Abstract
A systematic chemical investigation of the deep-sea-derived fungus Penicillium solitum MCCC 3A00215 resulted in the isolation of one novel polyketide (1), two new alkaloids (2 and 3), and 22 known (4–25) compounds. The structures of the new compounds were established mainly on the basis of exhaustive analysis of 1D and 2D NMR data. Viridicatol (13) displayed moderate anti-tumor activities against PANC-1, Hela, and A549 cells with IC50 values of around 20 μM. Moreover, 13 displayed potent in vitro anti-food allergic activity with an IC50 value of 13 μM, compared to that of 92 μM for the positive control, loratadine, while indole-3-acetic acid methyl ester (9) and penicopeptide A (10) showed moderate effects (IC50 = 50 and 58 μM, respectively).
1. Introduction
Penicillium solitum is a filamentous fungus associated with the decay of pomaceous fruits during storage [1]. As a matter of fact, it can infect fruit through wounds and cause significant economic losses [2]. Besides pome fruits such as apples and pears, this fungus was also isolated from other foods, including cheeses and processed meats [3,4]. Surprisingly, it can also be found under extremophilic circumstances: in the Berkeley Pit Lake (pH 2.7) [5] and the maritime Antarctic [6].
Chemical investigation of this fungus provided a broad spectrum of secondary metabolites, including compactin (known as mevastatin or ML-236B, which is utilized for the production of an important cholesterol-lowering drug, pravastatin) [7] and its analogues [8,9], in addition to sesquiterpenoids [5], alkaloids [10], and polyketides etc. [11].
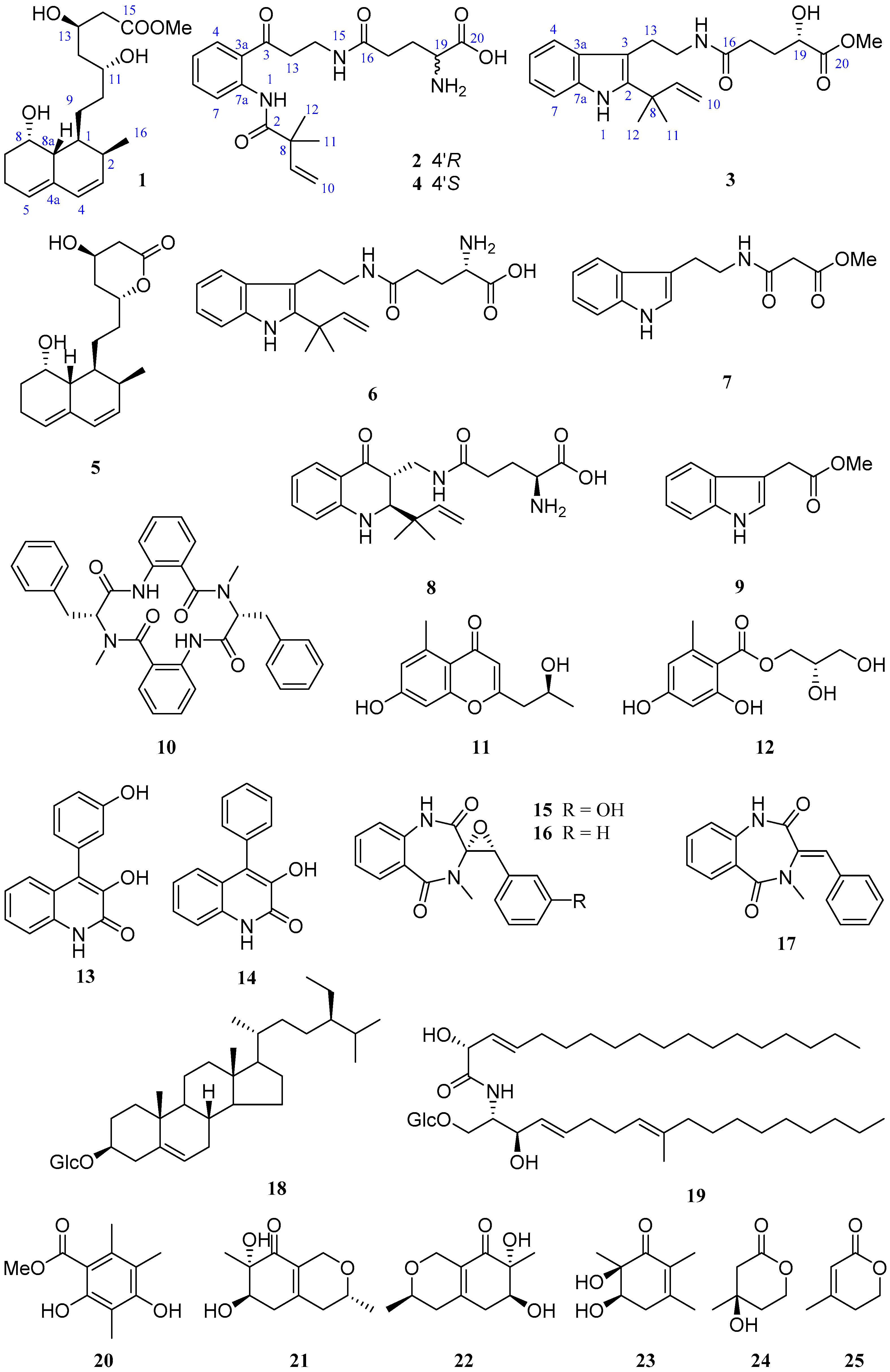
Penicillium solitum MCCC 3A00215 is a deep-sea-derived fungus from the Northwest Atlantic Ocean (−3034 m). A previous study on this strain provided a unique 6/6/6/6/5-pentacyclic steroid [12]. In order to discover more novel compounds, a further chemical investigation was conducted. As a result, three new (1–3) and 22 known (4–25) compounds (Figure 1) were obtained. By comparison of the NMR and MS data with those published in the literature, the known compounds were determined to be (−)-solitumidines D (4) [10], ML-236A (5) [13], solitumidine A (6) [10], methyl-2-([2-(1H-indol-3-yl)-ethyl]carbamoyl)acetate (7) [14], solitumine A (8) [10], indole-3-acetic acid methyl ester (9) [15], penicopeptide A (10) [16], (2′S)-7-hydroxy-2-(2-hydroxypropyl)-5-methylchromone (11) [17], hydroxypropan-2′,3′-diol orsellinate (12) [18], viridicatol (13) [19], viridicatin (14) [19], (−)-cyclopenol (15) [20], cyclopenin (16) [21], 3-benzylidene-3,4-dihydro-4-methyl-1H-1,4-benzodiazepine-2,5-dione (17) [16], β-sitosterol-3-O-β-d-glucopyranoside (18) [22], cerebrosides C (19) [23], methyl-2,4-dihydroxy-3,5,6-trimethylbenzoate (20) [24], felinone A (21) [25], xylariphilone (22) [26], 5,6-dihydroxy-2,3,6-trimethylcyclohex-2-enone (23) [27], (R)-mevalonolactone (24) [28], and 3-methyl-2-penten-5-olide (25) [29]. Here, we report the isolation, structure, and bioactivities of these 25 compounds.
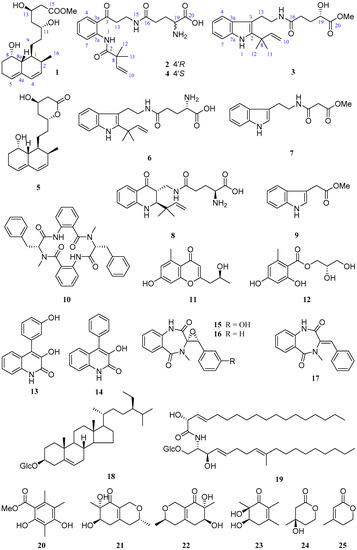
Figure 1.
Compounds 1–25 from Penicillium solitum MCCC 3A00215.
2. Results and Discussion
Compound 1 had a molecular formula C19H30O5, as established by its positive HRESIMS at m/z 361.1989 [M + Na]+, requiring five degrees of unsaturation. The 1H and 13C NMR spectroscopic data (Figures S1 and S2 from the Supplementary Materials, Table 1) revealed the presence of one methyl doublet [δH 0.89 (d, J = 6.8 Hz, H3-16); δC 14.3 (q, C-16)], one methoxyl [δH 3.70 (s, OMe); δC 52.1 (q, OMe)], six sp3 methylenes, nine methines including three aliphatic [δH 1.77 (m, H-1), 2.19 (brd, J = 11.8 Hz, H-8a), 2.37 (m, H-2); δC 32.1 (d, C-2), 37.8 (d, C-1), 40.0 (d, C-8a)], three olefinic [δH 5.47 (brs, H-5), 5.69 (dd, J = 9.4, 6.1 Hz, H-3), 5.91 (d, J = 9.4 Hz, H-4); δC 124.5 (d, C-5), 129.9 (d, C-4), 133.6 (d, C-3)] and three oxygenated [δH 3.80 (m, H-11), 4.19 (m, H-13), 4.22 (m, H-8); δC 65.2 (d, C-8), 68.1 (d, C-13), 71.1 (d, C-11)] ones, and two non-protonated carbons with one olefinic [δC 135.1 (s, C-4a)] and one carbonyl [δC 173.9 (s, C-15)] group. Altogether, the 1H and 13C NMR spectra provided 19 carbons, categorized as one methyl, one methoxyl, six methylenes, nine methines, and two quaternary carbons.

Table 1.
1H (400 Hz) and 13C (100 Hz) NMR data of 1–3 (δ in ppm, J in Hz within parentheses).
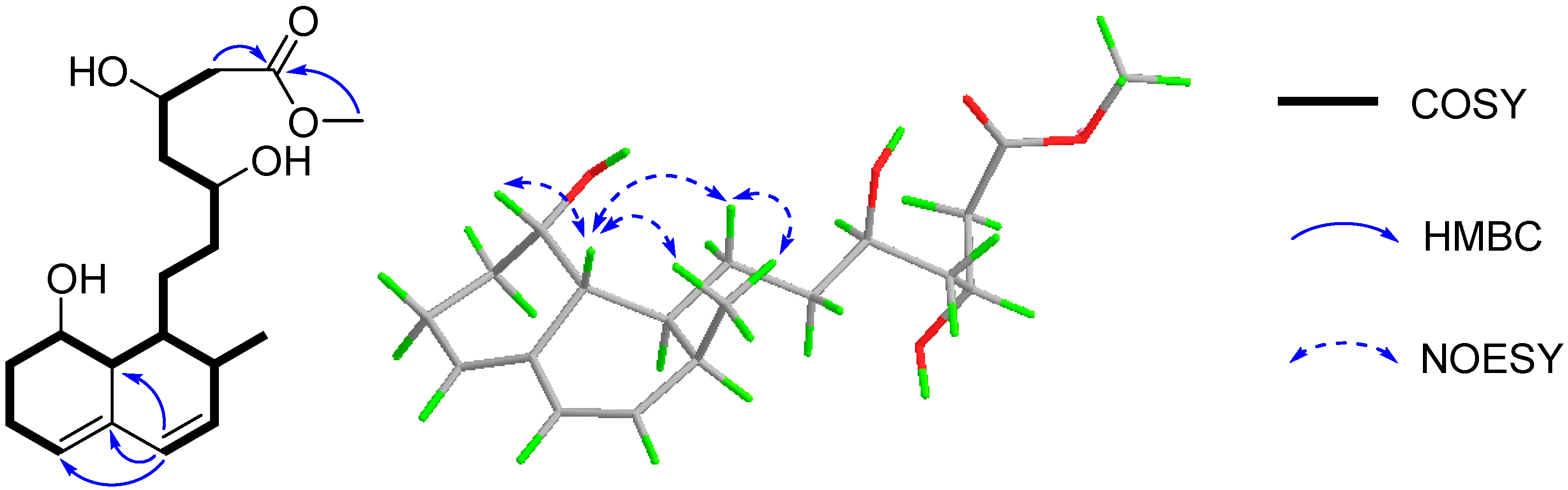
In the COSY spectrum, correlations were observed for H-5/H2-6/H2-7/H-8/H-8a/H-1/H2-9/H2-10/H-11/H2-12/H-13/H2-14 and H-1/H-2/H3-16/H-3/H-4, which constructed a long chain of C-5/C-6/C-7/C-8/C-8a/C-1/C-9/C-10/C-11/C-12/C-13/C-14 and C-1 via C-2 to C-16/C-3/C-4 (Figure 2). The segment and the methoxyl moiety could be connected on the basis of the HMBC correlations of H-4 to C-8a/C-4a/C-5, H2-14 and 15-OMe to C-15 (Figure 2). Therefore, the planar structure of 1 was established as a methyl ester of acyclic form of ML-236A (5) [13], which was previously prepared in the lab by the saponification of ML-236A in 0.1 N NaOH at 50 °C for 2 h [30].
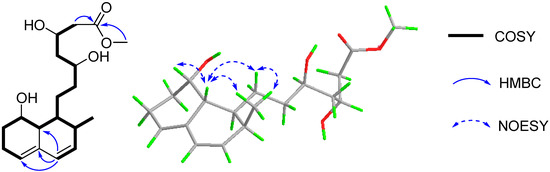
Figure 2.
The key COSY, HMBC, and NOESY correlations of 1.
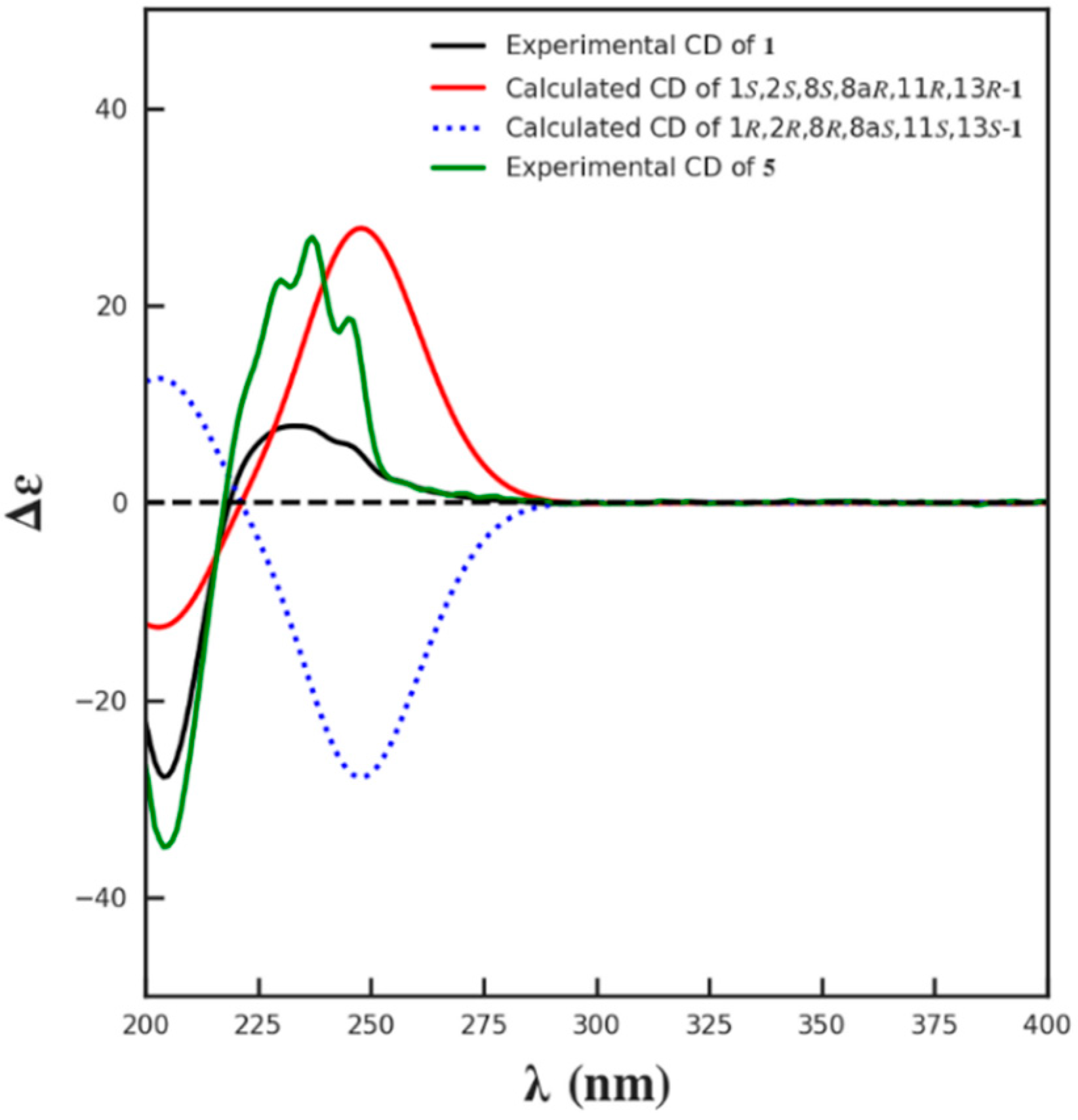
The relative configuration of 1 was supposed to be the same as that of 5, according to the NOESY correlations of H-8a to H-8/H-9a/H3-16 and H3-16 to H2-9. On the basis of the similar optical rotation values of 1 (+62.7) and 5 (+73.3), and further by comparison of their electronic circular dichroism (ECD) spectrum (Figure 3), 1 was then established to be 15-O-methyl ML-236A.
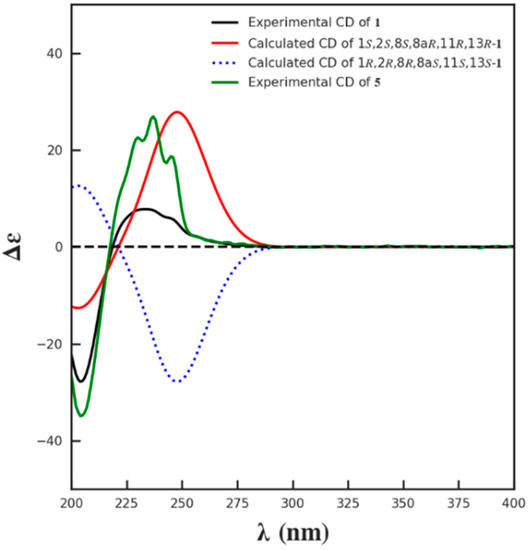
Figure 3.
The calculated ECD spectrum of 1 and the experimental ECD spectra of 1 and 5.
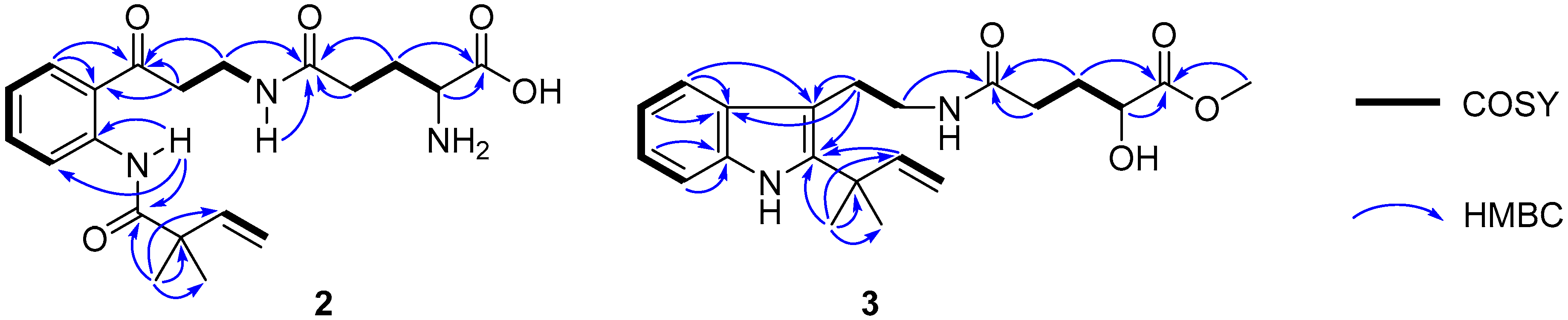
Compound 2 was assigned the molecular formula C20H27N3O5 on the basis of the [M − H]− ionic peak at m/z 388.2821 in its negative HRESIMS spectrum, suggesting nine degrees of unsaturation. The 1H and 13C NMR spectroscopic data, by the aide of the HSQC and 1H–1H COSY spectra, showed characteristics of a 1,2-disubsituted benzoic unit [δH 7.19 (t, J = 7.8 Hz, H-5), 7.60 (t, J = 7.8 Hz, H-6), 8.04 (t, J = 7.8 Hz, H-4), 8.54 (t, J = 7.8 Hz, H-7); δC 120.1 (d, C-7), 122.6 (s, C-7a), 122.8 (d, C-5), 131.4 (d, C-4), 134.5 (d, C-6), 139.9 (s, C-3a)], an isoprene [δH 1.32 (s × 2, C-11, 12), 5.25 (d, J = 17.4 Hz, H-10a), 5.29 (d, J = 10.6 Hz, H-10b),6.08 (d, J = 17.4, 10.6 Hz); δC 24.5 (q × 2, C-11, 12), 46.2 (s, C-8), 114.6 (t, C-10), 142.4 (d, C-9)], glutamic acid [δH 1.77−1.95 (m, H2-18), 2.23 (m, H2-17), 3.19 (m, H-19); δC 27.0 (t, C-18), 31.8 (t, C-17), 53.7 (d, C-19), 169.6 (s, C-20), 172.1 (s, C-16)], β-aminopropanone [δH 3.22 (t, J = 6.6 Hz, H2-13), 3.37 (m, H2-14), 8.16 (t, J = 4.9 Hz, H-15); δC 34.6 (t, C-14), 39.3 (t, C-13), 203.2 (s, C-3)], and one acylamide [δH 11.5 (s, H-1); δC 174.8 (s, C-2)]. These five fragments could be connected by the HMBC correlations of H3-11/H3-12 to C-2, H-1 to C-2/C-3a/C-7/C-7a, H2-4 to C-3, and H-14 to C-16 to construct the planar structure of 2 (Figure 4), the same as solitumidine D [10], namely 4, which was simultaneously obtained along with 2 by HPLC using the A4-5 chiral column. Since the specific optical rotation of 2 was +6, opposite to that of 4 (−7) in the same concentration of MeOH (c 0.10), 2 was then deduced to be the enantiomer of 4. Accordingly, 2 was determined as (+)-solitumidine D.
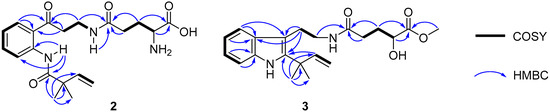
Figure 4.
Key 1H–1H COSY and HMBC correlations of 2 and 3.
Compound 3 presented its molecular formula asC21H28N2O4 by the positive HRESIMS at m/z 395.1947 [M + Na]+. The 1H and 13C NMR spectra consisted of signals almost the same as those of solitumidine B [10] except for an additional methoxyl unit. This was confirmed by the HMBC correlation of 20-OMe (δH 3.73, s) to C-20 (δC 176.0, s). Furthermore, by extensive analysis of the COSY and HMBC NMR spectra (Figure 4), 3 was determined as 20-O-methyl solitumidine B. Since the optical rotation value for solitumidine B was −55 in MeOH, while it was 0 for 3 in the same solvent, 3 was supposed to be a racemic mixture. As such, it was subjected to further isolation by HPLC with chiral columns. Yet, 3 seemed to be inseparable as it exhibited only one peak using several different mobile phases in A3-5 and A4-5 chiral columns, the latter of which was utilized to successfully isolate 2 from its enantiomer, 4. On the basis of the above evidence, 3 was then named as (±)-solitumidine E.
All isolates were tested for antiproliferative effect against 17 human tumor cell lines of A431, A549, MB231, MCF-7, PANC1, HepG2, HCT116, H460, H1299, QGY-7701, BGC823, SKGT4, A375, U2OS, HL-60, K562, and KYSE450 under the concentration of 20 μM and were tested for anti-food allergic activity under the concentration of 50 μM. Notably, viridicatol (13) showed significant cytotoxic activities against PANC1, Hela and A549 cells with IC50 values of 18, 19, and 24 μM, respectively.
Moreover, compounds 1–25 were also tested in vitro for anti-food allergic activity. Indole-3-acetic acid methyl ester (9) and penicopeptide A (10) showed modest activity (IC50 = 50 and 58 μM, respectively), while 13 displayed a potent effect with an IC50 value of 13 μM, compared to that of 92 μM for loratadine, an anti-food allergic medicine in clinic. In fact, viridicatol isolated from another deep-sea-derived fungus, Penicillium griseofulvum MCCC 3A00225, showed a significant anti-food allergic effect in the RBL-2H3 cell model and the ovalbumin-induced food allergy mouse [31]. Therefore, it may represent a novel therapeutic for allergic diseases.
3. Materials and Methods
3.1. General Experimental Procedures
NMR spectra were recorded on a Bruker 400 MHz spectrometer. The HRESIMS spectra were recorded on a Waters Q-TOF mass spectrometer (Xevo G2). Optical rotations were obtained with an Anton Paar polarimeter (MCP100). ECD spectra were measured on a Chirascan spectrometer. The semi-preparative HPLC was conducted on an Agilent instrument (1260) with different kinds of columns (COSMOSIL 5 C18-MS-II, Nacalai Tesque, Japan; ColumnTekTM Chiral A3-5 and A4-5). Column chromatography was performed on silica gel, Sephadex LH-20, and ODS.
3.2. Fungal Identification, Fermentation, and Extract
The fungus Penicillium solitum MCCC 3A00215 was isolated from a sediment sample of the Northwest Atlantic Ocean (−3034 m, W 44.9801°, N 14.7532°). For the large-scale fermentation procedure, see our recently published literature [12]. The crude extract (200 g) was subjected to column chromatography on silica gel using petroleum ether (PE), CH2Cl2, EtOAc to provide a CH2Cl2-soluble extract (11 g) and a EtOAc-soluble extract (114.5 g), respectively.
3.3. Isolation and Purification
The CH2Cl2 crude extract was separated into six fractions (Fr.A−Fr.F) via medium pressure liquid chromatography (MPLC, 460 mm × 36 mm) with gradient PE-EtOAc (5:1→1:5). Subfractions Fr.B-Fr.F were subsequently purified by column chromatography (CC) over Sephadex LH-20 (1.5 m × 3 cm; CH2Cl2-MeOH, 1:1) followed by semi-preparative HPLC with MeOH-H2O (40%→100%) to provide 10 (280 mg), 11 (2 mg), 17 (57 mg), 20 (3 mg), 21 (2 mg), 22 (1.5 mg), 23 (2 mg), and 25 (4.4 mg).
The EtOAc part was subjected to MPLC (460 mm × 46 mm) on silica gel with gradient CH2Cl2-MeOH (100%→50%) to obtain five fractions (Fr.1−Fr.5). Fraction Fr.1 (1 g) was separated by CC over Sephadex LH-20 (1.5 m × 3 cm, CH2Cl2-MeOH, 1:1) and subsequently purified by recrystallization to give 14 (60 mg). Fraction Fr.2 (4 g) was separated by CC over ODS (310 mm × 5 mm; MeOH-H2O, 10%→100%) and Sephadex LH-20 (1.5 m × 2 cm, MeOH), followed by semi-prep. HPLC (MeOH-H2O, 40%→100%) afforded 1 (22 mg), 3 (3 mg), 5 (6 mg), 7 (2 mg), 9 (12 mg), 12 (2 mg), 13 (1.4 g), 15 (3 mg), 16 (10 mg), 18 (3 mg), and 19 (50 mg), while 2 (4 mg), 4 (2 mg), 6 (30 mg), 8 (27 mg), and 24 (5 mg) were isolated from Fr.4 (8 g) by CC on ODS (310 mm × 5 mm; MeOH-H2O, 10%→100%) and Sephadex LH-20 (1.5 m × 2 cm, MeOH), followed by semi-prep. HPLC with chiral column A4-5 (MeOH-H2O, 40%→80%).
15-O-methyl ML-236A (1): colorless oil; [α]20D +62.7 (c 0.30, MeOH); UV (MeOH) λmax (logε) 237 (3.08) nm; CD (MeOH) (Δε) 204 (−1.90), 233 (+0.53), 236 (+0.53), 245 (+0.40) nm; 1H and 13C NMR data, see Table 1; HRESIMS m/z 361.1989 [M + Na]+ (calcd for C19H30O5Na, 361.1991).
(+)-solitumidine D (2): white amorphous solid; [α]20D +6 (c 0.10, MeOH); UV (MeOH) λmax (logε) 231 (4.73) nm, 261 (4.25) nm, 325 (3.85) nm; CD (MeOH) (Δε) 203 (+2.62) nm; 1H and 13C NMR data, see Table 1; HRESIMS m/z 388.1877 [M−H]− (calcd for C20H26N3O5, 388.1872).
(±)-Solitumidine E (3): white amorphous power; [α]20D 0 (c 0.44, MeOH); UV (MeOH) λmax (logε) 222 (4.10) nm, 261 (3.70) nm, 291 (3.52) nm; CD (MeOH) (Δε) 203 (−0.48), 299 (−0.04) nm; 1H and 13C NMR data, see Table 1; HRESIMS m/z 395.1945 [M + Na]+ (calcd for C21H28N2O4Na, 395.1947).
3.4. ECD Calculation
Conformational analysis was performed by the Sybyl-X 2.0 using the MMFF94S force field as reported [32]. Predominant conformers were relocated and confirmed at the B3LYP/6-31G(d) level. The theoretical ECD spectra were calculated with the time-dependent density functional theory (TD-DFT) in acetonitrile. The ECD spectrum was obtained by averaging each conformer using the Boltzmann distribution theory.
3.5. Cell Proliferation Assay
Cytotoxic activities of all isolates were conducted on 17 human tumor cell lines of A431, A549, MB231, MCF-7, PANC1, HepG2, HCT116, H460, H1299, QGY-7701, BGC823, SKGT4, A375, U2OS, HL-60, K562, and KYSE450 by the MTT method [33]. Paclitaxel was used as a positive control, and DMSO was used as a negative control. Different cancer cells were incubated on 96-well cell plates and cultured for 24 h. Thereafter, the cells were treated with different concentrations of tested compounds and controls. After 48 h, MTT (20 μL) was added to incubate for another 4 h. The supernatant was discarded softly, and the deposited formazan formed in the cells was dissolved with DMSO (100 μL). The absorbencies were measured at 490 nm.
3.6. Anti-Allergic Bioassay
The in vitro anti-food allergic experiment was performed as previously reported [32]. In brief, rat basophilic leukemia 2H3 (RBL-2H3) cells were incubated with dinitrophenyl (DNP)–immunoglobulin E (IgE) overnight. Then, the IgE-sensitized RBL-2H3 cells were pretreated with tested compounds and stimulated with DNP–bovine serum albumin (BSA). The bioactivity was quantified by measuring the fluorescence intensity of the hydrolyzed substrate in a fluorometer. Loratadine, a commercially available antiallergic medicine, was used as a positive control.
4. Conclusions
One new compactin analogue (1) and two previously unreported alkaloids (2 and 3), together with 22 known compounds (4–25), were isolated from the deep-sea-derived Penicillium solitum MCCC 3A00215. Viridicatol (13) exhibited weak cytotoxic activities against PANC-1, Hela, and A549 cells with IC50 values of 18, 19, and 24, respectively, while it showed remarkable anti-food allergic activity, with an IC50 value of 13 μM.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/md19100580/s1, Table S1 and Table S2, Figure S1–S65: One-dimensional and two-dimensional NMR spectra of all compounds.
Author Contributions
X.-W.Y. designed the project; Z.-H.H., L.X., M.-Y.H. and S.-J.L. isolated and purified all compounds. J.W. conducted the cytotoxic experiments. M.-M.X. and Y.-J.H. performed the fermentation. Z.-Z.S. provided the strain. Z.-H.H. and X.-W.Y. analyzed the data and wrote the paper, while critical revision of the publication was performed by all authors. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the National Natural Science Foundation of China (21877022) and the COMRA program (DY135-B2-08).
Institutional Review Board Statement
Not applicable.
Acknowledgments
The authors wish to thank Guangming Liu of the Jimei University for the anti-food allergic tests.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pitt, J.I.; Spotts, R.A.; Holmes, R.J.; Cruickshank, R.H. Penicillium solitum revived, and its role as a pathogen of pomaceous fruits. Phytopathology 1991, 81, 1108–1112. [Google Scholar] [CrossRef]
- Jurick, W.M., 2nd; Vico, I.; Gaskins, V.L.; Whitaker, B.D.; Garrett, W.M.; Janisiewicz, W.J.; Conway, W.S. Penicillium solitum produces a polygalacturonase isozyme in decayed Anjou pear fruit capable of macerating host tissue in vitro. Mycologia 2012, 104, 604–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, F.; Filtenborg, O.; Frisvad, J.C. Associated mycoflora of cheese. Food Microbiol. 1995, 12, 173–180. [Google Scholar] [CrossRef]
- Sørensen, L.M.; Jacobsen, T.; Nielsen, P.V.; Frisvad, J.C.; Koch, A.G. Mycobiota in the processing areas of two different meat products. Int. J. Food Microbiol. 2008, 124, 58–64. [Google Scholar] [CrossRef]
- Stierle, D.B.; Stierle, A.A.; Girtsman, T.; McIntyre, K.; Nichols, J. Caspase-1 and -3 inhibiting drimane sesquiterpenoids from the extremophilic fungus Penicillium solitum. J. Nat. Prod. 2012, 75, 262–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, V.N.; Campos, L.S.; Melo, I.S.; Pellizari, V.H.; Rosa, C.A.; Rosa, L.H. Penicillium solitum: A mesophilic, psychrotolerant fungus present in marine sediments from Antarctica. Polar Biol. 2013, 36, 1823–1831. [Google Scholar] [CrossRef]
- Boruta, T.; Przerywacz, P.; Ryngajllo, M.; Bizukojc, M. Bioprocess-related, morphological and bioinformatic perspectives on the biosynthesis of secondary metabolites produced by Penicillium solitum. Process Biochem. 2018, 68, 12–21. [Google Scholar] [CrossRef]
- Larsen, T.O.; Lange, L.; Schnorr, K.; Stender, S.; Frisvad, J.C. Solistatinol, a novel phenolic compactin analogue from Penicillium solitum. Tetrahedron Lett. 2007, 48, 1261–1264. [Google Scholar] [CrossRef]
- Sørensen, D.; Larsen, T.O.; Christophersen, C.; Nielsen, P.H.; Anthoni, U. Solistatin, an aromatic compactin analogue from Penicillium solitum. Phytochemistry 1999, 51, 1027–1029. [Google Scholar] [CrossRef]
- Rodriguez, J.P.G.; Bernardi, D.I.; Gubiani, J.R.; de Oliveira, J.M.; Morais-Urano, R.P.; Bertonha, A.F.; Bandeira, K.F.; Bulla, J.I.Q.; Sette, L.D.; Ferreira, A.G.; et al. Water-soluble glutamic acid derivatives produced in culture by Penicillium solitum IS1-A from King George Island, Maritime Antarctica. J. Nat. Prod. 2020, 83, 55–65. [Google Scholar] [CrossRef]
- Guo, W.; Kong, X.; Zhu, T.; Gu, Q.; Li, D. Penipyrols A-B and peniamidones A-D from the mangrove derived Penicillium solitum GWQ-143. Arch. Pharmacal Res. 2015, 38, 1449–1454. [Google Scholar] [CrossRef]
- He, Z.H.; Xie, C.L.; Hao, Y.J.; Xu, L.; Wang, C.F.; Hu, M.Y.; Li, S.J.; Zhong, T.H.; Yang, X.W. Solitumergosterol A, a unique 6/6/6/6/5 steroid from the deep-sea-derived Penicillium solitum MCCC 3A00215. Org. Biomol. Chem. 2021. [Google Scholar] [CrossRef]
- Endo, A.; Kuroda, M.; Tsujita, Y. ML-236A, ML-236B, and ML-236C, new inhibitors of cholesterogenesis produced by Penicillium citrinium. J. Antibiot. 1976, 29, 1346–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaala, L.A.; Youssef, D.T. Identification and bioactivity of compounds from the fungus Penicillium sp. CYE-87 isolated from a marine tunicate. Mar. Drugs 2015, 13, 1698–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evidente, A.; Iacobellis, N.S.; Sisto, A. Isolation of indole-3-acetic acid methyl ester, a metabolite of indole-3-acetic acid from Pseudomonas amygdali. Experientia 1993, 49, 182–183. [Google Scholar] [CrossRef]
- Sun, W.; Chen, X.; Tong, Q.; Zhu, H.; He, Y.; Lei, L.; Xue, Y.; Yao, G.; Luo, Z.; Wang, J.; et al. Novel small molecule 11beta-HSD1 inhibitor from the endophytic fungus Penicillium commune. Sci. Rep. 2016, 6, 26418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashiwada, Y.; Nonaka, G.; Nishioka, I. Studies on Rhubarb (Rhei Rhizoma). V. isolation and characterization of chromone and chromanone derivatives. Chem. Pharm. Bull. 1984, 32, 3493–3500. [Google Scholar] [CrossRef] [Green Version]
- Talontsi, F.M.; Facey, P.; Tatong, M.D.; Islam, M.T.; Frauendorf, H.; Draeger, S.; Tiedemann, A.; Laatsch, H. Zoosporicidal metabolites from an endophytic fungus Cryptosporiopsis sp. of Zanthoxylum leprieurii. Phytochemistry 2012, 83, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Harayama, T. A concise and versatile synthesis of Viridicatin alkaloids from cyanoacetanilides. Org. Lett. 2009, 11, 1603–1606. [Google Scholar] [CrossRef]
- Fremlin, L.J.; Piggott, A.M.; Lacey, E.; Capon, R.J. Cottoquinazoline A and cotteslosins A and B, metabolites from an Australian marine-derived strain of Aspergillus Wersicolor. J. Nat. Prod. 2009, 72, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Hodge, R.P.; Harris, C.M.; Harris, T.M. Verrucofortine, a major metabolite of Penicillium verrucosum var. Cyclopium, the fungus that produces the Mycotoxin Verrucosidin. J. Nat. Prod. 1988, 51, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Mizushina, Y.; Nakanishi, R.; Kuriyama, I.; Kamiya, K.; Satake, T.; Shimazaki, N.; Koiwai, O.; Uchiyama, Y.; Yonezawa, Y.; Takemura, M.; et al. β-sitosterol-3-O-β-D-glucopyranoside: A eukaryotic DNA polymerase lambda inhibitor. J. Steroid Biochem. Mol. Biol. 2006, 99, 100–107. [Google Scholar] [CrossRef]
- Koga, J.; Yamauchi, T.; Shimura, M.; Ogawa, N.; Oshima, K.; Umemura, K.; Kikuchi, M.; Ogasawara, N. Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J. Biol. Chem. 1998, 273, 31985–31991. [Google Scholar] [CrossRef] [Green Version]
- Soman, A.G.; Gloer, J.B.; Wicklow, D.T. Antifungal and antibacterial metabolites from a sclerotium-colonizing isolate of Mortierella vinacea. J. Nat. Prod. 1999, 62, 386–388. [Google Scholar] [CrossRef]
- Du, F.Y.; Li, X.M.; Zhang, P.; Li, C.S.; Wang, B.G. Cyclodepsipeptides and other O-containing heterocyclic metabolites from Beauveria felina EN-135, a marine-derived entomopathogenic fungus. Mar. Drugs 2014, 12, 2816–2826. [Google Scholar] [CrossRef] [Green Version]
- Arunpanichlert, J.; Rukachaisirikul, V.; Phongpaichit, S.; Supaphon, O.; Sakayaroj, J. Xylariphilone: A new azaphilone derivative from the seagrass-derived fungus Xylariales sp. PSU-ES163. Nat. Prod. Res. 2016, 30, 46–51. [Google Scholar] [CrossRef]
- Sommart, U.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Towatana, N.H.; Graidist, P.; Hajiwangoh, Z.; Sakayaroj, J. A cyclohexenone derivative from Diaporthaceous fungus PSU-H2. Arch. Pharmacal Res. 2009, 32, 1227–1231. [Google Scholar] [CrossRef]
- Kishida, M.; Yamauchi, N.; Sawada, K.; Ohashi, Y.; Eguchi, T.; Kakinuma, K. Diacetone-glucose architecture as a chirality template. Part 9.1 enantioselective synthesis of (R)-mevalonolactone and (R)-[2H9]mevalonolactone on carbohydrate template. J. Chem. Soc. Perkin Trans. 1997, 1, 891–896. [Google Scholar] [CrossRef]
- Shimomura, H.; Sashida, Y.; Mimaki, Y.; Adachi, T.; Yoshinari, K. A new mevalonolactone glucoside derivative from the bark of Prunus buergeriana. Chem. Pharm. Bull. 1989, 37, 829–830. [Google Scholar] [CrossRef] [Green Version]
- Endo, A.; Kuroda, M.; Tanzawa, K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 1976, 72, 323–326. [Google Scholar] [CrossRef] [Green Version]
- Shu, Z.; Liu, Q.; Xing, C.; Zhang, Y.; Zhou, Y.; Zhang, J.; Liu, H.; Cao, M.; Yang, X.; Liu, G. Viridicatol isolated from deep-sea Penicillium griseofulvum alleviates anaphylaxis and repairs the intestinal barrier in mice by suppressing mast cell activation. Mar. Drugs 2020, 18, 517. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.L.; Liu, Q.; He, Z.H.; Gai, Y.B.; Zou, Z.B.; Shao, Z.Z.; Liu, G.M.; Chen, H.F.; Yang, X.W. Discovery of andrastones from the deep-sea-derived Penicillium allii-sativi MCCC 3A00580 by OSMAC strategy. Bioorg. Chem. 2021, 108, 104671. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.F.; Huang, X.F.; Xiao, H.X.; Hao, Y.J.; Xu, L.; Yan, Q.X.; Zou, Z.B.; Xie, C.L.; Xu, Y.Q.; Yang, X.W. Chemical constituents of the marine fungus Penicillium sp. MCCC 3A00228. Chem. Biodivers. 2021, 18, e2100697. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).