Chitin Degradation Machinery and Secondary Metabolite Profiles in the Marine Bacterium Pseudoalteromonas rubra S4059
Abstract
1. Introduction
2. Results
2.1. In Silico Analysis of Chitin Degrading Machinery and Bioactive Potential in P. rubra S4059
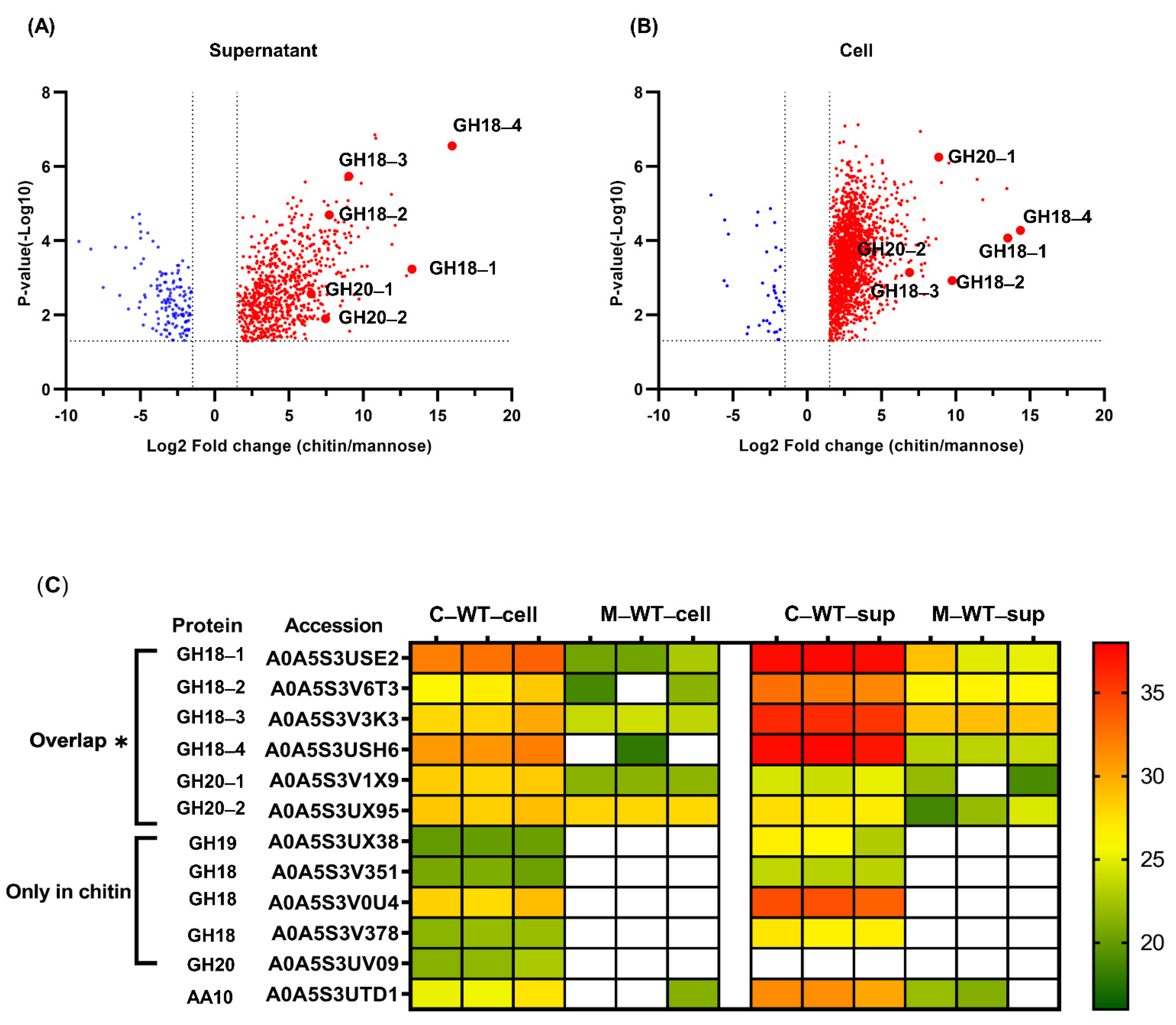
2.2. Global Proteome Profiles of P. rubra S4059 Grown on Chitin
2.3. Comparative Analysis of the Expression of Chitinolytic Enzymes in P. rubra S4059
2.4. Influence of Chitin on the Metabolome of S4059
2.5. The Deletion of GH19 Chitinase Does Not Affect Growth or Chitin Degradation
2.6. Biofilm Formation and Chitin Surface Attachment of P. rubra S4059 Was Not Affected by Deletion of GH19 Chitinase Gene
2.7. Deletion of GH19 Chitinase Does Not Significantly Influence the Proteome of S4059
2.8. Similar Secondary Metabolome Pattern in Wild Type Strain and GH19 Mutant
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
4.2. Whole Genome Sequencing and Assembly of P. rubra S4059
4.3. In Silico Analysis of Chitin Degrading Genes and Secondary Metabolites
4.4. Growing Bacteria and Sample Preparation for Proteomic Analyses
4.5. Proteomic Data Acquisition
4.6. DNA Manipulation
4.7. Construction of Suicide Plasmids for in-Frame Deletion of GH19 Chitinase in P. rubra S4059
4.8. Conjugation of P. rubra S4059
4.9. Confirmation of the First Crossing over Mutants and Deletion Mutants
4.10. Growth Curves of Wild Type and Mutant
4.11. Growth and Attachment on Shrimp Shells
4.12. Extraction of Metabolites for Chemical Analysis
4.13. Chemical Analysis by UHPLC-HRMS
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romano, G.; Costantini, M.; Sansone, C.; Lauritano, C.; Ruocco, N.; Ianora, A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar. Environ. Res. 2017, 128, 58–69. [Google Scholar] [CrossRef]
- Parte, S.; Sirisha, V.L.; D’Souza, J.S. Biotechnological applications of marine enzymes from algae, bacteria, fungi, and sponges. Adv. Food Nutr. Res. 2017, 80, 75–106. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 2007, 5, 220–241. [Google Scholar] [CrossRef]
- Offret, C.; Desriac, F.; Le Chevalier, P.; Mounier, J.; Jégou, C.; Fleury, Y. Spotlight on antimicrobial metabolites from the marine bacteria Pseudoalteromonas: Chemodiversity and ecological significance. Mar. Drugs 2016, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L.; Yang, J.; Chen, X.L.; Wang, P.; Zhao, H.L.; Su, H.N.; Li, C.Y.; Yu, Y.; Zhong, S.; Wang, L.; et al. A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria. Nat. Commun. 2020, 11, 285. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.L.; Chen, X.L.; Xie, B.B.; Zhou, M.Y.; Gao, X.; Zhang, X.Y.; Zhou, B.C.; Weiss, A.S.; Zhang, Y.Z. Elastolytic mechanism of a novel M23 metalloprotease pseudoalterin from deep-sea Pseudoalteromonas sp. CF6-2: Cleaving not only glycyl bonds in the hydrophobic regions but also peptide bonds in the hydrophilic regions involved in cross-linking. J. Biol. Chem. 2012, 287, 39710–39720. [Google Scholar] [CrossRef]
- Paulsen, S.S.; Strube, M.L.; Bech, P.K.; Gram, L.; Sonnenschein, E.C. Marine chitinolytic Pseudoalteromonas represents an untapped reservoir of bioactive potential. Msystems 2019, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Rudall, K.M.; Kenchington, W. The chitin system. Biol. Rev. 1973, 49, 597–636. [Google Scholar] [CrossRef]
- Beier, S.; Bertilsson, S. Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front. Microbiol. 2013, 4, 149. [Google Scholar] [CrossRef] [PubMed]
- Vaaje-Kolstad, G.; Horn, S.J.; Sørlie, M.; Eijsink, V.G.H. The chitinolytic machinery of Serratia marcescens—A model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 2013, 280, 3028–3049. [Google Scholar] [CrossRef] [PubMed]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 2018, 38, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Gooday, G.W. The ecology of chitin degradation. Adv. Microb. Ecol. 1990, 387–430. [Google Scholar] [CrossRef]
- Bai, Y.; Eijsink, V.G.H.; Kielak, A.M.; van Veen, J.A.; de Boer, W. Genomic comparison of chitinolytic enzyme systems from terrestrial and aquatic bacteria. Environ. Microbiol. 2016, 18, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Udaya Prakash, N.A.; Jayanthi, M.; Sabarinathan, R.; Kangueane, P.; Mathew, L.; Sekar, K. Evolution, homology conservation, and identification of unique sequence signatures in GH19 family chitinases. J. Mol. Evol. 2010, 70, 466–478. [Google Scholar] [CrossRef]
- Konno, N.; Takahashi, H.; Nakajima, M.; Takeda, T.; Sakamoto, Y. Characterization of β-N-acetylhexosaminidase (LeHex20A), a member of glycoside hydrolase family 20, from Lentinula edodes (shiitake mushroom). AMB Express 2012, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sørlie, M.; Eijsink, V.G.H. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef] [PubMed]
- García-Fraga, B.; da Silva, A.F.; López-Seijas, J.; Sieiro, C. A novel family 19 chitinase from the marine-derived Pseudoalteromonas tunicata CCUG 44952T: Heterologous expression, characterization and antifungal activity. Biochem. Eng. J. 2015, 93, 84–93. [Google Scholar] [CrossRef]
- Giubergia, S.; Phippen, C.; Gotfredsen, C.H.; Nielsen, K.F.; Gram, L. Influence of niche-specific nutrients on secondary metabolism in Vibrionaceae. Appl. Environ. Microbiol. 2016, 82, 4035–4044. [Google Scholar] [CrossRef] [PubMed]
- Giubergia, S.; Phippen, C.; Nielsen, K.F.; Gram, L. Growth on chitin impacts the transcriptome and metabolite profiles of antibiotic-producing Vibrio coralliilyticus S2052 and Photobacterium galatheae S2753. Msystems 2017, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wietz, M.; Duncan, K.; Patin, N.V.; Jensen, P.R. Antagonistic interactions mediated by marine bacteria: The role of small molecules. J. Chem. Ecol. 2013, 39, 879–891. [Google Scholar] [CrossRef]
- Nazari, B.; Kobayashi, M.; Saito, A.; Hassaninasab, A.; Miyashita, K.; Fujiia, T. Chitin-induced gene expression in secondary metabolic pathways of Streptomyces coelicolor A3(2) grown in soil. Appl. Environ. Microbiol. 2013, 79, 707–713. [Google Scholar] [CrossRef]
- Saravanan, P.; Nancharaiah, Y.V.; Venugopalan, V.P.; Rao, T.S.; Jayachandran, S. Biofilm formation by Pseudoalteromonas ruthenica and its removal by chlorine. Biofouling 2006, 22, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Margolis, J.J.; El-Etr, S.; Joubert, L.M.; Moore, E.; Robison, R.; Rasley, A.; Spormann, A.M.; Monack, D.M. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl. Environ. Microbiol. 2010, 76, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Veliz, E.A.; Martínez-Hidalgo, P.; Hirsch, A.M. Chitinase-producing bacteria and their role in biocontrol. AIMS Microbiol. 2017, 3, 689–705. [Google Scholar] [CrossRef] [PubMed]
- Reguera, G.; Kolter, R. Virulence and the environment: A novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 2005, 187, 3551–3555. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Tay, Q.X.M.; Kjelleberg, S.; Rice, S.A.; McDougald, D. Quorum sensing-regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J. 2015, 9, 1812–1820. [Google Scholar] [CrossRef]
- Pruzzo, C.; Vezzulli, L.; Colwell, R.R. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 2008, 10, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Tuveng, T.R.; Arntzen, M.Ø.; Bengtsson, O.; Gardner, J.G.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Proteomic investigation of the secretome of Cellvibrio japonicus during growth on chitin. Proteomics 2016, 16, 1904–1914. [Google Scholar] [CrossRef]
- Monge, E.C.; Tuveng, T.R.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Gardner, J.G. Systems analysis of the glycoside hydrolase family 18 enzymes from Cellvibrio japonicus characterizes essential chitin degradation functions. J. Biol. Chem. 2018, 293, 3849–3859. [Google Scholar] [CrossRef]
- Hayes, C.A.; Dalia, T.N.; Dalia, A.B. Systematic genetic dissection of chitin degradation and uptake in Vibrio cholerae. Environ. Microbiol. 2017, 19, 4154–4163. [Google Scholar] [CrossRef]
- Bengtsson, O.; Arntzen, M.; Mathiesen, G.; Skaugen, M.; Eijsink, V.G.H. A novel proteomics sample preparation method for secretome analysis of Hypocrea jecorina growing on insoluble substrates. J. Proteom. 2016, 131, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.W.; Sharma, P.K.; van der Mei, H.C.; Busscher, H.J. Bacterial cell surface damage due to centrifugal compaction. Appl. Environ. Microbiol. 2012, 78, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Caballero, S.; Cano-Sánchez, P.; Mares-Mejía, I.; Díaz-Sánchez, A.G.; Macías-Rubalcava, M.L.; Hermoso, J.A.; Rodríguez-Romero, A. Comparative study of two GH19 chitinase-like proteins from Hevea brasiliensis, one exhibiting a novel carbohydrate-binding domain. FEBS J. 2014, 281, 4535–4554. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Armand, S.; Hata, T.; Nikaidou, N.; Henrissat, B.; Mitsutomi, M.; Watanabe, T. A modular family 19 chitinase found in the prokaryotic organism Streptomyces griseus HUT 6037. J. Bacteriol. 1996, 178, 5065–5070. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Melchiorsen, J.; Bruhn, J.B. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar. Biotechnol. 2010, 12, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.D.; Santini, C.L.; Zhang, W.J.; Barbe, V.; Mangenot, S.; Guyomar, C.; Garel, M.; Chen, H.T.; Li, X.G.; Yin, Q.J.; et al. Genomic and physiological analysis reveals versatile metabolic capacity of deep-sea Photobacterium phosphoreum ANT-2200. Extremophiles 2016, 20, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Östling, J.; Goodman, A.; Kjelleberg, S. Behaviour of IncP-1 plasmids and a miniMu transposon in a marine Vibrio sp.: Isolation of starvation inducible lac operon fusions. FEMS Microbiol. Lett. 1991, 86, 83–94. [Google Scholar] [CrossRef]
- Paulsen, S.S.; Andersen, B.; Gram, L.; MacHado, H. Biological potential of chitinolytic marine bacteria. Mar. Drugs 2016, 14, 230. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Z.; Jia, R.; Hou, Y.; Yin, J.; Bian, X.; Li, A.; Müller, R.; Stewart, A.F.; Fu, J.; et al. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat. Protoc. 2016, 11, 1175–1190. [Google Scholar] [CrossRef] [PubMed]
- Dehio, C.; Meyer, M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Eescherichia coli. J. Bacteriol. 1997, 179, 538–540. [Google Scholar] [CrossRef] [PubMed]
- Milton, D.L.; O’Toole, R.; Hörstedt, P.; Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996, 178, 1310–1319. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Chevallier, V.; Schoof, E.M.; Malphettes, L.; Andersen, M.R.; Workman, C.T. Characterization of glutathione proteome in CHO cells and its relationship with productivity and cholesterol synthesis. Biotechnol. Bioeng. 2020, 117, 3448–3458. [Google Scholar] [CrossRef] [PubMed]
- Haddad Momeni, M.; Leth, M.L.; Sternberg, C.; Schoof, E.; Nielsen, M.W.; Holck, J.; Workman, C.T.; Hoof, J.B.; Abou Hachem, M. Loss of AA13 LPMOs impairs degradation of resistant starch and reduces the growth of Aspergillus nidulans. Biotechnol. Biofuels 2020, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Beyene, G.T.; Kalayou, S.; Riaz, T.; Tonjum, T. Comparative proteomic analysis of Neisseria meningitidis wildtype and dprA null mutant strains links DNA processing to pilus biogenesis. BMC Microbiol. 2017, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Yimer, S.A.; Birhanu, A.G.; Kalayou, S.; Riaz, T.; Zegeye, E.D.; Beyene, G.T.; Holm-Hansen, C.; Norheim, G.; Abebe, M.; Aseffa, A.; et al. Comparative proteomic analysis of Mycobacterium tuberculosis lineage 7 and lineage 4 strains reveals differentially abundant proteins linked to slow growth and virulence. Front. Microbiol. 2017, 8, 795. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.; Ferluga, S.; Sharma, V.; Futschik, M.; Hilton, D.A.; Adams, C.L.; Lasonder, E.; Hanemann, C.O. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine 2019, 40, 77–91. [Google Scholar] [CrossRef]
- Yu, Z.C.; Zhao, D.L.; Ran, L.Y.; Mi, Z.H.; Wu, Z.Y.; Pang, X.; Zhang, X.Y.; Su, H.N.; Shi, M.; Song, X.Y.; et al. Development of a genetic system for the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913. Microb. Cell Fact. 2014, 13, 1–9. [Google Scholar] [CrossRef]
- Wang, P.; Yu, Z.; Li, B.; Cai, X.; Zeng, Z.; Chen, X.; Wang, X. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microb. Cell Fact. 2015, 14, 1–11. [Google Scholar] [CrossRef]
- Kim, E.; Kinney, W.H.; Ovrutsky, A.R.; Vo, D.; Bai, X.; Honda, J.R.; Marx, G.; Peck, E.; Lindberg, L.; Falkinham, J.O.; et al. A surface with a biomimetic micropattern reduces colonization of Mycobacterium abscessus. FEMS Microbiol. Lett. 2014, 360, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Holm, D.K.; Petersen, L.M.; Klitgaard, A.; Knudsen, P.B.; Jarczynska, Z.D.; Nielsen, K.F.; Gotfredsen, C.H.; Larsen, T.O.; Mortensen, U.H. Molecular and chemical characterization of the biosynthesis of the 6-MSA-derived meroterpenoid yanuthone D in Aspergillus niger. Chem. Biol. 2014, 21, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Isbrandt, T.; Tolborg, G.; Ødum, A.; Workman, M.; Larsen, T.O. Atrorosins: A new subgroup of Monascus pigments from Talaromyces atroroseus. Appl. Microbiol. Biotechnol. 2020, 104, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, Y.S.; Park, S.; Kim, J.; Kang, S.J.; Lee, M.H.; Ryu, S.; Choi, J.M.; Oh, T.K.; Yoon, J.H. Exceptional production of both prodigiosin and cycloprodigiosin as major metabolic constituents by a novel marine bacterium, Zooshikella rubidus S1-1. Appl. Environ. Microbiol. 2011, 77, 4967–4973. [Google Scholar] [CrossRef] [PubMed]
- Couturier, M.; Bhalara, H.D.; Chawrai, S.R.; Monson, R.; Williamson, N.R.; Salmond, G.P.C.; Leeper, F.J. Substrate flexibility of the flavin-dependent dihydropyrrole oxidases Pigb and Hapb involved in antibiotic prodigiosin biosynthesis. ChemBioChem 2020, 21, 523–530. [Google Scholar] [CrossRef]


| Glycoside Hydrolase Type | Accession | Signal Peptide |
|---|---|---|
| GH18 | A0A5S3USE2 | Y |
| A0A5S3V351 | Y | |
| A0A5S3V6T3 | Y | |
| A0A5S3V0U4 | Y | |
| A0A5S3USH6 | Y | |
| A0A5S3V3K3 | Y | |
| A0A5S3V378 | Y | |
| GH19 | A0A5S3UX38 | Y |
| A0A5S3UPT5 | N | |
| GH20 | A0A5S3UX95 | Y |
| A0A5S3UV09 | Y | |
| A0A5S3V1X9 | Y | |
| LPMO | A0A5S3UTD1 | Y |
| A0A5S3V4S2 | Y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Isbrandt, T.; Strube, M.L.; Paulsen, S.S.; Nielsen, M.W.; Buijs, Y.; Schoof, E.M.; Larsen, T.O.; Gram, L.; Zhang, S.-D. Chitin Degradation Machinery and Secondary Metabolite Profiles in the Marine Bacterium Pseudoalteromonas rubra S4059. Mar. Drugs 2021, 19, 108. https://doi.org/10.3390/md19020108
Wang X, Isbrandt T, Strube ML, Paulsen SS, Nielsen MW, Buijs Y, Schoof EM, Larsen TO, Gram L, Zhang S-D. Chitin Degradation Machinery and Secondary Metabolite Profiles in the Marine Bacterium Pseudoalteromonas rubra S4059. Marine Drugs. 2021; 19(2):108. https://doi.org/10.3390/md19020108
Chicago/Turabian StyleWang, Xiyan, Thomas Isbrandt, Mikael Lenz Strube, Sara Skøtt Paulsen, Maike Wennekers Nielsen, Yannick Buijs, Erwin M. Schoof, Thomas Ostenfeld Larsen, Lone Gram, and Sheng-Da Zhang. 2021. "Chitin Degradation Machinery and Secondary Metabolite Profiles in the Marine Bacterium Pseudoalteromonas rubra S4059" Marine Drugs 19, no. 2: 108. https://doi.org/10.3390/md19020108
APA StyleWang, X., Isbrandt, T., Strube, M. L., Paulsen, S. S., Nielsen, M. W., Buijs, Y., Schoof, E. M., Larsen, T. O., Gram, L., & Zhang, S.-D. (2021). Chitin Degradation Machinery and Secondary Metabolite Profiles in the Marine Bacterium Pseudoalteromonas rubra S4059. Marine Drugs, 19(2), 108. https://doi.org/10.3390/md19020108







