An In-Silico Comparative Study of Lipases from the Antarctic Psychrophilic Ciliate Euplotes focardii and the Mesophilic Congeneric Species Euplotes crassus: Insight into Molecular Cold-Adaptation
Abstract
1. Introduction
2. Results
2.1. Lipase Sequence Characterization and Analysis
2.2. Amino Acid Composition Preferences
2.3. Secondary Structural Elements
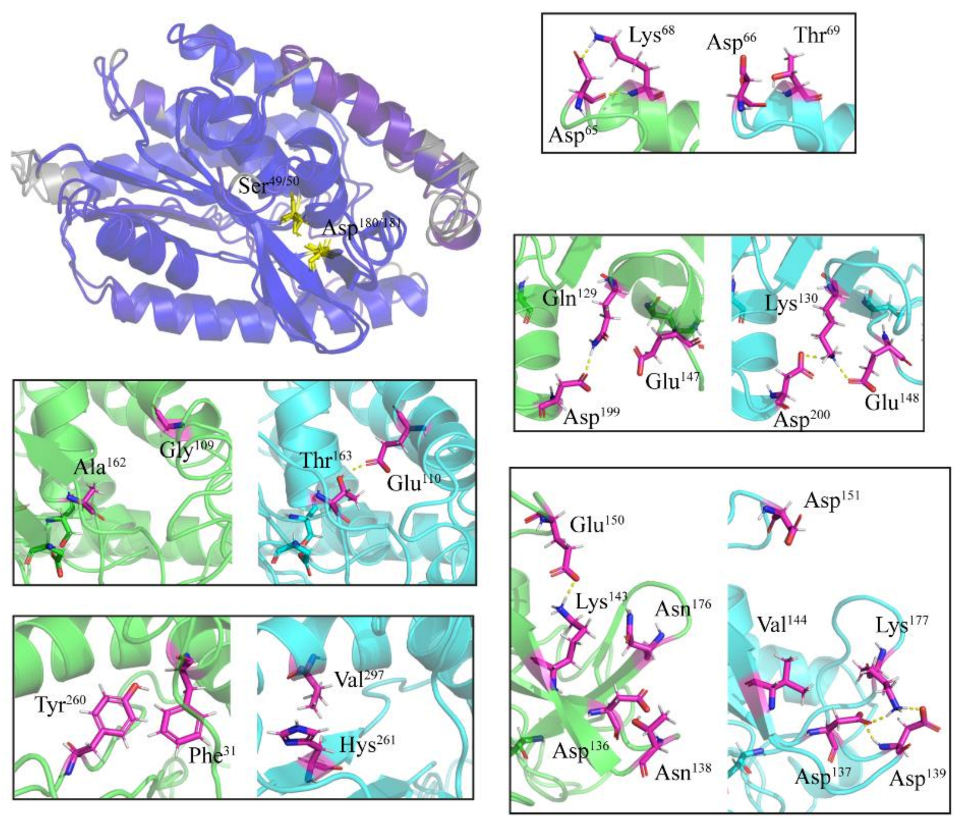
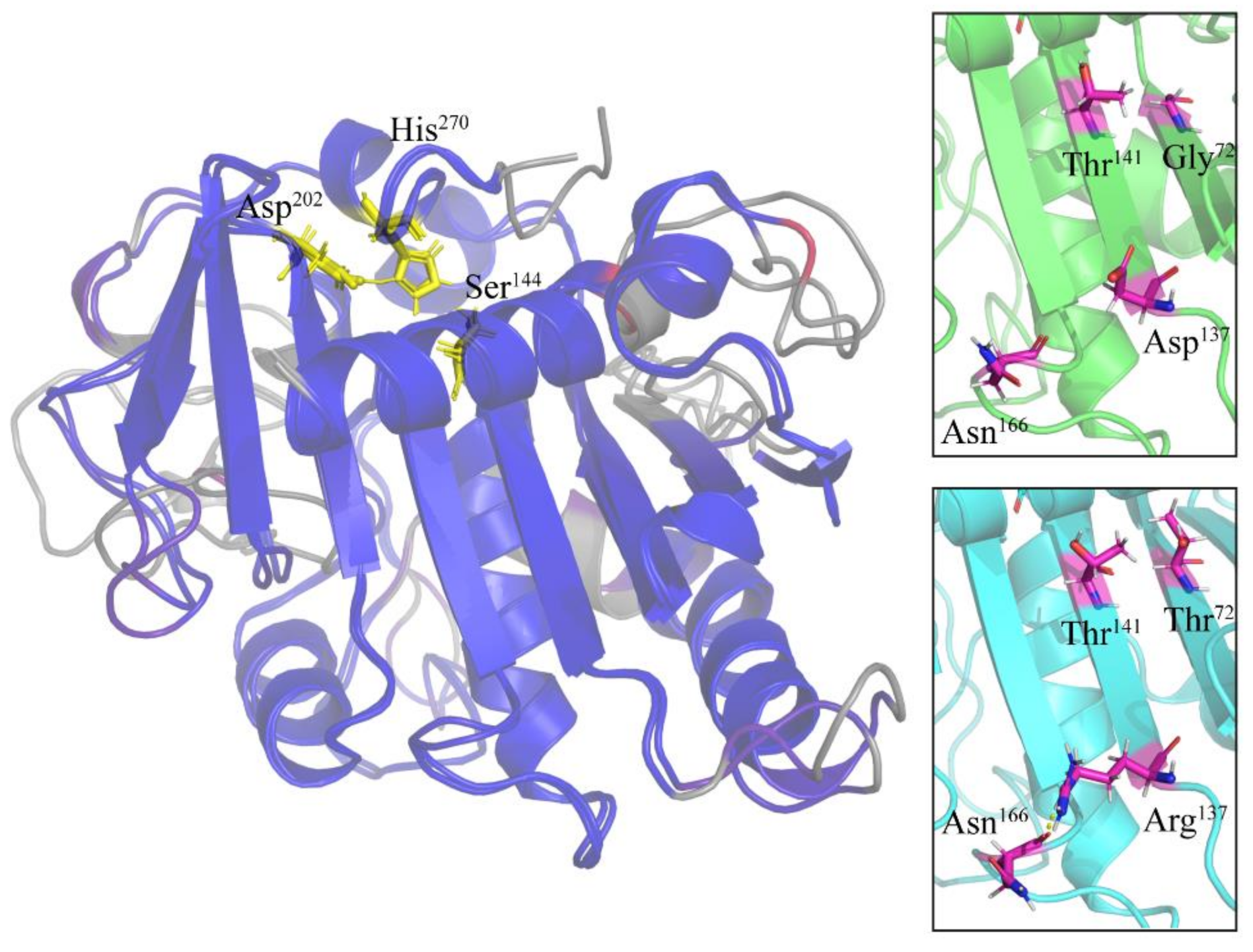
2.4. Specific Amino Acid Substitutions
2.5. Euplotes Lipases Codon Usage
3. Discussion
4. Materials and Methods
4.1. Sequence Collection and Analysis
4.2. Analysis of Amino Acid Composition
4.3. Secondary Structure Prediction
4.4. Amino Acid Substitution Bias
4.5. Tertiary Structure Prediction and Codon Usage Estimation
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, H.X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Margesin, R. Psychrophilic lifestyles: Mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Madigan, M.T.; Marrs, B.L. Extremophiles. Sci. Am. 1997, 276, 82–87. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Vance, T.D.R.; Bayer-Giraldi, M.; Davies, P.L.; Mangiagalli, M. Ice-binding proteins and the ‘domain of unknown function’ 3494 family. FEBS J. 2019, 286, 855–873. [Google Scholar] [CrossRef]
- Vallesi, A.; Pucciarelli, S.; Buonanno, F.; Fontana, A.; Mangiagalli, M. Bioactive molecules from protists: Perspectives in biotechnology. Eur. J. Protistol. 2020, 75, 125720. [Google Scholar] [CrossRef]
- Mangiagalli, M.; Brocca, S.; Orlando, M.; Lotti, M. The “cold revolution”. Present and future applications of cold-active enzymes and ice-binding proteins. New Biotechnol. 2020, 55, 5–11. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef]
- Bowman, J.P. Genomics of Psychrophilic Bacteria and Archaea. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Ed.; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Pucciarelli, S.; Devaraj, R.R.; Mancini, A.; Ballarini, P.; Castelli, M.; Schrallhammer, M.; Petroni, G.; Miceli, C. Microbial Consortium Associated with the Antarctic Marine Ciliate Euplotes focardii: An Investigation from Genomic Sequences. Microb. Ecol. 2015, 70, 484–497. [Google Scholar] [CrossRef]
- Ramasamy, K.P.; Telatin, A.; Mozzicafreddo, M.; Miceli, C.; Pucciarelli, S. Draft Genome Sequence of a New Pseudomonas sp. Strain, ef1, associated with the Psychrophilic Antarctic Ciliate Euplotes focardii. Microbiol. Resour. Announc. 2019, 8, e00867-19. [Google Scholar] [CrossRef] [PubMed]
- John, M.S.; Nagoth, J.A.; Ramasamy, K.P.; Ballarini, P.; Mozzicafreddo, M.; Mancini, A.; Telatin, A.; Liò, P.; Giuli, G.; Natalello, A.; et al. Horizontal gene transfer and silver nanoparticles production in a new Marinomonas strain isolated from the Antarctic Psychrophilic ciliate Euplotes focardii. Sci. Rep. 2020, 10, 10218. [Google Scholar] [CrossRef] [PubMed]
- Valbonesi, A.; Luporini, P. Biology of Euplotes focardii, an Antarctic Ciliate. Polar Biol. 1993, 13, 489–493. [Google Scholar] [CrossRef]
- Pucciarelli, S.; La Terza, A.; Ballarini, P.; Barchetta, S.; Yu, T.; Marziale, F.; Passini, V.; Methe, B.; Detrich, H.W., III; Miceli, C. Molecular cold-adaptation of protein function and gene regulation: The case for comparative genomic analyses in marine ciliated protozoa. Mar. Genom. 2009, 2, 57–66. [Google Scholar] [CrossRef]
- La Terza, A.; Papa, G.; Miceli, C.; Luporini, P. Divergence between two Antarctic species of the ciliate Euplotes, E. focardii and E. nobilii, in the expression of heat-shock protein 70 genes. Mol. Ecol. 2001, 10, 1061–1067. [Google Scholar] [CrossRef]
- Pischedda, A.; Ramasamy, K.P.; Mangiagalli, M.; Chiappori, F.; Milanesi, L.; Miceli, C.; Pucciarelli, S.; Lotti, M. Antarctic marine ciliates under stress: Superoxide dismutases from the psychrophilic Euplotes focardii are cold-active yet heat tolerant enzymes. Sci. Rep. 2018, 8, 14721. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yao, H.; Mozzicafreddo, M.; Ballarini, P.; Pucciarelli, S.; Miceli, C. Rational Engineering of a Cold-Adapted α-Amylase from the Antarctic Ciliate Euplotes focardii for Simultaneous Improvement of Thermostability and Catalytic Activity. Appl. Environ. Microbiol. 2017, 83, e00449-17. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, F.; Pucciarelli, S.; Merelli, I.; Ballarini, P.; Miceli, C.; Milanesi, L. Structural thermal adaptation of β-tubulins from the Antarctic psychrophilic protozoan Euplotes focardii. Proteins 2012, 80, 1154–1166. [Google Scholar] [CrossRef]
- Yang, G.; De Santi, C.; de Pascale, D.; Pucciarelli, S.; Pucciarelli, S.; Miceli, C. Characterization of the first eukaryotic cold-adapted patatin-like phospholipase from the psychrophilic Euplotes focardii: Identification of putative determinants of thermal-adaptation by comparison with the homologous protein from the mesophilic Euplotes crassus. Biochimie 2013, 95, 1795–1806. [Google Scholar]
- Marziale, F.; Pucciarelli, S.; Ballarini, P.; Melki, R.; Uzun, A.; Ilyin, V.A.; Detrich, H.W., 3rd; Miceli, C. Different roles of two gamma-tubulin isotypes in the cytoskeleton of the Antarctic ciliate Euplotes focardii: Remodelling of interaction surfaces may enhance microtubule nucleation at low temperature. FEBS J. 2008, 275, 5367–5382. [Google Scholar] [CrossRef]
- Pucciarelli, S.; Miceli, C. Characterization of the cold-adapted alpha-tubulin from the psychrophilic ciliate Euplotes focardii. Extremophiles 2002, 6, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Pucciarelli, S.; Marziale, F.; Di Giuseppe, G.; Barchetta, S.; Miceli, C. Ribosomal cold-adaptation: Characterization of the genes encoding the acidic ribosomal P0 and P2 proteins from the Antarctic ciliate Euplotes focardii. Gene 2005, 360, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, A.V.; Heaphy, S.M.; Turanov, A.A.; Gerashchenko, M.V.; Pucciarelli, S.; Devaraj, R.R.; Xie, F.; Petyuk, V.A.; Smith, R.D.; Klobutcher, L.A.; et al. Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nat. Struct. Mol. Biol. 2017, 24, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Gianese, G.; Bossa, F.; Pascarella, S. Comparative structural analysis of psychrophilic and meso- and thermophilic enzymes. Proteins 2002, 47, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.; Naderi-Manesh, H.; Zarrabi, M.; Ranjbar, B. Effective factors in thermostability of thermophilic proteins. Biophys. Chem. 2006, 119, 256–270. [Google Scholar] [CrossRef] [PubMed]
- Shokri, M.M.; Khajeh, K.; Alikhajeh, J.; Asoodeh, A.; Ranjbar, B.; Hosseinkhani, S.; Sadeghi, M. Comparison of the molten globule states of thermophilic and mesophilic alpha-amylases. Biophys. Chem. 2006, 122, 58–65. [Google Scholar] [CrossRef]
- Nishio, Y.; Nakamura, Y.; Kawarabayasi, Y.; Usuda, Y.; Kimura, E.S.; Matsui, K.; Yamagishi, A.; Kikuchi, H.; Ikeo, K.; Gojobori, T. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 2003, 13, 1572–1579. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, Molecular Mechanisms, and Industrial Applications of Cold-Active Enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- Pack, S.P.; Yoo, Y.J. Protein thermostability: Structure-based difference of amino acid between thermophilic and mesophilic proteins. J. Biotechnol. 2004, 111, 269–277. [Google Scholar] [CrossRef]
- Collins, T.; D’Amico, S.; Marx, J.-C.; Feller, G.; Gerday, C. Cold-Adapted Enzymes. In Physiology and Biochemistry of Extremophiles; Gerday, C., Glansdorff, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Feller, G. Molecular adaptations to cold in psychrophilic enzymes. CMLS Cell. Mol. Life Sci. 2003, 60, 648–662. [Google Scholar] [CrossRef]
- Lewis, P.N.; Go, N.; Go, M.; Kotelchuck, D.; Scheraga, H.A. Helix probability profiles of denatured proteins and their correlation with native structures. Proc. Natl. Acad. Sci. USA 1970, 65, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Metpally, R.P.; Reddy, B.V. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: Insights into the molecular basis of cold adaptation of proteins. BMC Genom. 2009, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Roussel, A.; Miled, N.; Berti-Dupuis, L.; Rivière, M.; Spinelli, S.; Berna, P.; Gruber, V.; Verger, R.; Cambillau, C. Crystal structure of the open form of dog gastric lipase in complex with a phosphonate inhibitor. J. Biol. Chem. 2002, 277, 2266–2274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yu, X.W.; Xu, Y.; Guo, R.T.; Swapna, G.V.T.; Szyperski, T.; Hunt, J.F.; Montelione, G.T. Structural Basis by Which the N-Terminal Polypeptide Segment of Rhizopus chinensis Lipase Regulates Its Substrate Binding Affinity. Biochemistry 2019, 58, 3943–3954. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Layer, R.M.; Skadron, K.; Robins, G.; Hall, I.M.; Quinlan, A.R. Binary Interval Search: A scalable algorithm for counting interval intersections. Bioinformatics 2013, 29, 1–7. [Google Scholar] [CrossRef]
- Piovesan, D.; Minervini, G.; Tosatto, S.C. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016, 44, W367–W374. [Google Scholar] [CrossRef]
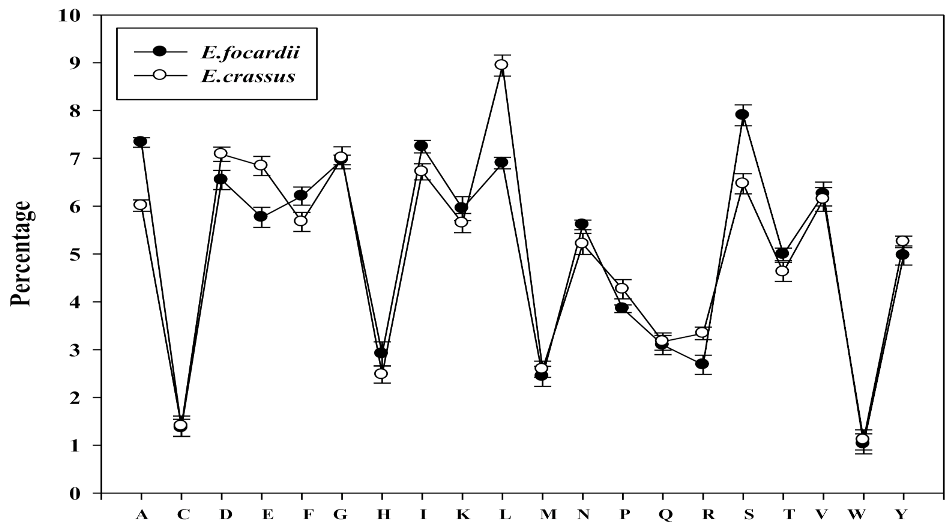

| Amino Acids | E. focardii | E. crassus | t-test p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ab | est | pat | Avg | ab | est | pat | Avg | ||
| A = Ala | 6.7 | 7.2 | 8.1 | 7.3 | 5.1 | 5.6 | 7.3 | 6.0 | 0.037 |
| C = Cys | 1.0 | 1.9 | 1.1 | 1.4 | 0.9 | 2.1 | 1.2 | 1.4 | 0.713 |
| D = Asp | 6.3 | 6.4 | 7.0 | 6.5 | 7.1 | 6.8 | 7.4 | 7.1 | 0.048 |
| E = Glu | 5.4 | 5.8 | 6.1 | 5.8 | 6.7 | 6.6 | 7.2 | 6.8 | 0.016 |
| F = Phe | 6.2 | 6.2 | 6.2 | 6.2 | 5.8 | 6.3 | 4.9 | 5.7 | 0.296 |
| G = Gly | 6.3 | 6.6 | 8.1 | 7.0 | 6.7 | 6.7 | 7.6 | 7.0 | 0.875 |
| H = His | 2.7 | 4.5 | 1.6 | 2.9 | 2.4 | 3.6 | 1.4 | 2.5 | 0.199 |
| I = Ile | 7.2 | 7.3 | 7.2 | 7.2 | 6.7 | 6.5 | 6.9 | 6.7 | 0.060 |
| K = Lys | 4.8 | 6.4 | 6.6 | 6.0 | 3.6 | 6.8 | 6.6 | 5.6 | 0.583 |
| L = Leu | 8.0 | 5.2 | 7.5 | 6.9 | 10.0 | 7.8 | 9.0 | 8.9 | 0.023 |
| M = Met | 2.9 | 2.0 | 2.4 | 2.4 | 3.1 | 2.0 | 2.6 | 2.6 | 0.116 |
| N = Asn | 5.5 | 6.0 | 5.3 | 5.6 | 5.2 | 5.3 | 5.2 | 5.2 | 0.173 |
| P = Pro | 4.4 | 3.9 | 3.2 | 3.9 | 4.9 | 4.3 | 3.6 | 4.3 | 0.001 |
| Q = Gln | 3.4 | 3.1 | 2.8 | 3.1 | 3.8 | 3.0 | 2.7 | 3.2 | 0.704 |
| R = Arg | 3.4 | 2.3 | 2.4 | 2.7 | 3.8 | 2.6 | 3.7 | 3.3 | 0.170 |
| S = Ser | 7.5 | 8.5 | 7.7 | 7.9 | 6.6 | 6.8 | 6.0 | 6.5 | 0.029 |
| T = Thr | 5.6 | 5.4 | 4.0 | 5.0 | 5.3 | 4.3 | 4.3 | 4.6 | 0.446 |
| V = Val | 5.6 | 5.9 | 7.3 | 6.3 | 5.2 | 6.3 | 6.9 | 6.1 | 0.698 |
| W = Trp | 1.7 | 0.1 | 1.3 | 1.0 | 1.4 | 0.5 | 1.5 | 1.1 | 0.714 |
| Y = Tyr | 5.4 | 5.2 | 4.2 | 5.0 | 5.7 | 6.0 | 4.1 | 5.3 | 0.427 |
| Amino acid property groups | |||||||||
| Tiny | 27.1 | 29.6 | 29.0 | 28.6 | 24.6 | 25.5 | 26.5 | 25.5 | 0.027 |
| Small | 48.9 | 51.8 | 51.8 | 50.8 | 46.9 | 48.2 | 49.5 | 48.2 | 0.036 |
| Aliphatic | 20.8 | 18.4 | 21.9 | 20.4 | 22.0 | 20.7 | 22.8 | 21.8 | 0.081 |
| Aromatic | 16.0 | 16.1 | 13.2 | 15.1 | 15.3 | 16.4 | 11.9 | 14.5 | 0.346 |
| Non-polar | 55.5 | 51.6 | 56.6 | 54.6 | 55.6 | 54.2 | 55.6 | 55.1 | 0.652 |
| Polar | 44.5 | 48.4 | 43.4 | 45.5 | 44.4 | 45.8 | 44.4 | 44.9 | 0.630 |
| Charged | 22.6 | 25.4 | 23.6 | 23.9 | 23.6 | 26.4 | 26.2 | 25.4 | 0.104 |
| Basic | 10.9 | 13.2 | 10.6 | 11.5 | 9.8 | 13.0 | 11.7 | 11.5 | 0.911 |
| Acidic | 11.7 | 12.2 | 13.1 | 12.3 | 13.8 | 13.4 | 14.6 | 13.9 | 0.022 |
| Hydrophilic | 28.7 | 30.0 | 30.2 | 29.7 | 30.1 | 31.1 | 32.7 | 31.3 | 0.067 |
| Hydrophobic | 44.8 | 41.1 | 45.3 | 43.7 | 44.0 | 43.2 | 44.3 | 43.8 | 0.933 |
| Neutral | 25.4 | 28.1 | 24.1 | 25.9 | 24.7 | 24.5 | 22.1 | 23.7 | 0.130 |
| A | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Amino Acids | α-helix | t-test p-Value | |||||||
| E. focardii | E. crassus | ||||||||
| ab | est | pat | Avg | ab | est | pat | Avg | ||
| A = Ala | 6.9 | 8.5 | 10.3 | 8.6 | 7.0 | 10.0 | 10.0 | 9.0 | 0.498 |
| C = Cys | 1.6 | 0.3 | 1.2 | 1.0 | 1.3 | 0.5 | 1.1 | 1.0 | 0.650 |
| D = Asp | 4.1 | 6.8 | 5.5 | 5.5 | 5.1 | 7.5 | 5.3 | 6.0 | 0.304 |
| E = Glu | 5.3 | 6.9 | 7.1 | 6.4 | 5.9 | 7.8 | 7.9 | 7.2 | 0.014 |
| F = Phe | 8.6 | 7.9 | 5.7 | 7.4 | 7.2 | 8.6 | 3.9 | 6.6 | 0.414 |
| G = Gly | 3.0 | 4.1 | 7.7 | 4.9 | 3.7 | 3.7 | 6.7 | 4.7 | 0.731 |
| H = His | 1.7 | 4.5 | 1.2 | 2.5 | 1.7 | 3.3 | 1.2 | 2.1 | 0.442 |
| I = Ile | 9.7 | 7.3 | 8.4 | 8.5 | 9.1 | 7.4 | 8.2 | 8.2 | 0.327 |
| K = Lys | 5.1 | 4.5 | 6.6 | 5.4 | 3.9 | 6.0 | 6.4 | 5.4 | 0.994 |
| L = Leu | 13.2 | 12.1 | 10.7 | 12.0 | 14.8 | 13.6 | 12.9 | 13.8 | 0.014 |
| M = Met | 3.4 | 2.1 | 3.0 | 2.8 | 3.6 | 2.3 | 2.9 | 2.9 | 0.488 |
| N = Asn | 4.6 | 5.0 | 4.0 | 4.5 | 3.9 | 4.0 | 4.1 | 4.0 | 0.267 |
| P = Pro | 2.4 | 1.1 | 0.8 | 1.4 | 3.0 | 0.9 | 1.3 | 1.7 | 0.328 |
| Q = Gln | 4.4 | 3.9 | 3.6 | 4.0 | 4.7 | 3.6 | 3.2 | 3.9 | 0.565 |
| R = Arg | 3.4 | 2.9 | 4.4 | 3.6 | 3.8 | 2.4 | 4.6 | 3.6 | 0.894 |
| S = Ser | 5.4 | 5.7 | 4.7 | 5.3 | 5.4 | 5.2 | 5.4 | 5.3 | 0.851 |
| T = Thr | 5.3 | 6.0 | 3.3 | 4.9 | 4.6 | 2.4 | 3.1 | 3.4 | 0.301 |
| V = Val | 6.1 | 5.0 | 6.2 | 5.8 | 6.0 | 4.6 | 5.9 | 5.5 | 0.080 |
| W = Trp | 2.0 | 0.2 | 1.1 | 1.1 | 1.5 | 0.3 | 1.6 | 1.1 | 0.879 |
| Y = Tyr | 4.0 | 5.3 | 4.3 | 4.5 | 3.7 | 5.9 | 4.1 | 4.6 | 0.875 |
| Amino acids property groups | |||||||||
| Tiny | 22.1 | 24.6 | 27.2 | 24.6 | 22.1 | 21.7 | 26.4 | 23.4 | 0.279 |
| Small | 39.3 | 42.4 | 43.8 | 41.9 | 40.1 | 38.8 | 43.1 | 40.6 | 0.443 |
| Aliphatic | 28.9 | 24.4 | 25.4 | 26.2 | 29.9 | 25.6 | 27.1 | 27.5 | 0.027 |
| Aromatic | 16.2 | 17.9 | 12.3 | 15.5 | 14.1 | 18.1 | 10.8 | 14.3 | 0.251 |
| Non-polar | 60.7 | 53.7 | 59.5 | 58.0 | 61.0 | 57.8 | 58.6 | 59.2 | 0.524 |
| Polar | 39.3 | 46.3 | 40.4 | 42.0 | 39.0 | 42.2 | 41.4 | 40.9 | 0.527 |
| Charged | 19.6 | 25.7 | 24.9 | 23.4 | 20.3 | 27.0 | 25.5 | 24.3 | 0.048 |
| Basic | 10.2 | 12.0 | 12.2 | 11.4 | 9.3 | 11.6 | 12.3 | 11.1 | 0.280 |
| Acidic | 9.4 | 13.7 | 12.6 | 11.9 | 11.0 | 15.4 | 13.2 | 13.2 | 0.063 |
| hydrophilic | 27.0 | 30.1 | 31.2 | 29.4 | 27.2 | 31.4 | 31.7 | 30.1 | 0.179 |
| hydrophobic | 55.4 | 48.6 | 51.0 | 51.7 | 54.3 | 53.2 | 50.7 | 52.7 | 0.617 |
| neutral | 19.7 | 24.3 | 20.5 | 21.5 | 20.1 | 18.2 | 19.7 | 19.3 | 0.393 |
| B | |||||||||
| Amino Acids | Coil | t-test p-Value | |||||||
| E. focardii | E. crassus | ||||||||
| ab | est | pat | Avg | ab | est | pat | Avg | ||
| A = Ala | 5.8 | 5.5 | 6.2 | 5.8 | 4.3 | 4.5 | 5.5 | 4.8 | 0.044 |
| C = Cys | 0.7 | 2.4 | 0.3 | 1.1 | 0.6 | 3.4 | 1.6 | 1.9 | 0.219 |
| D = Asp | 9.8 | 9.1 | 9.8 | 9.6 | 9.2 | 8.0 | 8.7 | 8.6 | 0.031 |
| E = Glu | 7.1 | 6.0 | 2.4 | 5.2 | 7.3 | 6.8 | 6.2 | 6.8 | 0.280 |
| F = Phe | 4.4 | 3.1 | 5.1 | 4.2 | 4.3 | 3.4 | 5.7 | 4.5 | 0.299 |
| G = Gly | 9.4 | 9.6 | 9.8 | 9.6 | 9.1 | 9.1 | 9.5 | 9.2 | 0.020 |
| H = His | 2.8 | 4.6 | 1.3 | 2.9 | 2.7 | 3.7 | 2.7 | 3.0 | 0.889 |
| I = Ile | 3.9 | 5.1 | 10.2 | 6.4 | 4.3 | 4.3 | 4.5 | 4.3 | 0.381 |
| K = Lys | 3.7 | 7.3 | 1.9 | 4.3 | 3.9 | 7.8 | 6.8 | 6.2 | 0.349 |
| L = Leu | 6.1 | 4.9 | 6.2 | 5.7 | 6.1 | 5.8 | 6.0 | 6.0 | 0.578 |
| M = Met | 1.5 | 2.2 | 2.4 | 2.0 | 2.4 | 1.6 | 2.1 | 2.1 | 0.949 |
| N = Asn | 7.5 | 8.3 | 2.4 | 6.1 | 6.8 | 7.4 | 7.0 | 7.1 | 0.636 |
| P = Pro | 6.1 | 5.3 | 1.2 | 4.2 | 7.0 | 7.1 | 3.2 | 5.8 | 0.043 |
| Q = Gln | 2.7 | 3.2 | 0.3 | 2.1 | 3.7 | 2.8 | 2.7 | 3.1 | 0.335 |
| R = Arg | 3.4 | 1.7 | 4.4 | 3.2 | 3.6 | 2.2 | 2.8 | 2.9 | 0.706 |
| S = Ser | 9.2 | 9.7 | 10.1 | 9.7 | 8.3 | 8.2 | 8.6 | 8.4 | 0.022 |
| T = Thr | 5.1 | 3.2 | 7.3 | 5.2 | 5.5 | 4.2 | 4.0 | 4.5 | 0.676 |
| V = Val | 3.2 | 4.9 | 11.7 | 6.6 | 3.3 | 4.7 | 7.4 | 5.1 | 0.399 |
| W = Trp | 1.3 | 0.2 | 1.3 | 0.9 | 1.1 | 0.4 | 1.4 | 0.9 | 0.998 |
| Y = Tyr | 6.3 | 3.8 | 5.7 | 5.3 | 6.7 | 4.6 | 3.6 | 5.0 | 0.781 |
| Amino acids property groups | |||||||||
| Tiny | 30.2 | 30.4 | 33.6 | 31.4 | 27.8 | 29.4 | 29.2 | 28.8 | 0.120 |
| Small | 56.8 | 58.0 | 58.8 | 57.9 | 54.0 | 56.6 | 55.5 | 55.4 | 0.047 |
| Aliphatic | 13.2 | 14.9 | 28.1 | 18.7 | 13.6 | 14.8 | 17.8 | 15.4 | 0.435 |
| Aromatic | 14.8 | 11.7 | 13.5 | 13.3 | 14.8 | 12.1 | 13.4 | 13.4 | 0.601 |
| Non-polar | 48.7 | 47.0 | 60.1 | 51.9 | 49.1 | 48.9 | 50.4 | 49.5 | 0.571 |
| Polar | 51.3 | 53.0 | 39.9 | 48.1 | 50.9 | 51.1 | 49.6 | 50.5 | 0.569 |
| Charged | 26.9 | 28.6 | 19.9 | 25.1 | 26.7 | 28.5 | 27.2 | 27.5 | 0.447 |
| Basic | 10.0 | 13.5 | 7.6 | 10.4 | 10.2 | 13.7 | 12.3 | 12.0 | 0.380 |
| Acidic | 16.9 | 15.1 | 12.2 | 14.7 | 16.5 | 14.8 | 14.9 | 15.4 | 0.567 |
| hydrophilic | 34.2 | 35.5 | 21.2 | 30.3 | 34.5 | 35.0 | 34.2 | 34.6 | 0.433 |
| hydrophobic | 33.2 | 32.1 | 49.1 | 38.1 | 33.0 | 32.7 | 37.7 | 34.5 | 0.446 |
| neutral | 29.2 | 30.3 | 28.8 | 29.4 | 29.2 | 28.0 | 27.5 | 28.2 | 0.215 |
| C | |||||||||
| Amino Acids | β-strand | t-test p-Value | |||||||
| E. focardii | E. crassus | ||||||||
| ab | est | pat | Avg | ab | est | pat | Avg | ||
| A = Ala | 6.2 | 2.2 | 6.2 | 4.9 | 5.3 | 1.6 | 6.2 | 4.4 | 0.198 |
| C = Cys | 0.9 | 0.7 | 0.3 | 0.6 | 0.8 | 0.7 | 0.3 | 0.6 | 0.349 |
| D = Asp | 0.6 | 1.5 | 7.5 | 3.2 | 2.1 | 1.6 | 8.0 | 3.9 | 0.213 |
| E = Glu | 4.3 | 4.6 | 2.4 | 3.8 | 3.6 | 3.9 | 2.4 | 3.3 | 0.178 |
| F = Phe | 7.9 | 13.4 | 6.5 | 9.3 | 9.1 | 11.0 | 5.6 | 8.5 | 0.562 |
| G = Gly | 2.9 | 2.7 | 4.0 | 3.2 | 3.0 | 3.9 | 3.9 | 3.6 | 0.445 |
| H = His | 5.2 | 4.6 | 1.3 | 3.7 | 3.2 | 3.9 | 1.1 | 2.7 | 0.194 |
| I = Ile | 13.6 | 15.1 | 15.6 | 14.8 | 10.8 | 12.6 | 12.5 | 12.0 | 0.004 |
| K = Lys | 3.4 | 3.7 | 1.9 | 3.0 | 2.9 | 4.9 | 2.1 | 3.3 | 0.572 |
| L = Leu | 10.5 | 6.3 | 9.2 | 8.7 | 13.7 | 6.2 | 10.3 | 10.1 | 0.281 |
| M = Met | 3.7 | 2.7 | 2.4 | 2.9 | 4.8 | 3.0 | 2.9 | 3.5 | 0.139 |
| N = Asn | 1.9 | 1.2 | 2.4 | 1.9 | 2.1 | 0.5 | 2.7 | 1.8 | 0.782 |
| P = Pro | 0.1 | 1.5 | 0.3 | 0.6 | 0.1 | 1.3 | 0.3 | 0.6 | 0.592 |
| Q = Gln | 3.4 | 1.5 | 0.3 | 1.7 | 3.4 | 2.6 | 0.5 | 2.1 | 0.363 |
| R = Arg | 2.9 | 4.4 | 5.4 | 4.2 | 2.7 | 4.1 | 5.4 | 4.1 | 0.258 |
| S = Ser | 3.3 | 3.2 | 3.0 | 3.1 | 3.3 | 3.4 | 2.7 | 3.1 | 0.912 |
| T = Thr | 5.2 | 10.2 | 7.3 | 7.6 | 6.7 | 8.2 | 8.0 | 7.6 | 0.948 |
| V = Val | 15.6 | 11.2 | 17.0 | 14.6 | 13.1 | 14.3 | 17.7 | 15.0 | 0.826 |
| W = Trp | 2.9 | 0.2 | 1.3 | 1.5 | 2.3 | 1.1 | 1.5 | 1.7 | 0.764 |
| Y = Tyr | 5.2 | 9.2 | 5.7 | 6.7 | 7.1 | 11.0 | 5.9 | 8.0 | 0.136 |
| Amino acids property groups | |||||||||
| Tiny | 18.5 | 19.0 | 20.8 | 19.4 | 19.1 | 17.9 | 21.1 | 19.4 | 0.960 |
| Small | 36.8 | 34.3 | 48.0 | 39.7 | 36.5 | 35.6 | 49.8 | 40.6 | 0.290 |
| Aliphatic | 39.8 | 32.6 | 41.8 | 38.1 | 37.6 | 33.1 | 40.5 | 37.1 | 0.338 |
| Aromatic | 21.2 | 27.5 | 14.8 | 21.2 | 21.6 | 27.1 | 14.0 | 20.9 | 0.528 |
| Non-polar | 69.7 | 65.2 | 68.5 | 67.8 | 70.1 | 66.7 | 67.1 | 68.0 | 0.863 |
| Polar | 30.3 | 34.8 | 31.5 | 32.2 | 29.9 | 33.3 | 32.9 | 32.0 | 0.863 |
| Charged | 16.5 | 18.7 | 18.6 | 17.9 | 14.5 | 18.5 | 19.0 | 17.3 | 0.500 |
| Basic | 11.5 | 12.7 | 8.6 | 10.9 | 8.8 | 13.0 | 8.6 | 10.1 | 0.485 |
| Acidic | 5.0 | 6.1 | 10.0 | 7.0 | 5.7 | 5.6 | 10.4 | 7.2 | 0.612 |
| hydrophilic | 16.7 | 16.8 | 19.9 | 17.8 | 16.7 | 17.7 | 21.1 | 18.5 | 0.165 |
| hydrophobic | 66.7 | 61.1 | 64.2 | 64.0 | 67.0 | 61.5 | 62.8 | 63.8 | 0.748 |
| neutral | 20.0 | 22.1 | 15.9 | 19.3 | 19.6 | 22.1 | 16.2 | 19.3 | 0.803 |
| E. focardii | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AΔ | C | DΔ | E∇ | F∇ | GΔ | H | I | K∇ | L | M | N | P | Q | R | SΔ | TΔ | V | W | Y∇ | ||
| E. crassus | A | 2.43 | 1.28 | 4.21 | −2.52 | −0.46 | 0.43 | 1.51 | −2.11 | −9.79 | −0.48 | 1.31 | −4.21 | −1.93 | 0.09 | −1.29 | 1.67 | 0.33 | −0.79 | 0.67 | −11.30 |
| C | 1.78 | 2.73 | 2.78 | −3.61 | −3.43 | 0.52 | 2.15 | 0.94 | −8.59 | 1.09 | −3.02 | 0.36 | 2.52 | 4.39 | 0.69 | 5.46 | 0.95 | −0.42 | 3.13 | −9.40 | |
| D | 3.69 | −1.38 | 3.46 | −1.55 | −6.53 | 0.95 | 1.64 | 0.37 | −0.51 | 1.38 | 0.94 | 0.91 | −0.54 | 0.74 | 0.48 | 3.94 | 1.23 | −0.38 | 0.74 | −7.34 | |
| E | 6.97 | −2.09 | 2.21 | −2.68 | −1.36 | 0.32 | 0.87 | 0.76 | −3.72 | 2.85 | 0.63 | 5.23 | 0.89 | −1.34 | 0.83 | 4.36 | 9.55 | 1.09 | 0.41 | −5.43 | |
| F | 4.73 | 0.43 | 7.53 | −4.22 | −0.53 | 0.65 | 0.31 | −0.52 | −8.96 | −0.92 | 0.35 | −4.38 | 0.48 | −0.46 | 0.76 | 1.29 | 0.32 | 0.25 | 1.94 | −0.33 | |
| G | 2.35 | −0.77 | 3.37 | −1.03 | −0.47 | 3.23 | −4.21 | 0.57 | −4.47 | −0.59 | −4.12 | −0.69 | −3.81 | −3.31 | −1.23 | 5.72 | 0.57 | 1.88 | 2.32 | −1.48 | |
| H | 5.83 | 0.32 | 3.77 | −7.23 | −5.65 | 0.41 | 2.50 | −1.82 | −9.46 | 0.35 | 0.39 | −3.63 | −2.09 | 1.06 | 1.93 | 5.83 | 1.49 | 0.82 | 0.18 | −2.80 | |
| I | 3.32 | 2.55 | 5.93 | −4.34 | −5.95 | 0.65 | 0.73 | −3.51 | −2.33 | 1.63 | 0.91 | −6.05 | 1.32 | −0.82 | 3.08 | 3.43 | 2.95 | −1.28 | 0.84 | −6.83 | |
| K | 5.53 | 1.82 | 1.25 | −6.53 | −4.92 | 2.45 | −2.35 | 2.36 | −0.38 | 1.37 | 1.02 | 4.38 | −1.03 | −0.41 | −5.61 | 9.81 | 5.18 | 4.14 | −4.22 | −3.06 | |
| L | 4.30 | 2.32 | 3.24 | −2.47 | −4.81 | 2.62 | −3.67 | −3.41 | −2.19 | 1.58 | 1.93 | 9.43 | 0.78 | 3.09 | −4.83 | 2.54 | 3.15 | 4.80 | −0.98 | −1.36 | |
| M | 5.04 | −2.08 | 4.21 | −5.32 | −3.19 | 1.31 | −1.43 | 0.33 | −0.37 | 1.06 | 2.64 | −0.09 | −3.96 | 8.31 | 0.33 | 3.09 | 0.82 | 0.22 | 5.33 | −0.94 | |
| N | 6.82 | 3.22 | 6.67 | −6.40 | −1.62 | 3.51 | 0.44 | 0.75 | −1.18 | −3.58 | −3.42 | −5.44 | 1.24 | −2.81 | −1.27 | 4.36 | 3.56 | 0.32 | −3.41 | −2.64 | |
| P | 5.38 | −3.72 | 4.28 | −4.37 | −4.71 | 0.58 | −5.55 | 0.23 | −4.78 | −3.96 | 0.59 | −2.36 | 0.99 | −7.03 | −0.57 | 3.68 | 5.47 | −1.75 | −1.82 | −5.99 | |
| Q | 8.82 | 3.49 | 4.74 | −4.51 | −5.31 | 0.28 | −2.39 | −0.43 | −7.21 | −2.35 | −1.36 | 5.23 | 0.39 | −0.06 | 0.38 | 6.71 | 0.83 | 4.39 | 2.99 | −5.69 | |
| R | 6.18 | 2.35 | 3.52 | −3.62 | −6.46 | 3.52 | 2.30 | 1.53 | −0.57 | 0.33 | −0.52 | 3.02 | 0.37 | 0.18 | 1.03 | 6.10 | 3.06 | 3.09 | −2.30 | −4.33 | |
| S | 7.05 | 0.64 | 7.12 | −1.83 | −3.67 | 4.12 | −5.51 | 4.37 | −1.16 | 1.22 | −2.10 | −2.34 | −0.05 | −2.26 | 2.08 | 1.05 | 0.97 | 4.59 | −8.20 | −8.52 | |
| T | 3.23 | 0.54 | 3.64 | −4.64 | −7.42 | 0.53 | 1.27 | −5.31 | −1.85 | −1.29 | −0.95 | −0.28 | 3.32 | 0.67 | 2.31 | 2.85 | 3.42 | −0.32 | −0.58 | −6.86 | |
| V | 2.06 | 0.73 | 6.42 | −9.45 | −5.51 | 0.48 | 0.31 | −0.69 | −9.32 | 0.31 | −0.64 | −2.93 | 0.28 | −0.70 | 0.73 | 2.92 | 1.89 | −3.22 | 0.72 | −3.18 | |
| W | 9.53 | 6.21 | 3.48 | −6.47 | −3.26 | 4.21 | 0.58 | 3.68 | −10.60 | 0.47 | 1.08 | −4.03 | 0.40 | 3.46 | −2.18 | 7.53 | 3.91 | 1.20 | 3.47 | −5.77 | |
| Y | 3.46 | 0.69 | 2.14 | −3.27 | −2.53 | 4.74 | 1.91 | 0.84 | −6.83 | −3.04 | −0.58 | −1.58 | −2.19 | 8.95 | −1.24 | 6.71 | 0.39 | 0.33 | −3.95 | −1.48 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Mozzicafreddo, M.; Ballarini, P.; Pucciarelli, S.; Miceli, C. An In-Silico Comparative Study of Lipases from the Antarctic Psychrophilic Ciliate Euplotes focardii and the Mesophilic Congeneric Species Euplotes crassus: Insight into Molecular Cold-Adaptation. Mar. Drugs 2021, 19, 67. https://doi.org/10.3390/md19020067
Yang G, Mozzicafreddo M, Ballarini P, Pucciarelli S, Miceli C. An In-Silico Comparative Study of Lipases from the Antarctic Psychrophilic Ciliate Euplotes focardii and the Mesophilic Congeneric Species Euplotes crassus: Insight into Molecular Cold-Adaptation. Marine Drugs. 2021; 19(2):67. https://doi.org/10.3390/md19020067
Chicago/Turabian StyleYang, Guang, Matteo Mozzicafreddo, Patrizia Ballarini, Sandra Pucciarelli, and Cristina Miceli. 2021. "An In-Silico Comparative Study of Lipases from the Antarctic Psychrophilic Ciliate Euplotes focardii and the Mesophilic Congeneric Species Euplotes crassus: Insight into Molecular Cold-Adaptation" Marine Drugs 19, no. 2: 67. https://doi.org/10.3390/md19020067
APA StyleYang, G., Mozzicafreddo, M., Ballarini, P., Pucciarelli, S., & Miceli, C. (2021). An In-Silico Comparative Study of Lipases from the Antarctic Psychrophilic Ciliate Euplotes focardii and the Mesophilic Congeneric Species Euplotes crassus: Insight into Molecular Cold-Adaptation. Marine Drugs, 19(2), 67. https://doi.org/10.3390/md19020067







