Identification and Heterologous Expression of the Kendomycin B Biosynthetic Gene Cluster from Verrucosispora sp. SCSIO 07399
Abstract
:1. Introduction
2. Results and Discussion
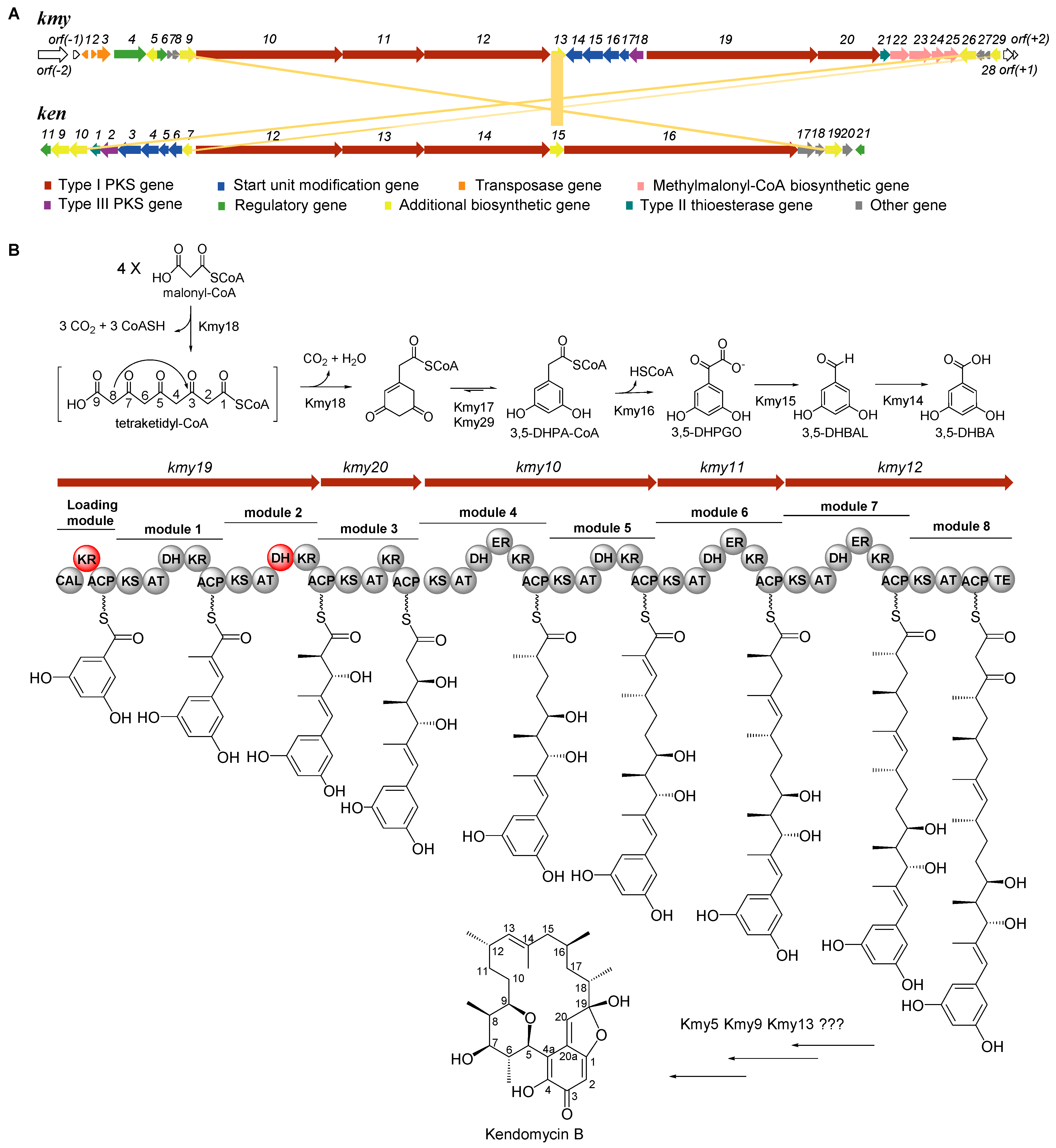
2.1. Identification of Kendomycin B Biosynthetic Gene Cluster (BGC)
2.2. Determination of the Boundaries of Kendomycin B BGC
2.3. Verification of the Starter Unit 3,5-DHBA for Kendomycin B
2.4. Analysis of Type I PKS and Tailoring Genes for Polyketide Skeleton Assembly and Cyclization
2.5. Characterization of a Positive Regulatory Genes
2.6. Heterologous Expression of Kendomycin B BGC
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Genome Sequencing Assembly and DNA Sequence Analysis
3.3. Construction of Gene Mutant Strains
3.4. Gene Complementation of Δkmy4 and Δkmy27-orf(+2) Mutant Strains
3.5. Small-Scale Fermentation and Analyses
3.6. Chemical Complementation of Type III PKS Gene Mutant Strain Δkmy18
3.7. Phylogenetic Analysis and Sequence Alignment
3.8. RNA Extraction of Wild-Type and Δkmy4 Mutant Strains, cDNA Synthesis, and Real-Time Polymerase Chain Reaction
3.9. Heterologous Expression of the kmy Gene Cluster
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wenzel, S.C.; Bode, H.B.; Kochems, I.; Müller, R. A type I/type III polyketide synthase hybrid biosynthetic pathway for the structurally unique ansa compound kendomycin. ChemBioChem 2008, 9, 2711–2721. [Google Scholar] [CrossRef]
- Elnakady, Y.A.; Chatterjee, I.; Bischoff, M.; Rohde, M.; Josten, M.; Sahl, H.; Herrmann, M.; Müller, R. Investigations to the antibacterial mechanism of action of kendomycin. PLoS ONE 2016, 11, e0146165. [Google Scholar] [CrossRef] [Green Version]
- Elnakady, Y.A.; Rohde, M.; Sasse, F.; Backes, C.; Keller, A.; Lenhof, H.; Weissman, K.J.; Müller, R. Evidence for the mode of action of the highly cytotoxic Streptomyces polyketide kendomycin. ChemBioChem 2007, 8, 1261–1272. [Google Scholar] [CrossRef]
- Tranter, D.; Filipuzzi, I.; Lochmann, T.; Knapp, B.; Kellosalo, J.; Estoppey, D.; Pistorius, D.; Meissner, A.; Paavilainen, V.O.; Hoepfner, D. Kendomycin cytotoxicity against bacterial, fungal, and mammalian cells is due to cation chelation. J. Nat. Prod. 2020, 83, 965–971. [Google Scholar] [CrossRef]
- Xu, S.; Arimoto, H. Strategies for construction of the all-carbon macrocyclic skeleton of the ansamycin antibiotic—Kendomycin. J. Antibiot. 2016, 69, 203–212. [Google Scholar] [CrossRef]
- Bode, H.B.; Zeeck, A. Biosynthesis of kendomycin: Origin of the oxygen atoms and further investigations. J. Chem. Soc. Perkin Trans. 2000, 1, 2665–2670. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, Q.; Sun, C.; Tian, X.; Gui, C.; Qin, X.; Zhang, H.; Ju, J. Cytotoxic kendomycins containing the carbacylic ansa scaffold from the marine-derived Verrucosispora sp. SCSIO 07399. J. Nat. Prod. 2019, 82, 3366–3371. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tseng, C.C.; Hubbard, B.K.; Walsh, C.T. Glycopeptide antibiotic biosynthesis: Enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenylglycine. Proc. Natl. Acad. Sci. USA 2001, 98, 14901–14906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeifer, V.; Nicholson, G.J.; Ries, J.; Recktenwald, J.; Schefer, A.B.; Shawky, R.M.; Schröder, J.; Wohlleben, W.; Pelzer, S. A polyketide synthase in glycopeptide biosynthesis: The biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J. Biol. Chem. 2001, 276, 38370–38377. [Google Scholar] [CrossRef] [Green Version]
- Keatinge-Clay, A.T. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 2007, 14, 898–908. [Google Scholar] [CrossRef] [Green Version]
- Caffrey, P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. ChemBioChem 2003, 4, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Migita, A.; Watanabe, M.; Hirose, Y.; Watanabe, K.; Tokiwano, T.; Kinashi, H.; Oikawa, H. Identification of a gene cluster of polyether antibiotic lasalocid from Streptomyces lasaliensis. Biosci. Biotechnol. Biochem. 2009, 73, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Lin, L.; Sun, Y.; Pang, C. Research progress of pathways and mechanisms of decarboxylation. Chem. Bioeng. 2008, 25, 1–6. [Google Scholar]
- Huijbers, M.M.E.; Montersino, S.; Westphal, A.H.; Tischler, D.; Berkel, W.J.H. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Van Berkel, W.J.; Kamerbeek, N.M.; Fraaije, M.W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006, 124, 670–689. [Google Scholar] [CrossRef] [Green Version]
- Wilson, D.J.; Xue, Y.; Reynolds, K.A.; Sherman, D.H. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 2001, 183, 3468–3475. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Lei, J.; Liu, Y.; Wang, Y. The LuxR family members GdmRI and GdmRII are positive regulators of geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Arch. Microbiol. 2008, 189, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Kong, L.; Li, Y.; Zheng, X.; Wang, Q.; Yang, W.; Deng, Z.; You, D. A LuxR family transcriptional regulator AniF promotes the production of anisomycin and its derivatives in Streptomyces hygrospinosus var. beijingensis. Synth. Syst. Biotechnol. 2019, 4, 40–48. [Google Scholar] [CrossRef]
- Guerra, S.M.; Rodríguez-García, A.; Santos-Aberturas, J.; Vicente, C.M.; Payero, T.D.; Martín, J.F.; Aparicio, J.F. LAL regulators SCO0877 and SCO7173 as pleiotropic modulators of phosphate starvation response and actinorhodin biosynthesis in Streptomyces coelicolor. PLoS ONE 2012, 7, e31475. [Google Scholar] [CrossRef] [Green Version]
- Routh, M.D.; Su, C.C.; Zhang, Q.; Yu, E.W. Structures of AcrR and CmeR: Insight into the mechanisms of transcriptional repression and multi-drug recognition in the TetR family of regulators. Biochim. Biophys. Acta 2009, 1794, 844–851. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Li, C.; Xie, J. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell Signal. 2013, 25, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Hug, J.; Fu, C.; Bian, X.; Zhang, Y.; Müller, R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019, 36, 1412–1436. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Kieser, T.; Chater, K. PCR-targeting system in Streptomyces coelicolor A3(2). John. Innes. Centre 2002, 3, 1–39. [Google Scholar]
- Luo, M.; Wang, Y.H.; Frisch, D.; Joobeur, T.; Wing, R.A.; Dean, R.A. Melon bacterial artificial chromosome (BAC) library construction using improved methods and identification of clones linked to the locus conferring resistance to melon Fusarium wilt (Fom-2). Genome 2001, 44, 154–162. [Google Scholar] [CrossRef]
- Tu, J.; Li, S.; Chen, J.; Song, Y.; Fu, S.; Ju, J. Characterization and heterologous expression of the neoabyssomicin/abyssomicin biosynthetic gene cluster from Streptomyces koyangensis SCSIO 5802. Microb. Cell Fact. 2018, 17, 28. [Google Scholar] [CrossRef] [Green Version]


| ORF | Size a | Proposed Function | Closest Homolog, Origin (Protein ID); ID/SI (%) | ken Homolog |
|---|---|---|---|---|
| orf(-2) | 778 | DUF262 domain-containing protein | DUF262 domain-containing protein, Salinispora arenicola (WP_028680247.1); 98.2/100 | - |
| orf(-1) | 95 | Hypothetical protein | Hypothetical protein BCD48_40600, Frankia sp. BMG5.36 (OHV60767.1); 57.8/47 | - |
| kmy1 | 87 | Transposase | Transposase, Micromonospora marina (WP_141708283.1); 96.5/100 | - |
| kmy2 | 78 | Transposase | Transposase, Salinispora arenicola (WP_028673963.1); 98.6/91 | - |
| kmy3 | 375 | Transposase | Transposase (or an inactivated derivative), Micromonospora inyonensis (SCL17972.1); 96.2/90 | - |
| kmy4 | 921 | Transcriptional regulator | LuxR family transcriptional regulator, Candidatus Frankia datiscae (WP_131764890.1); 38.9/99 | - |
| kmy5 | 274 | NAD(P)-dependent oxidoreductase | SPR family NAD(P)-dependent oxidoreductase, Actinoplanes derwentensis (WP_157751350.1); 71.2/100 | - |
| kmy6 | 205 | Transcriptional regulator | DNA-binding transcriptional regulator, AcrR family, Actinoplanes derwentensis (SDS69921.1); 67.3/98 | - |
| kmy7 | 74 | Hypothetical protein | Hypothetical protein, Micromonospora sp. M71_S20 (WP_121401669.1); 67.6/100 | - |
| kmy8 | 149 | Hypothetical protein | Hypothetical protein, Streptomyces violaceoruber (CAQ52628.1); 63.4/94 | ken18 |
| kmy9 | 517 | FAD-dependent oxidoreductase | FAD-dependent oxidoreductase, Streptomyces violaceoruber (CAQ52629.1); 72.9/97 | ken19 |
| kmy10 | 3859 | PKS I (module 4: KS, AT, DH, ER, KR, and ACP; module 5: KS, AT, DH, KR, and ACP) | Type I polyketide synthase, modules 4–5, Streptomyces violaceoruber (CAQ52622.1); 58.3/99 | ken12 |
| kmy11 | 2143 | PKS I (module 6: KS, AT, DH, ER, KR, and ACP) | Type I polyketide synthase, modules 6, Streptomyces violaceoruber (CAQ52623.1); 60.4/99 | ken13 |
| kmy12 | 3346 | PKS I (module 7: KS, AT, DH, ER, KR, and ACP; module 8: KS, AT, ACP, and TE) | Type I polyketide synthase, modules 7–8, Streptomyces violaceoruber (CAQ52624.1); 62.2/99 | ken14 |
| kmy13 | 390 | FAD-dependent monooxygenase | FAD-dependent monooxygenase, Streptomyces violaceoruber (CAQ52625.1); 68.9/98 | ken15 |
| kmy14 | 486 | Benzaldehyde dehydrogenase | Benzaldehyde dehydrogenase, Streptomyces libani (WP_190842308.1); 70.0/100 | ken6 |
| kmy15 | 551 | Benzoylformate decarboxylase | Thiamine pyrophosphate-binding protein, Streptomyces albus subsp. albus (KUJ59733.1); 75.7/98 | ken5 |
| kmy16 | 444 | Dioxygenase | Enoyl-CoA hydratase/isomerase family protein, Streptomyces griseus (WP_069170596.1); 66.7/90 | ken4 |
| kmy17 | 227 | Enoyl-CoA hydratase/isomerase | Enoyl-CoA hydratase/isomerase family protein, Streptomyces inusitatus (WP_190127028.1); 56.6/92 | ken3 |
| kmy18 | 372 | PKS III | Type III polyketide synthase, Amycolatopsis anabasis (WP_158891515.1); 72.8/95 | ken2 |
| kmy19 | 4494 | PKS I (loading module: CAL, KR, and ACP; module 1: KS, AT, DH, KR, and ACP; module 2: KS, AT, DH, and KR) | Type I polyketide synthase, loading module and modules 1–3, Streptomyces violaceoruber (CAQ52626.1); 54.9/98 | ken16 |
| kmy20 | 1640 | PKS I (module 3: ACP, KS, AT, KR, and ACP) | Type I polyketide synthase, loading module and modules 1–3, Streptomyces violaceoruber (CAQ52626.1); 58.8/99 | ken16 |
| kmy21 | 261 | Type II Thioesterase | Thioesterase, Streptacidiphilus melanoqenes (WP 042389076.1); 56.3/93 | - |
| kmy22 | 644 | Methylmalonyl-CoA mutase | Methylmalonyl-CoA mutase small subunit, Micromonospora krabiensis (WP 091592824.1); 56.3/94 | - |
| kmy23 | 723 | Methylmalonyl-CoA mutase | Methylmalonyl-CoA mutase, Micromonospora fluostatini (TDB93065.1); 77.8/99 | - |
| kmy24 | 343 | Methylmalonyl-CoA mutase-associated GTPase | Methylmalonyl-CoA mutase-associated GTPase MeaB, Nonomuraea rubra (WP_185106477.1); 55.7/92 | - |
| kmy25 | 433 | Propionyl-CoA carboxylase | Acyl-CoA carboxylase subunit beta, Amycolatopsis sp. YIM10 (WP_153035332.1); 75.6/100 | - |
| kmy26 | 547 | PQQ-dependent enzyme | Polyvinyl alcohol dehydrogenase (cytochrome), Allokutzneria albata (SDN 68233.1); 69.5/90 | ken10 |
| kmy27 | 174 | Hypothetical protein | Hypothetical protein, Streptomyces abikoensis (WP_18945037.1); 40.4/83 | - |
| kmy28 | 125 | DoxX family protein | DoxX family protein, Streptomyces odonnellii (WP 046501079.1); 45.6/100 | - |
| kmy29 | 268 | Enoyl-CoA hydratase/isomerase | Enoyl-CoA hydratase/isomerase family protein, Phytohabitans houttuyneae (WP_173055440.1); 78.7/100 | ken7 |
| orf(+1) | 207 | Hypothetical protein | Hypothetical protein, Phytoactinopolyspora sp. HAJB-30 (WP_166351888.1); 48.1/100 | - |
| orf(+2) | 67 | Hypothetical protein | Hypothetical protein, Phytoactinopolyspora sp. HAJB-30 (WP_166351888.1); 62.5/95 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Zhang, S.; Chen, Y.; Tian, X.; Gu, Y.; Ju, J. Identification and Heterologous Expression of the Kendomycin B Biosynthetic Gene Cluster from Verrucosispora sp. SCSIO 07399. Mar. Drugs 2021, 19, 673. https://doi.org/10.3390/md19120673
Chen J, Zhang S, Chen Y, Tian X, Gu Y, Ju J. Identification and Heterologous Expression of the Kendomycin B Biosynthetic Gene Cluster from Verrucosispora sp. SCSIO 07399. Marine Drugs. 2021; 19(12):673. https://doi.org/10.3390/md19120673
Chicago/Turabian StyleChen, Jiang, Shanwen Zhang, Yingying Chen, Xinpeng Tian, Yucheng Gu, and Jianhua Ju. 2021. "Identification and Heterologous Expression of the Kendomycin B Biosynthetic Gene Cluster from Verrucosispora sp. SCSIO 07399" Marine Drugs 19, no. 12: 673. https://doi.org/10.3390/md19120673
APA StyleChen, J., Zhang, S., Chen, Y., Tian, X., Gu, Y., & Ju, J. (2021). Identification and Heterologous Expression of the Kendomycin B Biosynthetic Gene Cluster from Verrucosispora sp. SCSIO 07399. Marine Drugs, 19(12), 673. https://doi.org/10.3390/md19120673








