5969 | 790.5968 | 0.13 | 283.5096, 73.2201 | [27,32] |
| 45 | 11.47 | C16:0/C16:0 sulfoquinovosyl diacylglyceride (SQDG) | ESI− | C41H78O12S | N.D | 793.5125 | 793.5141 | −2.02 | 227.20132 | [33] |
| 46 | 1.58 | Crocetin dialdehyde | ESI+ | C20H24O2 | N.D | 297.1844 | 297.1849 | −1.68 | 213.1372 | [34,35] |
| 47 | 5.42 | N-butyl-N(2)-[ 2-hydroxy-2-phenylacetyl]-l-serinamide | ESI+ | C15H22N2O4 | N.D | 295.1634 | 295.1652 | −6.1 | 277.2147, 221.118 | [6] |
| 48 | 1.81 | Formyl 2,4,6-decatrienoate | ESI+ | C11H16O2 | N.D | 181.1217 | 181.1223 | −3.31 | 163.111, 135.1164 | [36] |
| 49 | 1.29 | Dipentyl phthalate | ESI+ | C18H26O4 | N.D | 307.1889 | 307.1904 | −4.88 | 163.111, 135.1164 | [6] |
| 50 | 3.98 | Diisooctyl phtalate | ESI+ | C24H38O4 | N.D | 413.2677 | 413.2671 | 1.45 | 381.2407, 363.2321 | [6] |
| 51 | 3.57 | 9,11 pentadecadienal | ESI+ | C15H26O | N.D | 223.2047 | 223.2056 | −4.03 | 205.1954, 193.1005 | [37] |
| 52 | 10.26 | N-stearoyl taurine | ESI+ | C20H41NO4S | N.D | 392.2864 | 392.2829 | 8.92 | 150.0262, 149.0229 | [6] |
| 53 | 1.07 | (2,4)-N-isobutyl-6-phenyl-hexa-2,4-dienamide | ESI+ | C16H21NO | N.D | 244.1696 | 244.1696 | 0 | 226.1438, 184.0968 | [6] |
| 54 | 6.97 | N-palmitoyl proline | ESI− | C21H39NO3 | N.D | 352.2859 | 352.2857 | 0.56 | 116.0705 | [6] |
| Compounds | Retention Time (Rt) | Linearity (R2) | Slope | LOQ (mg/mL) | LOD (mg/mL) |
|---|---|---|---|---|---|
| Fucoxanthin | 8.15 | 0.9994 | 1.84 × 10−5 | 0.0000162 | 0.0000049 |
| Astaxanthin | 9.51 | 0.9998 | 0.00065 | 0.0001579 | 0.0000474 |
| Zeaxanthin | 10.52 | 0.9967 | 0.00015 | 0.0000019 | 0.0000006 |
| Lutein | 10.53 | 0.9991 | 0.01421 | 0.0029388 | 0.0008816 |
| Extract | Metabolites | Concentration in Diatom in 100 mg of Extracts (µg/mg) |
|---|---|---|
| Chloroform | Fucoxanthin | 1.2537 ± 0.0896 |
| Astaxanthin | 0.3794 ± 0.0263 | |
| Zeaxanthin | 0.1503 ± 0.0001 | |
| Lutein | 0.1513 ± 0.0584 | |
| Acetone | Fucoxanthin | 1.3031 ± 0.0521 |
| Astaxanthin | 0.4346 ± 0.0438 | |
| Zeaxanthin | 0.1582 ± 0.0003 | |
| Lutein | 0.2414 ± 0.0325 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azizan, A.; Maulidiani, M.; R., R.; Shaari, K.; Ismail, I.S.; Nagao, N.; Abas, F. Mass Spectrometry-Based Metabolomics Combined with Quantitative Analysis of the Microalgal Diatom (Chaetoceros calcitrans). Mar. Drugs 2020, 18, 403. https://doi.org/10.3390/md18080403
Azizan A, Maulidiani M, R. R, Shaari K, Ismail IS, Nagao N, Abas F. Mass Spectrometry-Based Metabolomics Combined with Quantitative Analysis of the Microalgal Diatom (Chaetoceros calcitrans). Marine Drugs. 2020; 18(8):403. https://doi.org/10.3390/md18080403
Chicago/Turabian StyleAzizan, Awanis, M. Maulidiani, Rudiyanto R., Khozirah Shaari, Intan Safinar Ismail, Norio Nagao, and Faridah Abas. 2020. "Mass Spectrometry-Based Metabolomics Combined with Quantitative Analysis of the Microalgal Diatom (Chaetoceros calcitrans)" Marine Drugs 18, no. 8: 403. https://doi.org/10.3390/md18080403
APA StyleAzizan, A., Maulidiani, M., R., R., Shaari, K., Ismail, I. S., Nagao, N., & Abas, F. (2020). Mass Spectrometry-Based Metabolomics Combined with Quantitative Analysis of the Microalgal Diatom (Chaetoceros calcitrans). Marine Drugs, 18(8), 403. https://doi.org/10.3390/md18080403





