Structure Characteristics, Biochemical Properties, and Pharmaceutical Applications of Alginate Lyases
Abstract
1. Introduction
2. Alginate Lyases
2.1. Sources of Alginate Lyases
2.2. Classification of Alginate Lyases
2.3. Catalytic Mechanism
- (1)
- Neutralization of carboxyl group on the substrate by a salt bridge;
- (2)
- Abstract the proton on C5 by a general base reaction. The H-5 proton has to be accepted by a Brønsted base residue, and a Brønsted acid residue serves as a proton donor;
- (3)
- Transfer of electrons from the carboxyl group to cleave the 4-O-glycosidic bond, which generates a double bond between C4 and C5. When the 4-O-glycosidic bond is eliminated by the alginate lyases, oligosaccharides containing 4-deoxy-L-erythro-hex-4-enopyranosyluronic acid as the unsaturated non-reducing terminal are created simultaneously, which can be called unsaturated alginate oligosaccharide [1,18,27]. By analysis of configurations at C-4 and C-5, which takes the relative position between abstracted proton and the C-4-bridging oxygen into consideration, the products can be classified as two different configurations: syn-configuration and anti-configuration. For M residues, the C-5 proton and the C-4-bridging oxygen lie syn relative to each other, which is in contrast to G residues, whose C-5 proton and C-4-bridging oxygen lie anti relative to each other [28].
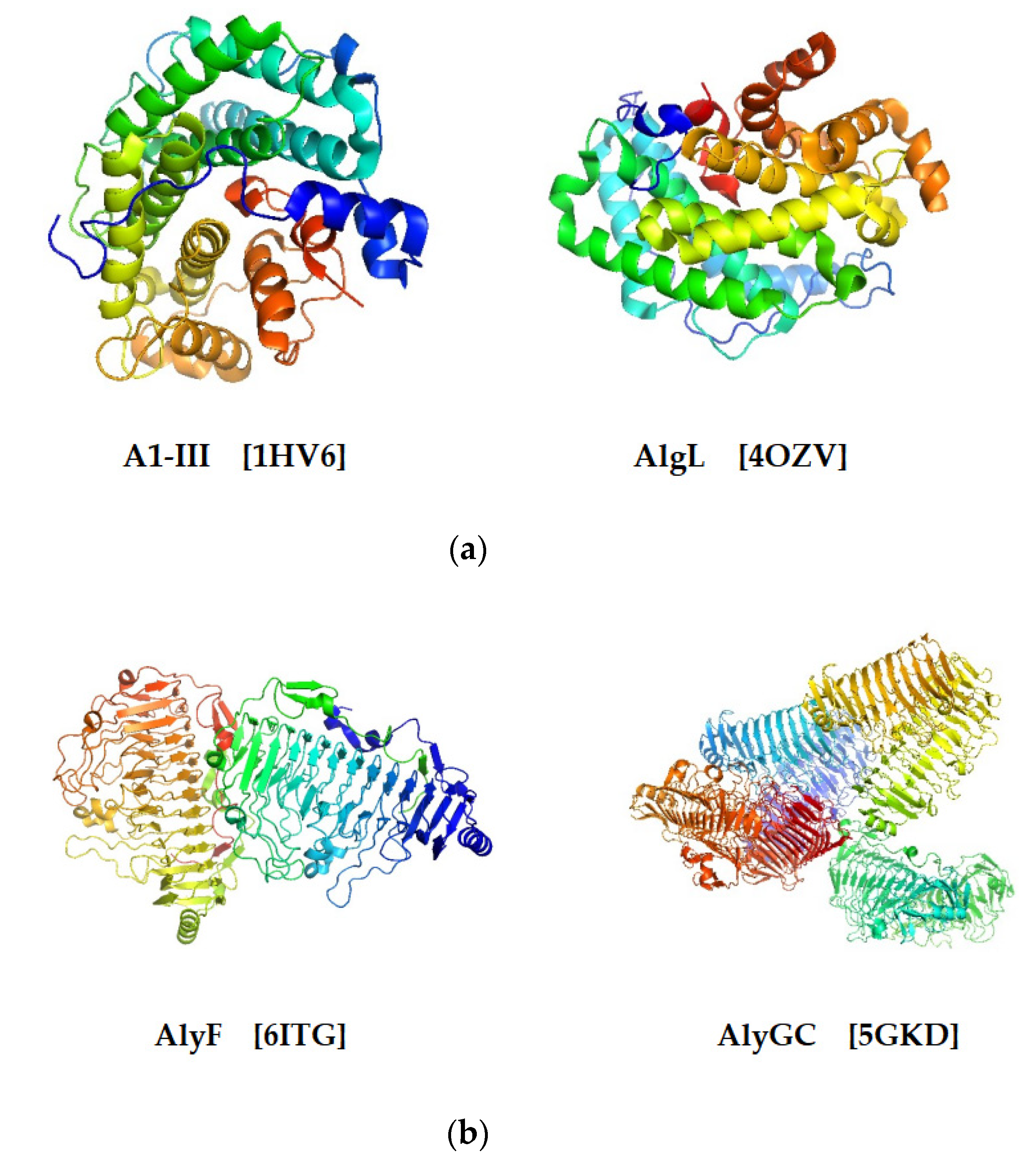
2.4. Structure Characteristics
- (1)
- (α/α)n toroid fold (Figure 4a). This type of three-dimensional structure is constructed by several antiparallel α-helices. The arrangement of helices toroid is counterclockwise looking from the top of it. The majority of alginate lyases from the PL5 family contain only one catalytic domain that adopts a tunnel-like barrel architecture formed by several antiparallel α-helices [28]. For example, the active site of A1-III from PL5 is formed by 12 α-helices, which form an α6/α5 barrel fold with a deep tunnel-like cleft [39]. The structure suggests a mode of action between substrates and catalytic sites, in which substrate molecules penetrate into the tunnel-like cleft and further interact with the catalytic site [39].
- (2)
- β-helix (Figure 4b). Alginate lyases from PL6 and PL31 adopt such a structure, which consists of three β-sheets (PB) and three turns (T) [28]. The turns (T) are located between two β-sheets, respectively, which form a coil of the β-helix with PB together: PB1-T1-PB2-T2-PB3-T3 [40]. Several alginate lyases adopting β-helix structure have been investigated clearly with the help of structural biology techniques, such as X-ray diffraction. The PL6 alginate lyase BcAlyPL6 is a monomer in solution, and it is comprised of two domains, the N-terminal domain (NTD) and the C-terminal domain (CTD), both of which adopt the right-handed parallel β-helix fold [15]. Although the monomer character in solution is different from another PL6 lyase, AlyGC, which forms a homodimer in solution [29], the two domains’ structure is similar to AlyGC and distinct from other structure-determined lyases that adopt this fold but contain only a single domain, such as AlyF [38] and BcelPL6 [14]. Biochemical analysis indicates that the substrate-binding affinity is mainly contributed by the NTD, while CTD of BcAlyPL6 is involved in fixing the substrates into appropriate conformation. However, CTD has weak alginate lyase activity, which may cooperate with the PL6 domain for more effective catalysis. Furthermore, CTD is involved in shaping a closed catalytic pocket, and deletion of it leads to increased activity towards highly polymerized substrate [15].
- (3)
- β-jelly roll (Figure 4c). The β-jelly roll class, also named as β-Sandwich jelly roll, is the most common and most thoroughly investigated fold structure [28]. Up to the time of this publication, four PL families (PL7, 14, 18, 36) adopt such structure, including 12 alginate lyases from PL7, 2 from PL14, 2 from PL18, and 1 from PL36 (Table 1). The structure can be divided into two curved antiparallel β-sheets that are linked with each other: the inner concave sheet (SA) and the outer concave sheet (SB) [40]. They further bent in the middle to form a globular shape by nearly 90°. The inner concave sheet plays an indispensable role in catalyzing the reaction, which forms a cleft containing catalytic sites and binding the substrates [28].
- (4)
- (α/α)n toroid fold + anti-parallel β-sheet (Figure 4d). The last fold structure is more complex than the other three kinds of structures mentioned before. This multidomain alginate lyases combine (α/α)n toroid fold domains with anti-parallel β-sheet domains. It is reported that PL15, 17, 39 adopt (α/α)n toroid fold + anti-parallel β-sheet fold structure, including 1 from PL15, 2 from PL17, and 1 from PL39 (Table 1). Ji et al. determined the structure of PL39 whose catalytic domains can be divided into three parts: NTD formed from an incomplete (α/α)6 toroid, central domain constructed by 16 antiparallel strands arranged in two sheets together with a distorted α-helix, and CTD consisting of a typical β-sandwich [41]. Atu3025 from PL15 adopts α/α barrel + anti-parallel β-sheet fold. Additionally, a pocket-like structure, which is formed by the conformational change at the interface between the central and CTD, has been discovered in Atu3025, which is essential for the exolytic mode of action. This kind of structure is special because it is not reported in other alginate lyases, and lyases that have been reported to have conformational change do not show such pocket-link structure either [35]. The crystal structure of AlyA3 from PL17 has been well investigated [42]. It is similar to another PL17 family lyase, Alg17c, which was the first structure-determined alginate lyase of the PL17 family and organized in two domains, an N-terminal (α/α)6 barrel fused to a C-terminal β-sheet domain [36].
2.5. Structure–Substrate Specificity Relationship
2.6. Special Properties of Alginate Lyases
2.6.1. Salt-Activated Property
2.6.2. Wide pH Adaptation Range and Alkaline Property
2.6.3. Thermostable Property
2.6.4. Cold-Adapted Property
Basic Characteristics of Cold-Adapted Property
Advantages of Cold-Adapted Alginate Lyases
Cold-Adapted Alginate Lyases Excreting Bacteria
2.7. Strategies for Improving Application Ability
2.8. Applications of Alginate Lyases
2.8.1. Antibiotic Applications
2.8.2. Preparation of Alginate Oligosaccharides
2.8.3. Preparation of Pharmaceutical Intermediate
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhu, B.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, Y.; Qin, Y.; Shen, P.; Peng, Q. Structures, properties and application of alginic acid: A review. Int. J. Biol. Macromol. 2020, 162, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, S.; Li, X.; Yan, Q.; Reaney, M.J.T.; Jiang, Z. Alginate Oligosaccharides: Production, Biological Activities, and Potential Applications. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1859–1881. [Google Scholar] [CrossRef] [PubMed]
- Peteiro, C. Alginate Production from Marine Macroalgae, with Emphasis on Kelp Farming. In Alginates and Their Biomedical Applications; Springer Nature: Cham, Switzerland, 2018; pp. 27–66. [Google Scholar] [CrossRef]
- Mrudulakumari Vasudevan, U.; Lee, O.K.; Lee, E.Y. Alginate derived functional oligosaccharides: Recent developments, barriers, and future outlooks. Carbohydr. Polym. 2021, 267, 118158. [Google Scholar] [CrossRef]
- Nakata, S.; Murata, K.; Hashimoto, W.; Kawai, S. Uncovering the reactive nature of 4-deoxy-L-erythro-5-hexoseulose uronate for the utilization of alginate, a promising marine biopolymer. Sci. Rep. 2019, 9, 17147. [Google Scholar] [CrossRef]
- Takagi, T.; Sasaki, Y.; Motone, K.; Shibata, T.; Tanaka, R.; Miyake, H.; Mori, T.; Kuroda, K.; Ueda, M. Construction of bioengineered yeast platform for direct bioethanol production from alginate and mannitol. Appl. Microbiol. Biotechnol. 2017, 101, 6627–6636. [Google Scholar] [CrossRef]
- Matsuoka, F.; Hirayama, M.; Kashihara, T.; Tanaka, H.; Hashimoto, W.; Murata, K.; Kawai, S. Crucial role of 4-deoxy-L-erythro-5-hexoseulose uronate reductase for alginate utilization revealed by adaptive evolution in engineered Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 4206. [Google Scholar] [CrossRef]
- Ryu, M.; Lee, E.Y. Saccharification of alginate by using exolytic oligoalginate lyase from marine bacterium Sphingomonas sp. MJ-3. J. Ind. Eng. Chem. 2011, 17, 853–858. [Google Scholar] [CrossRef]
- Camus, C.; Ballerino, P.; Delgado, R.; Olivera-Nappa, Á.; Leyton, C.; Buschmann, A.H. Scaling up bioethanol production from the farmed brown macroalga Macrocystis pyriferain in Chile. Biofuels Bioprod. Biorefining 2016, 10, 673–685. [Google Scholar] [CrossRef]
- Dong, S.; Yang, J.; Zhang, X.Y.; Shi, M.; Song, X.Y.; Chen, X.L.; Zhang, Y.Z. Cultivable alginate lyase-excreting bacteria associated with the Arctic brown alga Laminaria. Mar. Drugs 2012, 10, 2481–2491. [Google Scholar] [CrossRef]
- Inoue, A.; Mashino, C.; Uji, T.; Saga, N.; Mikami, K.; Ojima, T. Characterization of an Eukaryotic PL-7 Alginate Lyase in the Marine Red Alga Pyropia yezoensis. Curr. Biotechnol. 2015, 4, 240–248. [Google Scholar] [CrossRef]
- Xue, X.; Zhou, Y.; Gao, X.; Yan, P. Advances in Application of Alginate Lyase And Its Enzymatic Hydrolysate. IOP Conf. Ser. Mater. Sci. Eng. 2019, 612, 022005. [Google Scholar] [CrossRef]
- Stender, E.G.P.; Dybdahl Andersen, C.; Fredslund, F.; Holck, J.; Solberg, A.; Teze, D.; Peters, G.H.J.; Christensen, B.E.; Aachmann, F.L.; Welner, D.H.; et al. Structural and functional aspects of mannuronic acid-specific PL6 alginate lyase from the human gut microbe Bacteroides cellulosilyticus. J. Biol. Chem. 2019, 294, 17915–17930. [Google Scholar] [CrossRef]
- Wang, B.; Dong, S.; Li, F.L.; Ma, X.Q. Structural basis for the exolytic activity of polysaccharide lyase family 6 alginate lyase BcAlyPL6 from human gut microbe Bacteroides clarus. Biochem. Biophys. Res. Commun. 2021, 547, 111–117. [Google Scholar] [CrossRef]
- Zeng, J.; An, D.; Jiao, C.; Xiao, Q.; Weng, H.; Yang, Q.; Xiao, A. Cloning, expression, and characterization of a new pH- and heat-stable alginate lyase from Pseudoalteromonas carrageenovora ASY5. J. Food Biochem. 2019, 43, e12886. [Google Scholar] [CrossRef]
- Kim, H.-S. Enhancing the Alginate Degrading Activity of Streptomyces sp. Strain M3 Alginate Lyase by Mutation. J. Life Sci. 2012, 22, 7–15. [Google Scholar] [CrossRef]
- Cheng, D.; Jiang, C.; Xu, J.; Liu, Z.; Mao, X. Characteristics and applications of alginate lyases: A review. Int. J. Biol. Macromol. 2020, 164, 1304–1320. [Google Scholar] [CrossRef]
- Chao, Y.; Wang, S.; Wu, S.; Wei, J.; Chen, H. Cloning and characterization of a new endo-type polyG-specific alginate lyase from bacteria Vibrio sp. QD-5. Acta Oceanol. Sin. 2019, 38, 65–74. [Google Scholar] [CrossRef]
- Lee, S.I.; Choi, S.H.; Lee, E.Y.; Kim, H.S. Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 2012, 95, 1643–1653. [Google Scholar] [CrossRef]
- Li, J.W.; Dong, S.; Song, J.; Li, C.B.; Chen, X.L.; Xie, B.B.; Zhang, Y.Z. Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 2011, 9, 109–123. [Google Scholar] [CrossRef]
- Zhu, B.; Ni, F.; Sun, Y.; Ning, L.; Yao, Z. Elucidation of degrading pattern and substrate recognition of a novel bifunctional alginate lyase from Flammeovirga sp. NJ-04 and its use for preparation alginate oligosaccharides. Biotechnol. Biofuels 2019, 12, 13. [Google Scholar] [CrossRef]
- Hirayama, M.; Hashimoto, W.; Murata, K.; Kawai, S. Comparative characterization of three bacterial exo-type alginate lyases. Int. J. Biol. Macromol. 2016, 86, 519–524. [Google Scholar] [CrossRef][Green Version]
- Lu, D.; Zhang, Q.; Wang, S.; Guan, J.; Jiao, R.; Han, N.; Han, W.; Li, F. Biochemical characteristics and synergistic effect of two novel alginate lyases from Photobacterium sp. FC615. Biotechnol. Biofuels 2019, 12, 260. [Google Scholar] [CrossRef]
- Yang, J.; Cui, D.; Ma, S.; Chen, W.; Chen, D.; Shen, H. Characterization of a novel PL 17 family alginate lyase with exolytic and endolytic cleavage activity from marine bacterium Microbulbifer sp. SH-1. Int. J. Biol. Macromol. 2021, 169, 551–563. [Google Scholar] [CrossRef]
- Gacesa, P. Enzymic degradation of alginates. Int. J. Biochem. 1992, 24, 545–552. [Google Scholar] [CrossRef]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. ALGINATE LYASE: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef]
- Xu, F.; Wang, P.; Zhang, Y.Z.; Chen, X.L. Diversity of Three-Dimensional Structures and Catalytic Mechanisms of Alginate Lyases. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- Xu, F.; Dong, F.; Wang, P.; Cao, H.Y.; Li, C.Y.; Li, P.Y.; Pang, X.H.; Zhang, Y.Z.; Chen, X.L. Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6. J. Biol. Chem. 2017, 292, 4457–4468. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Xu, F.; Chen, X.L.; Li, P.Y.; Li, C.Y.; Li, F.C.; Chen, Y.; Wang, P.; Zhang, Y.Z. Alginate Lyase Aly36B is a New Bacterial Member of the Polysaccharide Lyase Family 36 and Catalyzes by a Novel Mechanism With Lysine as Both the Catalytic Base and Catalytic Acid. J. Mol. Biol. 2019, 431, 4897–4909. [Google Scholar] [CrossRef] [PubMed]
- Mikami, B.; Ban, M.; Suzuki, S.; Yoon, H.J.; Miyake, O.; Yamasaki, M.; Ogura, K.; Maruyama, Y.; Hashimoto, W.; Murata, K. Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Ogura, K.; Yamasaki, M.; Mikami, B.; Hashimoto, W.; Murata, K. Substrate recognition by family 7 alginate lyase from Sphingomonas sp. A1. J. Mol. Biol. 2008, 380, 373–385. [Google Scholar] [CrossRef]
- Narsico, J.; Inoue, A.; Oka, S.; Ojima, T. Production of a novel dimeric 4-deoxy-L-erythro-5-hexoseulose uronic acid by a PL-17 exolytic alginate lyase from Hydrogenophaga sp. UMI-18. Biochem. Biophys. Res. Commun. 2020, 525, 982–988. [Google Scholar] [CrossRef]
- Garron, M.L.; Cygler, M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 2010, 20, 1547–1573. [Google Scholar] [CrossRef]
- Ochiai, A.; Yamasaki, M.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens. J. Biol. Chem. 2010, 285, 24519–24528. [Google Scholar] [CrossRef]
- Park, D.; Jagtap, S.; Nair, S.K. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J. Biol. Chem. 2014, 289, 8645–8655. [Google Scholar] [CrossRef]
- Rozeboom, H.J.; Bjerkan, T.M.; Kalk, K.H.; Ertesvag, H.; Holtan, S.; Aachmann, F.L.; Valla, S.; Dijkstra, B.W. Structural and mutational characterization of the catalytic A-module of the mannuronan C-5-epimerase AlgE4 from Azotobacter vinelandii. J. Biol. Chem. 2008, 283, 23819–23828. [Google Scholar] [CrossRef]
- Lyu, Q.; Zhang, K.; Shi, Y.; Li, W.; Diao, X.; Liu, W. Structural insights into a novel Ca(2+)-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1167–1176. [Google Scholar] [CrossRef]
- Yoon, H.J.; Hashimoto, W.; Miyake, O.; Murata, K.; Mikami, B. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution. J. Mol. Biol. 2001, 307, 9–16. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, L.; Guo, Z.; Tang, T.; Zhu, B. Alginate degrading enzymes: An updated comprehensive review of the structure, catalytic mechanism, modification method and applications of alginate lyases. Crit. Rev. Biotechnol. 2021, 41, 953–968. [Google Scholar] [CrossRef]
- Ji, S.; Dix, S.R.; Aziz, A.A.; Sedelnikova, S.E.; Baker, P.J.; Rafferty, J.B.; Bullough, P.A.; Tzokov, S.B.; Agirre, J.; Li, F.L.; et al. The molecular basis of endolytic activity of a multidomain alginate lyase from Defluviitalea phaphyphila, a representative of a new lyase family, PL39. J. Biol. Chem. 2019, 294, 18077–18091. [Google Scholar] [CrossRef]
- Jouanneau, D.; Klau, L.J.; Larocque, R.; Jaffrennou, A.; Duval, G.; Le Duff, N.; Roret, T.; Jeudy, A.; Aachmann, F.L.; Czjzek, M.; et al. Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT. Glycobiology 2021, 23, 1638–1655. [Google Scholar] [CrossRef]
- Osawa, T.; Matsubara, Y.; Muramatsu, T.; Kimura, M.; Kakuta, Y. Crystal structure of the alginate (poly alpha-l-guluronate) lyase from Corynebacterium sp. at 1.2 A resolution. J. Mol. Biol. 2005, 345, 1111–1118. [Google Scholar] [CrossRef]
- Qin, H.M.; Miyakawa, T.; Inoue, A.; Nishiyama, R.; Nakamura, A.; Asano, A.; Ojima, T.; Tanokura, M. Structural basis for controlling the enzymatic properties of polymannuronate preferred alginate lyase FlAlyA from the PL-7 family. Chem. Commun. 2018, 54, 555–558. [Google Scholar] [CrossRef]
- Sim, P.F.; Furusawa, G.; Teh, A.H. Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2. Sci. Rep. 2017, 7, 13656. [Google Scholar] [CrossRef]
- Yamasaki, M.; Moriwaki, S.; Miyake, O.; Hashimoto, W.; Murata, K.; Mikami, B. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: A novel alginate lyase with a beta-sandwich fold. J. Biol. Chem. 2004, 279, 31863–31872. [Google Scholar] [CrossRef]
- Xu, F.; Chen, X.L.; Sun, X.H.; Dong, F.; Li, C.Y.; Li, P.Y.; Ding, H.; Chen, Y.; Zhang, Y.Z.; Wang, P. Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic. J. Biol. Chem. 2020, 295, 16380–16392. [Google Scholar] [CrossRef]
- Yamasaki, M.; Ogura, K.; Hashimoto, W.; Mikami, B.; Murata, K. A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J. Mol. Biol. 2005, 352, 11–21. [Google Scholar] [CrossRef]
- Thomas, F.; Lundqvist, L.C.; Jam, M.; Jeudy, A.; Barbeyron, T.; Sandstrom, C.; Michel, G.; Czjzek, M. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 2013, 288, 23021–23037. [Google Scholar] [CrossRef]
- Pilgaard, B.; Vuillemin, M.; Holck, J.; Wilkens, C.; Meyer, A.S. Specificities and Synergistic Actions of Novel PL8 and PL7 Alginate Lyases from the Marine Fungus Paradendryphiella salina. J. Fungi 2021, 7, 80. [Google Scholar] [CrossRef]
- Ogura, K.; Yamasaki, M.; Yamada, T.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action. J. Biol. Chem. 2009, 284, 35572–35579. [Google Scholar] [CrossRef]
- Qin, H.M.; Miyakawa, T.; Inoue, A.; Nishiyama, R.; Nakamura, A.; Asano, A.; Sawano, Y.; Ojima, T.; Tanokura, M. Structure and Polymannuronate Specificity of a Eukaryotic Member of Polysaccharide Lyase Family 14. J. Biol. Chem. 2017, 292, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Wei, T.D.; Chen, X.L.; Li, C.Y.; Wang, P.; Xie, B.B.; Qin, Q.L.; Zhang, X.Y.; Pang, X.H.; Zhou, B.C.; et al. Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18. J. Biol. Chem. 2014, 289, 29558–29569. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Nakagawa, E.; Yoda, M.; Nakaichi, A.; Hibi, T.; Kimoto, H. Structural and biochemical characterisation of a novel alginate lyase from Paenibacillus sp. str. FPU-7. Sci. Rep. 2019, 9, 14870. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, C.-G.; Lee, E.Y. Alginate lyase: Structure, property, and application. Biotechnol. Bioprocess Eng. 2011, 16, 843–851. [Google Scholar] [CrossRef]
- Yang, J.; Cui, D.; Chen, D.; Chen, W.; Ma, S.; Shen, H. Purification and Characterization of a Novel Endolytic Alginate Lyase from Microbulbifer sp. SH-1 and Its Agricultural Application. Mar. Drugs 2020, 18, 184. [Google Scholar] [CrossRef]
- Zhuang, J.; Zhang, K.; Liu, X.; Liu, W.; Lyu, Q.; Ji, A. Characterization of a Novel PolyM-Preferred Alginate Lyase from Marine Vibrio splendidus OU02. Mar. Drugs 2018, 16, 295. [Google Scholar] [CrossRef]
- Chen, X.L.; Dong, S.; Xu, F.; Dong, F.; Li, P.Y.; Zhang, X.Y.; Zhou, B.C.; Zhang, Y.Z.; Xie, B.B. Characterization of a New Cold-Adapted and Salt-Activated Polysaccharide Lyase Family 7 Alginate Lyase from Pseudoalteromonas sp. SM0524. Front. Microbiol. 2016, 7, 1120. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, T.; Liu, W.; Lyu, Q. Structural insights into the substrate-binding cleft of AlyF reveal the first long-chain alginate-binding mode. Acta Crystallogr. Sect. D Struct. Biol. 2021, 77, 336–346. [Google Scholar] [CrossRef]
- Xu, X.; Zeng, D.; Wu, D.; Lin, J. Single-Point Mutation Near Active Center Increases Substrate Affinity of Alginate Lyase AlgL-CD. Appl. Biochem. Biotechnol. 2021, 193, 1513–1531. [Google Scholar] [CrossRef]
- Huang, G.; Wang, Q.; Lu, M.; Xu, C.; Li, F.; Zhang, R.; Liao, W.; Huang, S. AlgM4: A New Salt-Activated Alginate Lyase of the PL7 Family with Endolytic Activity. Mar. Drugs 2018, 16, 120. [Google Scholar] [CrossRef]
- Uchimura, K.; Miyazaki, M.; Nogi, Y.; Kobayashi, T.; Horikoshi, K. Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. 2010, 12, 526–533. [Google Scholar] [CrossRef]
- Kobayashi, T.; Uchimura, K.; Miyazaki, M.; Nogi, Y.; Horikoshi, K. A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp. Extremophiles 2009, 13, 121–129. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Bi, X.; Ren, Y.; Han, Q.; Zhou, Y.; Han, Y.; Yao, R.; Li, S. Characterization of an Alkaline Alginate Lyase with pH-Stable and Thermo-Tolerance Property. Mar. Drugs 2019, 17, 308. [Google Scholar] [CrossRef]
- Zhu, B.; Ni, F.; Ning, L.; Sun, Y.; Yao, Z. Cloning and characterization of a new pH-stable alginate lyase with high salt tolerance from marine Vibrio sp. NJ-04. Int. J. Biol. Macromol. 2018, 115, 1063–1070. [Google Scholar] [CrossRef]
- Ma, Y.; Li, J.; Zhang, X.Y.; Ni, H.D.; Wang, F.B.; Wang, H.Y.; Wang, Z.P. Characterization of a New Intracellular Alginate Lyase with Metal Ions-Tolerant and pH-Stable Properties. Mar. Drugs 2020, 18, 416. [Google Scholar] [CrossRef]
- Yan, J.; Chen, P.; Zeng, Y.; Men, Y.; Mu, S.; Zhu, Y.; Chen, Y.; Sun, Y. The Characterization and Modification of a Novel Bifunctional and Robust Alginate Lyase Derived from Marinimicrobium sp. H1. Mar. Drugs 2019, 17, 545. [Google Scholar] [CrossRef]
- Zhu, B.; Hu, F.; Yuan, H.; Sun, Y.; Yao, Z. Biochemical Characterization and Degradation Pattern of a Unique pH-Stable PolyM-Specific Alginate Lyase from Newly Isolated Serratia marcescens NJ-07. Mar. Drugs 2018, 16, 129. [Google Scholar] [CrossRef]
- Suda, K.; Tanji, Y.; Hori, K.; Unno, H. Evidence for a novel Chlorella virus-encoded alginate lyase. FEMS Microbiol. Lett. 1999, 180, 45–53. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, Z.; Li, S.; Su, H.; Tang, L.; Tan, Y.; Yu, W.; Han, F. Characterization of a new endo-type polysaccharide lyase (PL) family 6 alginate lyase with cold-adapted and metal ions-resisted property. Int. J. Biol. Macromol. 2018, 120, 729–735. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Zhang, Y.; Chen, L. High-Level Expression of a Thermally Stable Alginate Lyase Using Pichia pastoris, Characterization and Application in Producing Brown Alginate Oligosaccharide. Mar. Drugs 2018, 16, 158. [Google Scholar] [CrossRef]
- Zhu, B.; Ni, F.; Sun, Y.; Yao, Z. Expression and characterization of a new heat-stable endo-type alginate lyase from deep-sea bacterium Flammeovirga sp. NJ-04. Extremophiles 2017, 21, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Arntzen, M.O.; Pedersen, B.; Klau, L.J.; Stokke, R.; Oftebro, M.; Antonsen, S.G.; Fredriksen, L.; Sletta, H.; Aarstad, O.A.; Aachmann, F.L.; et al. Alginate Degradation: Insights Obtained through Characterization of a Thermophilic Exolytic Alginate Lyase. Appl. Environ. Microbiol. 2021, 87. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Anraku, M.; Nakagawa, S.; Ojima, T. Discovery of a Novel Alginate Lyase from Nitratiruptor sp. SB155-2 Thriving at Deep-sea Hydrothermal Vents and Identification of the Residues Responsible for Its Heat Stability. J. Biol. Chem. 2016, 291, 15551–15563. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Fujise, A.; Itabashi, N.; Ohshiro, T. Purification and characterization of a novel alginate lyase from the marine bacterium Cobetia sp. NAP1 isolated from brown algae. Biosci. Biotechnol. Biochem. 2016, 80, 2338–2346. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, L.; Chen, Y.; Ni, H.; Xiao, A.; Cai, H. Characterization of an extracellular biofunctional alginate lyase from marine Microbulbifer sp. ALW1 and antioxidant activity of enzymatic hydrolysates. Microbiol. Res. 2016, 182, 49–58. [Google Scholar] [CrossRef]
- Wang, Z.P.; Cao, M.; Li, B.; Ji, X.F.; Zhang, X.Y.; Zhang, Y.Q.; Wang, H.Y. Cloning, Secretory Expression and Characterization of a Unique pH-Stable and Cold-Adapted Alginate Lyase. Mar. Drugs 2020, 18, 189. [Google Scholar] [CrossRef]
- Yin, R.; Yi, Y.J.; Chen, Z.; Wang, B.X.; Li, X.H.; Zhou, Y.X. Characterization of a New Biofunctional, Exolytic Alginate Lyase from Tamlana sp. s12 with High Catalytic Activity and Cold-Adapted Features. Mar. Drugs 2021, 19, 191. [Google Scholar] [CrossRef]
- Zhou, H.X.; Xu, S.S.; Yin, X.J.; Wang, F.L.; Li, Y. Characterization of a New Bifunctional and Cold-Adapted Polysaccharide Lyase (PL) Family 7 Alginate Lyase from Flavobacterium sp. Mar. Drugs 2020, 18, 388. [Google Scholar] [CrossRef]
- Huang, H.; Li, S.; Bao, S.; Mo, K.; Sun, D.; Hu, Y. Expression and Characterization of a Cold-Adapted Alginate Lyase with Exo/Endo-Type Activity from a Novel Marine Bacterium Alteromonas portus HB161718T. Mar. Drugs 2021, 19, 155. [Google Scholar] [CrossRef]
- Li, S.; Yang, X.; Zhang, L.; Yu, W.; Han, F. Cloning, Expression, and Characterization of a Cold-Adapted and Surfactant-Stable Alginate Lyase from Marine Bacterium Agarivorans sp. L11. J. Microbiol. Biotechnol. 2015, 25, 681–686. [Google Scholar] [CrossRef]
- He, M.; Guo, M.; Zhang, X.; Chen, K.; Yan, J.; Irbis, C. Purification and characterization of alginate lyase from Sphingomonas sp. ZH0. J. Biosci. Bioeng. 2018, 126, 310–316. [Google Scholar] [CrossRef]
- Zhu, B.; Sun, Y.; Ni, F.; Ning, L.; Yao, Z. Characterization of a new endo-type alginate lyase from Vibrio sp. NJU-03. Int. J. Biol. Macromol. 2018, 108, 1140–1147. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, L.; Bao, M.; Liu, Z.; Yu, W.; Han, F. Functional Characterization of Carbohydrate-Binding Modules in a New Alginate Lyase, TsAly7B, from Thalassomonas sp. LD5. Mar. Drugs 2019, 18, 25. [Google Scholar] [CrossRef]
- Bernal, C.; Rodriguez, K.; Martinez, R. Integrating enzyme immobilization and protein engineering: An alternative path for the development of novel and improved industrial biocatalysts. Biotechnol. Adv. 2018, 36, 1470–1480. [Google Scholar] [CrossRef]
- Yang, M.; Yang, S.X.; Liu, Z.M.; Li, N.N.; Li, L.; Mou, H.J. Rational Design of Alginate Lyase from Microbulbifer sp. Q7 to Improve Thermal Stability. Mar. Drugs 2019, 17, 378. [Google Scholar] [CrossRef]
- Yang, S.; Liu, Z.; Fu, X.; Zhu, C.; Kong, Q.; Yang, M.; Mou, H. Expression and Characterization of an Alginate Lyase and Its Thermostable Mutant in Pichia pastoris. Mar. Drugs 2020, 18, 305. [Google Scholar] [CrossRef]
- Liu, T.; Wang, Y.; Luo, X.; Li, J.; Reed, S.A.; Xiao, H.; Young, T.S.; Schultz, P.G. Enhancing protein stability with extended disulfide bonds. Proc. Natl. Acad. Sci. USA 2016, 113, 5910–5915. [Google Scholar] [CrossRef]
- Inoue, A. Characterization of PL-7 Family Alginate Lyases from Marine Organisms and Their Applications. Methods Enzymol. 2018, 605, 499–524. [Google Scholar] [CrossRef]
- Patel, K.K.; Tripathi, M.; Pandey, N.; Agrawal, A.K.; Gade, S.; Anjum, M.M.; Tilak, R.; Singh, S. Alginate lyase immobilized chitosan nanoparticles of ciprofloxacin for the improved antimicrobial activity against the biofilm associated mucoid P. aeruginosa infection in cystic fibrosis. Int. J. Pharm. 2019, 563, 30–42. [Google Scholar] [CrossRef]
- Wan, B.; Zhu, Y.; Tao, J.; Zhu, F.; Chen, J.; Li, L.; Zhao, J.; Wang, L.; Sun, S.; Yang, Y.; et al. Alginate Lyase Guided Silver Nanocomposites for Eradicating Pseudomonas aeruginosa from Lungs. ACS Appl. Mater. Interfaces 2020, 12, 9050–9061. [Google Scholar] [CrossRef]
- Blanco-Cabra, N.; Paetzold, B.; Ferrar, T.; Mazzolini, R.; Torrents, E.; Serrano, L.; LLuch-Senar, M. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci. Rep. 2020, 10, 9390. [Google Scholar] [CrossRef]
- Tavafi, H.; Ali, A.A.; Ghadam, P.; Gharavi, S. Screening, cloning and expression of a novel alginate lyase gene from P. aeruginosa TAG 48 and its antibiofilm effects on P. aeruginosa biofilm. Microb. Pathog. 2018, 124, 356–364. [Google Scholar] [CrossRef]
- Mahajan, S.; Sunsunwal, S.; Gautam, V.; Singh, M.; Ramya, T.N.C. Biofilm inhibitory effect of alginate lyases on mucoid P. aeruginosa from a cystic fibrosis patient. Biochem. Biophys. Rep. 2021, 26, 101028. [Google Scholar] [CrossRef]
- Kawada, A.; Hiura, N.; Shiraiwa, M.; Tajima, S.; Hiruma, M.; Hara, K.; Ishibashi, A.; Takahara, H. Stimulation of human keratinocyte growth by alginate oligosaccharides, a possible co-factor for epidermal growth factor in cell culture. FEBS Lett. 1997, 408, 43–46. [Google Scholar] [CrossRef]
- Wilcox, M.D.; Brownlee, I.A.; Richardson, J.C.; Dettmar, P.W.; Pearson, J.P. The modulation of pancreatic lipase activity by alginates. Food Chem. 2014, 146, 479–484. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Y.; Zhang, L.; Wang, Y.; Wang, S.; Zhang, Y.; Guo, H.; Ji, D.; Wang, Y. Alginate Oligosaccharide DP5 Exhibits Antitumor Effects in Osteosarcoma Patients following Surgery. Front. Pharmacol. 2017, 8, 623. [Google Scholar] [CrossRef]
- Hao, J.; Hao, C.; Zhang, L.; Liu, X.; Zhou, X.; Dun, Y.; Li, H.; Li, G.; Zhao, X.; An, Y.; et al. OM2, a Novel Oligomannuronate-Chromium(III) Complex, Promotes Mitochondrial Biogenesis and Lipid Metabolism in 3T3-L1 Adipocytes via the AMPK-PGC1alpha Pathway. PLoS ONE 2015, 10, e0131930. [Google Scholar] [CrossRef]
- Ueno, M.; Tamura, Y.; Toda, N.; Yoshinaga, M.; Terakado, S.; Otsuka, K.; Numabe, A.; Kawabata, Y.; Murota, I.; Sato, N.; et al. Sodium alginate oligosaccharides attenuate hypertension in spontaneously hypertensive rats fed a low-salt diet. Clin. Exp. Hypertens 2012, 34, 305–310. [Google Scholar] [CrossRef]
- Khan, S.; Tondervik, A.; Sletta, H.; Klinkenberg, G.; Emanuel, C.; Onsoyen, E.; Myrvold, R.; Howe, R.A.; Walsh, T.R.; Hill, K.E.; et al. Overcoming drug resistance with alginate oligosaccharides able to potentiate the action of selected antibiotics. Antimicrob. Agents Chemother. 2012, 56, 5134–5141. [Google Scholar] [CrossRef]
- Saigusa, M.; Nishizawa, M.; Shimizu, Y.; Saeki, H. In vitro and in vivo anti-inflammatory activity of digested peptides derived from salmon myofibrillar protein conjugated with a small quantity of alginate oligosaccharide. Biosci. Biotechnol. Biochem. 2015, 79, 1518–1527. [Google Scholar] [CrossRef]
- Powell, L.C.; Pritchard, M.F.; Emanuel, C.; Onsoyen, E.; Rye, P.D.; Wright, C.J.; Hill, K.E.; Thomas, D.W. A nanoscale characterization of the interaction of a novel alginate oligomer with the cell surface and motility of Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 2014, 50, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Tusi, S.K.; Khalaj, L.; Ashabi, G.; Kiaei, M.; Khodagholi, F. Alginate oligosaccharide protects against endoplasmic reticulum- and mitochondrial-mediated apoptotic cell death and oxidative stress. Biomaterials 2011, 32, 5438–5458. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Kurachi, M.; Nakashima, T.; Kim, D.; Yamaguchi, K.; Oda, T.; Iwamoto, Y.; Muramatsu, T. Structure-activity relationship of alginate oligosaccharides in the induction of cytokine production from RAW264.7 cells. FEBS Lett. 2005, 579, 4423–4429. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, L.; Zhang, Z. Potential applications of alginate oligosaccharides for biomedicine—A mini review. Carbohydr. Polym. 2021, 271, 118408. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, X.; Wu, L.; Li, H.; Chen, Y.; Li, L.; Ni, H.; Li, Q.; Zhu, Y. Exolytic products of alginate by the immobilized alginate lyase confer antioxidant and antiapoptotic bioactivities in human umbilical vein endothelial cells. Carbohydr. Polym. 2021, 251, 116976. [Google Scholar] [CrossRef]
- Zhu, B.; Li, K.; Wang, W.; Ning, L.; Tan, H.; Zhao, X.; Yin, H. Preparation of trisaccharides from alginate by a novel alginate lyase Alg7A from marine bacterium Vibrio sp. W13. Int. J. Biol. Macromol. 2019, 139, 879–885. [Google Scholar] [CrossRef]
- Chen, Y.; Dou, W.; Li, H.; Shi, J.; Xu, Z. The alginate lyase from Isoptericola halotolerans CGMCC 5336 as a new tool for the production of alginate oligosaccharides with guluronic acid as reducing end. Carbohydr. Res. 2018, 470, 36–41. [Google Scholar] [CrossRef]
- Li, Q.; Hu, F.; Wang, M.; Zhu, B.; Ni, F.; Yao, Z. Elucidation of degradation pattern and immobilization of a novel alginate lyase for preparation of alginate oligosaccharides. Int. J. Biol. Macromol. 2020, 146, 579–587. [Google Scholar] [CrossRef]
- Li, Y.; Chen, J.; Lun, S.Y. Biotechnological production of pyruvic acid. Appl. Microbiol. Biotechnol. 2001, 57, 451–459. [Google Scholar] [CrossRef]
- Anh, H.T.L.; Kawata, Y.; Tam, L.T.; Thom, L.T.; Ha, N.C.; Hien, H.T.M.; Thu, N.T.H.; Huy, P.Q.; Hong, D.D. Production of pyruvate from Ulva reticulata using the alkaliphilic, halophilic bacterium Halomonas sp. BL6. J. Appl. Phycol. 2020, 32, 2283–2293. [Google Scholar] [CrossRef]
- Wang, D.; Aarstad, O.A.; Li, J.; McKee, L.S.; Saetrom, G.I.; Vyas, A.; Srivastava, V.; Aachmann, F.L.; Bulone, V.; Hsieh, Y.S. Preparation of 4-Deoxy-L-erythro-5-hexoseulose Uronic Acid (DEH) and Guluronic Acid Rich Alginate Using a Unique exo-Alginate Lyase from Thalassotalea crassostreae. J. Agric. Food Chem. 2018, 66, 1435–1443. [Google Scholar] [CrossRef]
- Kawai, S.; Ohashi, K.; Yoshida, S.; Fujii, M.; Mikami, S.; Sato, N.; Murata, K. Bacterial pyruvate production from alginate, a promising carbon source from marine brown macroalgae. J. Biosci. Bioeng. 2014, 117, 269–274. [Google Scholar] [CrossRef]






| Name | PL Family | Structure | EC | PDB | Refence |
|---|---|---|---|---|---|
| AlgL | PL5 | (α/α)n toroid fold | 4.2.2.3 | 4OZV | [40] |
| A1-III | PL5 | (α/α)n toroid fold | 4.2.2.3 | 1HV6 | [39] |
| BcelPL6 | PL6 | β-helix | 4.2.2.3 | 6QPS | [14] |
| AlyGC | PL6 | β-helix | 4.2.2.11 | 5GKD | [29] |
| AlyF | PL6 | β-helix | 4.2.2.11 | 5Z9T | [38] |
| AlyPG | PL7 | β-jelly roll | 4.2.2.11 | 1UAI | [43] |
| AlgAT5 | PL7 | β-jelly roll | NA | 5ZQI | NA |
| FlAlyA | PL7 | β-jelly roll | 4.2.2.3 | 5Y33 | [44] |
| AlyA | PL7 | β-jelly roll | 4.2.2.11 | 4OZX | NA |
| AlyQ | PL7 | β-jelly roll | 4.2.2.3 | 5XNR | [45] |
| PA1167 | PL7 | β-jelly roll | 4.2.2.- | 1VAV | [46] |
| AlyC3 | PL7 | β-jelly roll | 4.2.2.3 | 7C8G | [47] |
| A1-II’ | PL7 | β-jelly roll | 4.2.2.11 | 2CWS | [48] |
| AlyA5 | PL7 | β-jelly roll | 4.2.2.26 | 4BE3 | [49] |
| AlyA1 | PL7 | β-jelly roll | 4.2.2.11 | 3ZPY | [49] |
| Psalg7A | PL7 | β-jelly roll | 4.2.2.3 | 6YWF | NA |
| PsMan8A | PL8 | NA | NA | NA | [50] |
| vAL-1 | PL14 | β-jelly roll | 4.2.2.14 | 3A0N | [51] |
| AkAly30 | PL14 | β-jelly roll | 4.2.2.3 | 5GMT | [52] |
| Atu3025 | PL15 | (α/α)n toroid fold + anti-parallel β-sheet | 4.2.2.26 | 3A0O | [35] |
| AlyA3 | PL17 | (α/α)n toroid fold + anti-parallel β-sheet | 4.2.2.3 | 7BJT | [42] |
| Alg17c | PL17 | (α/α)n toroid fold + anti-parallelβ-sheet | 4.2.2.26 | 4OK2 | [36] |
| Aly-SJ02 | PL18 | β-jelly roll | 4.2.2.- | 4Q8K | [53] |
| P84143 | PL18 | β-jelly roll | 4.2.2.- | 1J1T | NA |
| PsAly | PL31 | β-helix | 4.2.2.3 | 6KFN | [54] |
| Aly36B | PL36 | β-jelly roll | 4.2.2.3 | 6KCV | [30] |
| Dp0100 | PL39 | (α/α)n toroid fold + anti-parallelβ-sheet | 4.2.2.- | 6JP4 | [41] |
| Name | PL | Source | Optimal Concentration of NaCl | Optimal Enzymatic Condition | Km | Vmax | kcat | Enzyme Activity a | Reference |
|---|---|---|---|---|---|---|---|---|---|
| AlgM4 | PL7 | Marine bacterium Vibrio weizhoudaoensis M0101 | 1 mol/L | 30 °C; pH = 8.5 | Km = 2.72 mg/mL for sodium alginate | Vmax = 2.75 nmol/s for sodium alginate | kcat = 30.25 S−1 for sodium alginate | 7-fold increase | [61] |
| AlyPM | PL7 | Marine bacterium Pseudoalteromonas sp. SM0524 | 0.5–1.2 mol/L | 30 °C; pH = 8.5 | Km = 3.15 mg/mL (in 0.5 M NaCl) for sodium alginate Km = 74.39 mg/mL (in 0 M NaCl) for sodium alginate | NA | NA | 6-fold increase | [58] |
| rA9mT | PL7 | Deep-sea bacterium Vibrio sp. JAM-A9m | 0.4 mol/L | 30 °C in the presence of 0.2 M NaCl at pH 7.5 | NA | NA | NA | 24-fold increase | [62] |
| A1m | PL7 | Agarivorans sp. JAM-A1m from a deep-sea sediment | 0.6–0.8 mol/L | 30 °C either in the presence of 0.2 M NaCl at pH 9 or in its absence at pH 10 | NA | Vmax values are 38.4, 285.7, 416.7, and 526.3 units mg−1 protein in the presence of 0, 0.1, 0.2, and 0.5 M NaCl, respectively for sodium alginate | NA | 20-fold increase | [63] |
| AlyC3 | PL7 | Psychromonas sp. C-3 isolated from the Arctic brown alga Laminaria | 0.5 mol/L | 20 °C; pH = 8.0 | Km = 0.24 ± 0.05 mg/mL, for polyM | Vmax = 19,704.73 ± 1865.49 U/mg for polyM | NA | 2.9-fold increase | [47] |
| Aly08 | PL7 | Marine bacterium Vibrio sp. SY01 | 0.3 mol/L | 45 °C; pH = 8.35 | NA | NA | NA | 8-fold increase | [64] |
| Name | Origin/Strain | PL | Optimal pH | Relative Activity at Various pH Values | Km | Vmax | kcat | Reference |
|---|---|---|---|---|---|---|---|---|
| Alyw202 | Vibrio sp. W2 | PL7 | 9.0 | >80% a (pH 5.0–9.0) >60% a (pH 3.0–10.0) | NA | NA | NA | [66] |
| Aly08 | Vibrio sp. SY01 | PL7 | 8.35 | >80% a (pH 4.0–10.0) >60% a (pH 7.0–11.0) | NA | NA | NA | [64] |
| AlgNJ–04 | Vibrio sp. NJ04 | PL7 | 7.0 | >60% b (pH 4.0–10.0) | Km = 0.49, 0.86, 0.24 mM, respectively, for alginate, polyM and polyG | Vmax = 72, 95, 35 pmol/s, respectively, for alginate, polyM and polyG | kcat = 59, 77, 29 s−1, respectively, for alginate, polyM and polyG | [65] |
| AlgH | Marinimicrobium sp. H1 | PL7 | 10.0 | >60% a (pH 6.0–10.0) | Km = 6.6 ± 2.2, 7.6 ± 1.6, 9.1 ± 2.4 mg·mL−1, respectively, for sodium alginate, polyG and polyM | Vmax = 224.6 ± 33.6, 146.6 ± 15.6, 62.6 ± 8.8 U·mg of protein−1, respectively, for sodium alginate, polyG and polyM | kcat = 260.6 ± 36.2, 155.7 ± 17.1, 66.8 ± 6.7 s−1, respectively, for sodium alginate, polyG and polyM | [67] |
| Alg823 | Pseudoalteromonas carrageenovora ASY5 | PL6 | 8.0 | >80% b (pH 6.0–10.0) | Km = 0.15 mg/mL for sodium alginate | Vmax = 1.84 U/g for sodium alginate | 1.19 × 106 s−1 for sodium alginate | [16] |
| AlgNJ–07 | Serratia marcescens NJ–07 | NA | 9.0 | >80% c (pH 7.0–10.0) | Km = 0.53, 0.27 mM, respectively, for sodium alginate and polyM | Vmax = 74, 67 nmol/s, respectively, for sodium alginate and polyM | kcat = 34, 31 s−1, respectively, for sodium alginate and polyM | [68] |
| Name | Origin | PL | Optimal Temperature | Thermal Stability | Km | Vmax | kcat | Reference |
|---|---|---|---|---|---|---|---|---|
| rSAGL | Flavobacterium sp. H63 | NA | 45 °C | Retained 49% of activity at 50 °C for 72 h | Km = 4.63 mg/mL (from P. pastoris), 4.64 mg/mL (from E. coli) for sodium alginate | NA | NA | [71] |
| rNitAly | Nitratiruptor sp. SB155-2 | PL7 | 70 °C | Retained 50% of activity at 67 °C for 30 min | NA | NA | NA | [74] |
| Alg823 | Pseudoalteromonas carrageenovora ASY5 | PL6 | 55 °C | Retained over 75% of the maximum activity at 50 °C for 30 min | Km = 0.15 mg/mL for sodium alginate | Vmax = 1.84 U/g for sodium alginate | 1.19 × 106 s−1 for sodium alginate | [16] |
| AlgC-PL7 | Cobetia sp. NAP1 | PL7 | 45 °C | Retained 60 and 30% of activity at 80 and 90 °C for 1 h | NA | NA | NA | [75] |
| AMOR_PL17A | Arctic Mid-Ocean Ridge (AMOR) metagenomics data set | PL17 | High temperature (>50 °C) | Retained 100% of activity at 60 °C for 24h (in the absence of substrate) | NA | NA | NA | [73] |
| ALW1 | Microbulbifer sp. ALW1 | NA | 45 °C | Retained 68% of activity at 45 °C for 1 h | Km = 1.03 mg/mL for sodium alginate | Vmax = 4.63 U/mg for sodium alginate | kcat = 69.38 s−1 for sodium alginate | [76] |
| Name | PL | Source | Substrate Preference | Action Mode | Optimal Temperature | Cold-Adapted Property | Km | Vmax | kcat | Main Products | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AlyC3 | PL7 | Psychromonas sp. C-3 isolated from the Arctic brown alga Laminaria | polyM | Endolytic lyase | 20 °C | Retained 48.2% of maximum activity at 1 °C; Unstable at 30 °Cand above | Km = 0.24 ± 0.05 mg/mL, polyM | Vmax = 19,704.73 ± 1865.49 U/mg for polyM | NA | ΔMM (Δ represents 4-deoxy-L-erythro-hex-4- enopyranosyluronic acid) | [47] |
| AlgSH7 | PL7 | Microbulbifer sp. SH-1 | polyM | Endolytic lyase | 40 °C | Retained 80% of the maximum activity over at 25 °C; Unstable at temperatures beyond 30 °C | NA | NA | NA | Disaccharides, tri-saccharides, and tetrasaccharides | [56] |
| TsAly6A | PL6 | Thalassomonas sp. LD5 | polyG | Endolytic lyase | 35 °C | Retained 73.1% and 21.1% of the maximum activity at 20 °C and 10 °C; Retained over 90% of initial activity after incubation at 30 °C for 1 h, but only 29% remained at 40 °C | NA | NA | NA | Disaccharides and trisaccharides | [70] |
| Alys1 | PL7 | Tamlana sp. s12 | polyM | Exolytic lyase | 35 °C | Activity reduced at temperatures above 35 °C; Retained more than 50% of the maximum activity at 10 °C; No detectable activity at 60 °C | Km = 0.20 ± 0.01 mM for sodium alginate | NA | kcat = 4.43 ± 0.027 s−1 for sodium alginate | Monosaccharides, disaccharides, trisaccharides and some lower monomers | [78] |
| AlyS02 | PL7 | Flavobacterium sp. S02 | bifunctional | Endolytic lyase | 30 °C | Retained more than 70% of relative activity in the range of 20–40 °C (retained more than 90% of maximum activity at 25 °C); Activity reduced at temperatures above 40 °C | NA | NA | NA | Disaccharides and trisaccharides | [79] |
| AlgM4 | PL7 | Vibrio weizhoudaoensis M0101 | bifunctional | Endolytic lyase | 30 °C | Retained 92% of its initial activity after a 30 min incubation at 30 or 35 °C (in the presence of 1 mol/L NaCl); Decreased rapidly at temperatures exceeding 40 °C and decreased by 63% at 45 °C | Km = 2.72 mg/mL , for sodium alginate | Vmax = 2.75 nmol/s for sodium alginate | kcat = 30.25 S−1 for sodium alginate | Oligosaccharides with DP of 2–9 | [61] |
| Alg2951 | PL7 | Alteromonas portus HB161718T | polyG | Endolytic and exolytic lyase | 25 °C | Retained over 60% of the maximum activity at the temperature range of 15–40 °C; No detectable activity at 60 °C | NA | NA | NA | Monosaccharides and trisaccharides | [80] |
| AlyL1 | PL7 | Agarivorans sp. L11 | bifunctional | Endolytic lyase | 40 °C | Retained 54.5% and 72.1% of the maximum activity at 15 °C and 20 °C | NA | NA | NA | Disaccharides and trisaccharides | [81] |
| AlyPM | PL7 | Pseudoalteromonas sp. SM0524 | polyM | Endolytic lyase | 30 °C | Retained 19% of the maximum activity at 5 °C; No detectable activity at 45 °C | Km = 3.15 mg/ml (in 0.5 M NaCl) for sodium alginate Km = 74.39 mg/ml (in 0 M NaCl) for sodium alginate | NA | NA | Oligosaccharides with DP of 2–3 | [58] |
| Alyw201 | PL7 | Vibrio sp. W2 | polyG | Endolytic lyase | 30 °C | Retained more than 80% activity at 25–40 °C; Retained 72.9% and 38.4% of the highest activity at 10 °C and 20 °C | NA | NA | NA | Oligosaccharides with DP of 2–6 | [77] |
| ZH0-IV | NA | Sphingomonas sp. ZH0 | bifunctional | Exolytic lyase | 37 °C | Retained more than 80% of activity after incubating in phosphate buffer with an optimum pH for 10 min between 25 and 42 °C | Km = 0.41 mg/ml for sodium alginate | Vmax = 5.53 U/ml for sodium alginate | NA | Monosaccharides | [82] |
| AlyGC | PL6 | Glaciecola chathamensis S18K6T | polyG | Exolytic lyase | 30 °C | Activity rapidly decreased above 30 °C; Retained relatively high activity between 10 and 30 °C | NA | NA | NA | Monosaccharides | [29] |
| AlgNJU-03 | PL7 | Vibrio sp. NJU-03 | bifunctional | Endolytic lyase | 30 °C | Retained approximately 40% activity after incubation at 40 °C for 30 min and was gradually inactivated as temperature increased | Km = 8.50, 10.94, 4.00 mM, respectively for sodium alginate, polyM and polyG | Vmax = 1.67, 0.30, 2.50 nmol/s, respectively for sodium alginate, polyM and polyG | kcat = 30.64, 5.50, 45.87 s−1, respectively for sodium alginate, polyM and polyG | Disaccharides, trisaccharides and tetrasaccharides | [83] |
| rA9mT | PL7 | Vibrio sp. JAM-A9m | polyM | NA | 30 °C | Relative activities at 10 °C and 2 °C were around 45% and 30% of the maximal activity; enzyme was rapidly inactivated after incubating at above 40 °C in the absence of NaCl. | NA | NA | NA | NA | [62] |
| TsAly7B | PL7 | Thalassomonas sp. LD5 | bifunctional | Endolytic lyase | 30 °C | Retained approximately 60% of relative activity at 20 °C; Unstable at temperatures beyond 30 °C | NA | NA | NA | Disaccharides, trisaccharides, and tetrasaccharides | [84] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, S.-K.; Yin, R.; Wang, X.-C.; Jiang, H.-N.; Liu, X.-X.; Lv, W.; Ma, Y.; Zhou, Y.-X. Structure Characteristics, Biochemical Properties, and Pharmaceutical Applications of Alginate Lyases. Mar. Drugs 2021, 19, 628. https://doi.org/10.3390/md19110628
Gao S-K, Yin R, Wang X-C, Jiang H-N, Liu X-X, Lv W, Ma Y, Zhou Y-X. Structure Characteristics, Biochemical Properties, and Pharmaceutical Applications of Alginate Lyases. Marine Drugs. 2021; 19(11):628. https://doi.org/10.3390/md19110628
Chicago/Turabian StyleGao, Shu-Kun, Rui Yin, Xiao-Chen Wang, Hui-Ning Jiang, Xiao-Xiao Liu, Wei Lv, Yu Ma, and Yan-Xia Zhou. 2021. "Structure Characteristics, Biochemical Properties, and Pharmaceutical Applications of Alginate Lyases" Marine Drugs 19, no. 11: 628. https://doi.org/10.3390/md19110628
APA StyleGao, S.-K., Yin, R., Wang, X.-C., Jiang, H.-N., Liu, X.-X., Lv, W., Ma, Y., & Zhou, Y.-X. (2021). Structure Characteristics, Biochemical Properties, and Pharmaceutical Applications of Alginate Lyases. Marine Drugs, 19(11), 628. https://doi.org/10.3390/md19110628






