Anti-Food Allergic Compounds from Penicillium griseofulvum MCCC 3A00225, a Deep-Sea-Derived Fungus
Abstract
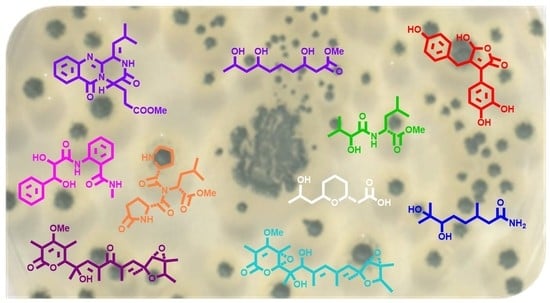
1. Introduction
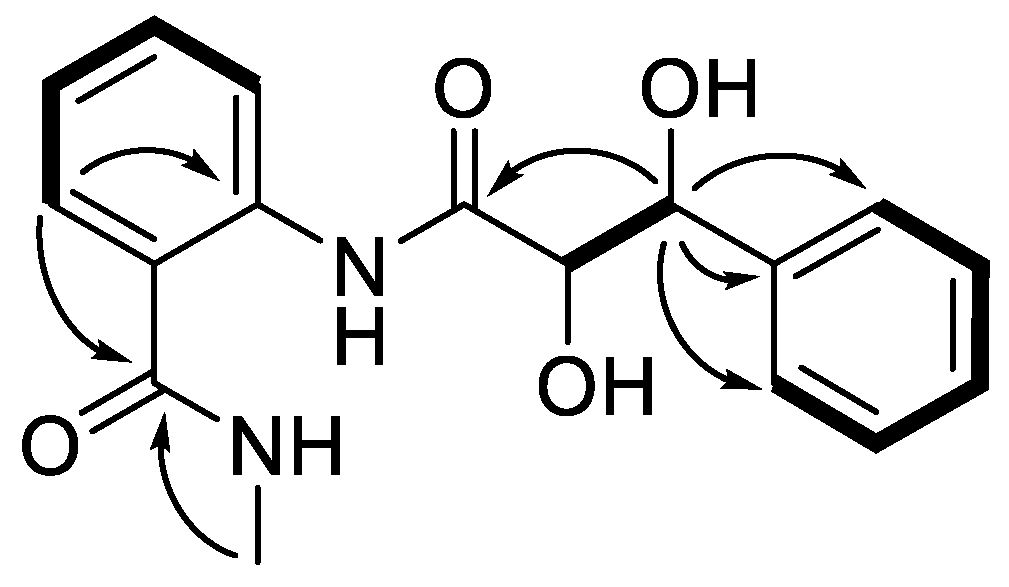
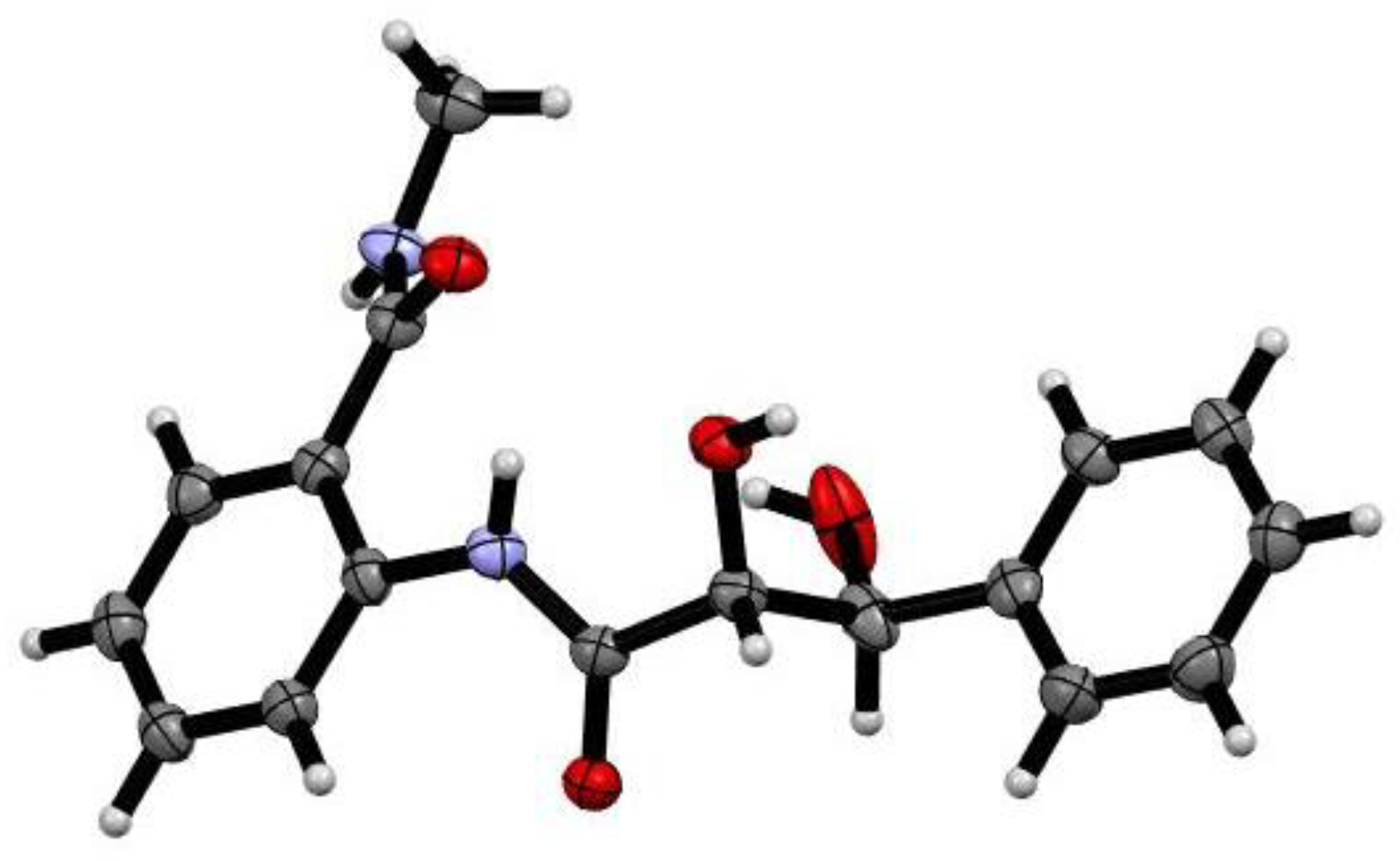
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures and Fungal Fermentation
3.2. Isolation and Purification
3.3. X-ray Crystallography of 1
3.4. Maryer’s Method
3.5. Induced CD (ICD) Experiment
3.6. Anti-Food Allergic Experiment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, Y.; Ruan, Q.; Zhao, M.; Zhao, Z.; Cui, H. Aspermeroterpenes A–C: Three meroterpenoids from the marine-derived fungus Aspergillus terreus GZU-31-1. Org. Lett. 2020, 22, 1336–1339. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.H.; Xu, Q.H.; Ge, G.B.; Shang, R.Y.; Zhu, H.R.; Liu, H.Y.; Cui, J.; Sun, F.; Lin, H.W. Flavipesides A–C, PKS-NRPS hybrids as pancreatic lipase inhibitors from a marine sponge symbiotic fungus Aspergillus flavipes 164013. Org. Lett. 2020, 22, 1825–1829. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef]
- Xie, C.L.; Zhang, D.; Lin, T.; He, Z.H.; Yan, Q.X.; Cai, Q.; Zhang, X.K.; Yang, X.W.; Chen, H.F. Antiproliferative sorbicillinoids from the deep-sea-derived Penicillium allii-sativi. Front. Microbiol. 2021, 11, 636948. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.D.; Fan, P.; Zhou, L.M.; Ma, Q.Y.; Xie, Q.Y.; Zheng, H.Z.; Zheng, Z.H.; Zhang, R.S.; Yuan, J.Z.; Dai, H.F.; et al. Penerpenes A–D, four indole terpenoids with potent protein tyrosine phosphatase inhibitory activity from the marine-derived fungus Penicillium sp. KFD28. Org. Lett. 2019, 21, 4864–4867. [Google Scholar] [CrossRef]
- Frank, M.; Hartmann, R.; Plenker, M.; Mándi, A.; Kurtán, T.; Özkaya, F.C.; Müller, W.E.G.; Kassack, M.U.; Hamacher, A.; Lin, W.; et al. Brominated azaphilones from the sponge-associated fungus Penicillium canescens strain 4.14.6a. J. Nat. Prod. 2019, 82, 2159–2166. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Fan, Z.; Xie, C.L.; Liu, Q.; Luo, Z.H.; Liu, G.; Yang, X.W. Spirograterpene A, a tetracyclic spiro-diterpene with a fused 5/5/5/5 ring system from the deep-sea-derived fungus Penicillium granulatum MCCC 3A00475. J. Nat. Prod. 2017, 80, 2174–2177. [Google Scholar] [CrossRef]
- Zou, Z.B.; Zhang, G.; Li, S.M.; He, Z.H.; Yan, Q.X.; Lin, Y.K.; Xie, C.L.; Xia, J.M.; Luo, Z.H.; Luo, L.Z.; et al. Asperochratides A–J, Ten new polyketides from the deep-sea-derived Aspergillus ochraceus. Bioorg. Chem. 2020, 105, 104349. [Google Scholar] [CrossRef]
- Niu, S.; Xie, C.L.; Xia, J.M.; Liu, Q.M.; Peng, G.; Liu, G.M.; Yang, X.W. Botryotins A–H, Tetracyclic diterpenoids representing three carbon skeletons from a deep-sea-derived Botryotinia fuckeliana. Org. Lett. 2019, 22, 580–583. [Google Scholar] [CrossRef]
- Xie, C.L.; Liu, Q.; He, Z.H.; Gai, Y.B.; Zou, Z.B.; Shao, Z.Z.; Liu, G.M.; Chen, H.F.; Yang, X.W. Discovery of andrastones from the deep-sea-derived Penicillium allii-sativi MCCC 3A00580 by OSMAC strategy. Bioorg. Chem. 2021, 108, 104671. [Google Scholar] [CrossRef]
- Xing, C.P.; Xie, C.L.; Xia, J.M.; Liu, Q.M.; Lin, W.X.; Ye, D.Z.; Liu, G.M.; Yang, X.W. Penigrisacids A–D, four new sesquiterpenes from the deep-sea-derived Penicillium griseofulvum. Mar. Drugs 2019, 17, 507. [Google Scholar] [CrossRef] [PubMed]
- Shu, Z.; Liu, Q.; Xing, C.; Zhang, Y.; Zhou, Y.; Zhang, J.; Liu, H.; Cao, M.; Yang, X.; Liu, G. Viridicatol isolated from deep-sea Penicillium griseofulvum alleviates anaphylaxis and repairs the intestinal barrier in mice by suppressing mast cell activation. Mar. Drugs 2020, 18, 517. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.H.; Fang, Y.C.; Du, L.; Zhu, T.J.; Duan, L.; Chen, J.; Gu, Q.Q.; Zhu, W.M. Aurantiomides A-C, quinazoline alkaloids from the sponge-derived fungus Penicillium aurantiogriseum SP0-19. J. Nat. Prod. 2007, 70, 853–855. [Google Scholar] [CrossRef] [PubMed]
- Orlowska, A.; Witkowska, E.; Izdebski, J. Sequence dependence in the formation of pyroglutamyl peptides in solid phase peptide synthesis. Int. J. Pept. Protein Res. 2009, 30, 141–144. [Google Scholar] [CrossRef]
- Bu, Y.Y.; Yamazaki, H.; Takahashi, O.; Kirikoshi, R.; Ukai, K.; Namikoshi, M. Penicyrones A and B, an epimeric pair of α-pyrone-type polyketides produced by the marine-derived Penicillium sp. J. Antibiot. 2015, 69, 57–61. [Google Scholar] [CrossRef]
- Yu, K.; Ren, B.; Wei, J.; Chen, C.; Sun, J.; Song, F.; Dai, H.; Zhang, L. Verrucisidinol and verrucosidinol acetate, two pyrone-type polyketides isolated from a marine derived fungus, Penicillium aurantiogriseum. Mar. Drugs 2010, 8, 2744–2754. [Google Scholar] [CrossRef]
- Furukawa, T.; Fukuda, T.; Nagai, K.; Uchida, R.; Tomoda, H. Helvafuranone produced by the fungus Aspergillus nidulans BF0142 isolated from hot spring-derived soil. Nat. Prod. Commun. 2016, 11, 1001–1003. [Google Scholar] [CrossRef]
- Hodge, R.P.; Harris, C.M.; Harris, T.M. Verrucofortine, a major metabolite of Penicillium verrucosum var. cyclopium, the fungus that produces the mycotoxin verrucosidin. J. Nat. Prod. 1988, 51, 66–73. [Google Scholar] [CrossRef]
- Fremlin, L.J.; Piggott, A.M.; Lacey, E.; Capon, R.J. Cottoquinazoline A and cotteslosins A and B, metabolites from an Australian marine-derived strain of Aspergillus versicolor. J. Nat. Prod. 2009, 72, 666–670. [Google Scholar] [CrossRef]
- Sugimori, T.; Okawa, T.; Eguchi, S.; Kakehi, A.; Yashima, E.; Okamoto, Y. The first total synthesis of (-)-benzomalvin A and benzomalvin B via the intramolecular aza-wittig reactions. Tetrahedron 1998, 54, 7997–8008. [Google Scholar] [CrossRef]
- Sun, W.N.; Chen, X.T.; Tong, Q.Y.; Zhu, H.C.; He, Y.; Lei, L.; Xue, Y.B.; Yao, G.M.; Luo, Z.W.; Wang, J.P.; et al. Novel small molecule 11 β-HSD1 inhibitor from the endophytic fungus Penicillium commune. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Spulak, M.; Pourova, J.; Voprsalova, M.; Mikusek, J.; Kunes, J.; Vacek, J.; Ghavre, M.; Gathergood, N.; Pour, M. Novel bronchodilatory quinazolines and quinoxalines: Synthesis and biological evaluation. Eur. J. Med. Chem. 2014, 74, 65–72. [Google Scholar] [CrossRef]
- Moehrle, H.; Seidel, C.M. ChemInform Abstract: Dehydrogenation of N-secondary cyclic aminals and acylaminals. Chem. Inf. 1976, 7, 471–479. [Google Scholar] [CrossRef]
- Park, J.; Lee, J.; Chang, S. Iterative C−H functionalization leading to multiple amidations of anilides. Angew. Chem. Int. Ed. 2017, 56, 4256–4260. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Peng, C.; Dai, O.; Geng, Z.; Guo, Y.P.; Xie, X.F.; He, C.J.; Li, X.H. Two new pyrazines from the parent roots of Aconitum carmichaelii. Biochem. Syst. Ecol. 2013, 48, 92–95. [Google Scholar] [CrossRef]
- Kimura, Y.; Tani, K.; Kojima, A.; Sotoma, G.; Okada, K.; Shimada, A. Cyclo-(l-tryptophyl-l-phenylalanyl), a plant growth regulator produced by the fungus Penicillium sp. Phytochemistry 1996, 41, 665–669. [Google Scholar] [CrossRef]
- Arai, K.; Kimura, K.; Mushiroda, T.; Yamamoto, Y. Structures of fructigenines A and B, new alkaloids isolated from Penicillium fructigenum takeuchi. Chem. Pharm. Bull. 1989, 37, 2937–2939. [Google Scholar] [CrossRef]
- Kusano, M.; Sotoma, G.; Koshino, H.; Uzawa, J.; Chijimatsu, M.; Fujioka, S.; Kawano, T.; Kimura, Y. Brevicompanines A and B: New plant growth regulators produced by the fungus, Penicillium brevicompactum. J. Chem. Soc. Perkin Trans. 1 1998, 1, 2823–2826. [Google Scholar] [CrossRef]
- Jian, Y.J.; Wu, Y. On the structure of penipratynolene and WA. Tetrahedron 2010, 66, 637–640. [Google Scholar] [CrossRef]
- Oh, H.; Swenson, D.C.; Gloer, J.B.; Shearer, C.A. New bioactive rosigenin analogues and aromatic polyketide metabolites from the freshwater aquatic fungus Massarina tunicata. J. Nat. Prod. 2003, 66, 73–79. [Google Scholar] [CrossRef]
- Xin, Z.H.; Zhu, W.M.; Gu, Q.Q.; Fang, Y.C.; Duan, L.; Cui, C.B. A new cytotoxic compound from Penicillium auratiogriseum, symbiotic or epiphytic fungus of sponge Mycale plumose. Chin. Chem. Lett. 2005, 16, 1227–1229. [Google Scholar]
- Mandava, N.; Finegold, H. 1H and 13C Nuclear magnetic resonance spectra of phenylacetic acid derivatives. Spectrosc. Lett. 1980, 13, 59–68. [Google Scholar] [CrossRef]
- Gutierrez-Lugo, M.T.; Woldemichael, G.M.; Singh, M.P.; Suarez, P.A.; Maiese, W.M.; Montenegro, G.; Timmermann, B.N. Isolation of three new naturally occurring compounds from the culture of Micromonospora sp. P1068. Nat. Prod. Res. 2005, 19, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Kardono, L.B.; Tsauri, S.; Padmawinata, K.; Pezzuto, J.M.; Kinghorn, A.D. Phenylacetic acid derivatives and a thioamide glycoside from Entada phaseoloides. Phytochemistry 1991, 30, 3749–3752. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, N.; Zhao, Y.; Gao, H.; Hu, Y.H.; Hu, J.F. Fructose-derived carbohydrates from Alisma orientalis. Nat. Prod. Res. 2009, 23, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Davison, J.; Al Fahad, A.; Cai, M.; Song, Z.; Yehia, S.Y.; Lazarus, C.M.; Bailey, A.M.; Simpson, T.J.; Cox, R.J. Genetic, molecular, and biochemical basis of fungal tropolone biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 7642–7647. [Google Scholar] [CrossRef]
- Shimomura, H.; Sashida, Y.; Mimaki, Y.; Adachi, T.; Yoshinari, K. A new mevalonolactone glucoside derivative from the bark of Prunus buergeriana. Chem. Pharm. Bull. 1989, 37, 829–830. [Google Scholar] [CrossRef]
- Kishida, M.; Yamauchi, N.; Sawada, K.; Ohashi, Y.; Eguchi, T.; Kakinuma, K. Diacetone-glucose architecture as a chirality template. Part 9.1 Enantioselective synthesis of (R)-mevalonolactone and (R)-[2H9]mevalonolactone on carbohydrate template. J. Chem. Soc. Perkin Trans. 1 1997, 1, 891–896. [Google Scholar] [CrossRef]
- Krasnoff, S.B.; Keresztes, I.; Gillilan, R.E.; Szebenyi, D.M.E.; Donzelli, B.G.G.; Churchill, A.C.L.; Gibson, N.M. Serinocyclins A and B, cyclic heptapeptides from Metarhizium anisopliae. J. Nat. Prod. 2007, 70, 1919–1924. [Google Scholar] [CrossRef]
- Xia, M.W.; Cui, C.B.; Li, C.W.; Wu, C.J. Three new and eleven known unusual C25 steroids: Activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Mar. Drugs 2014, 12, 1545–1568. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Peng, G.; Xia, J.M.; Xie, C.L.; Li, Z.; Yang, X.W. A new pimarane diterpenoid from the Botryotinia fuckeliana fungus isolated from deep-sea water. Chem. Biodivers. 2019, 16, e1900519. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.L.; Liu, Q.; Xia, J.M.; Gao, Y.; Yang, Q.; Shao, Z.Z.; Liu, G.; Yang, X.W. Anti-allergic compounds from the deep-sea-derived actinomycete Nesterenkonia flava MCCC 1K00610. Mar. Drugs 2017, 15, 71. [Google Scholar] [CrossRef] [PubMed]
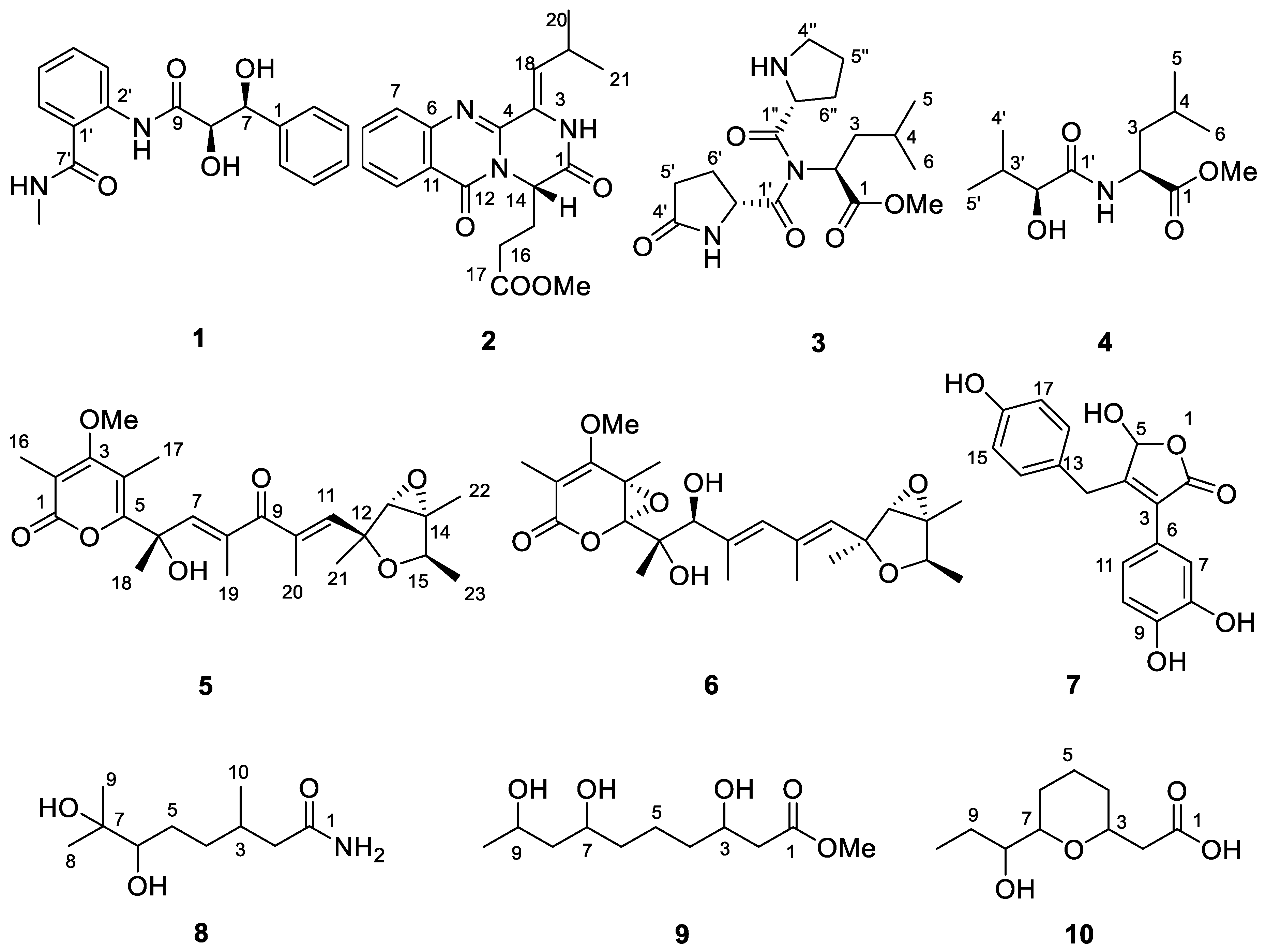

| No. | 1 | 3 | 4 | 8 | 9 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | δC | δH | |
| 1 | 143.2 C | 174.7 C | 174.5 C | 178.8 C | 173.7 C | |||||
| 2 | 127.5 CH | 7.46 (d, 7.8) | 52.3 CH | 4.41 (dd, 8.9, 6.2) | 51.6 CH | 4.52 (dd, 9.8, 4.8) | 44.0 CH2 | 2.24 (dd, 12.8, 5.2)1.98 m | 42.2 CH2 | 2.49 (dd, 15.1, 4.3)2.42 (dd, 15.1, 8.9) |
| 3 | 129.1 CH | 7.33 (dd, 7.8, 7.3) | 41.4 CH2 | 1.60 m | 41.6 CH2 | 1.67 m | 32.1 CH | 1.94 m | 75.9 CH | 3.79 (tdd, 8.9, 4.4, 2.0) |
| 4 | 128.3 CH | 7.24 (br t, 7.4) | 25.9 CH | 1.74 m | 26.0 CH | 1.68 m | 35.4 CH2 | 1.64 m; 1.22 m | 32.1 CH2 | 1.62 m; 1.22 m |
| 5 | 129.1 CH | 7.33 (dd, 7.8, 7.3) | 23.3 CH3 | 0.95 (d, 6.6) | 23.3 CH3 | 0.95 (d, 6.2) | 29.6 CH2 | 1.66 m; 1.23 m | 24.3 CH2 | 1.82 m; 1.58 m |
| 6 | 127.5 CH | 7.46 (d, 7.8) | 21.9 CH3 | 0.91 (d, 6.6) | 21.7 CH3 | 0.92 (d, 6.2) | 79.8 CH | 3.21 (d, 9.5) | 32.5 CH2 | 1.57 m; 1.21 m |
| 7 | 75.6 CH | 5.16 (d, 2.0) | 73.8 C | 78.5 CH | 3.54 (tdd, 10.2, 3.7, 1.7) | |||||
| 8 | 77.8 CH | 4.25 (d, 2.3) | 25.8 CH3 | 1.16 s | 45.9 CH2 | 1.61 m; 1.48 (dt, 14.0, 4.4) | ||||
| 9 | 174.0 C | 24.8 CH3 | 1.12 s | 67.4 CH | 3.94 m | |||||
| 10 | 20.2 CH3 | 0.96 (d, 6.2) | 23.1 CH3 | 1.14 (d, 6.2) | ||||||
| 1′ | 124.2 C | 172.9 C | 176.7 C | |||||||
| 2′ | 138.8 C | 56.2 CH | 4.55 (dd, 8.8, 4.0) | 77.0 CH | 3.86 (d, 3.7) | |||||
| 3′ | 122.4 CH | 8.51 (d, 8.1) | 33.0 CH | 2.07 m | ||||||
| 4′ | 132.7 CH | 7.47 (td, 7.8, 1.5) | 181.6 C | 19.5 CH3 | 1.00 (d, 7.0) | |||||
| 5′ | 124.7 CH | 7.16 (td, 7.6, 1.0) | 30.3 CH2 | 2.36 m; 2.30 m | 16.3 CH3 | 0.84 (d, 6.8) | ||||
| 6′ | 128.8 CH | 7.60 (dd, 7.8, 1.4) | 25.5 CH2 | 2.47 m; 2.16 m | ||||||
| 7′ (1″) | 171.3 C | 174.4 C | ||||||||
| 2″ | 61.3 CH | 4.47 (dd, 8.4, 2.8) | ||||||||
| 4″ | 48.1 CH2 | 3.63 m | ||||||||
| 5″ | 25.9 CH2 | 2.02 m | ||||||||
| 6″ | 30.3 CH2 | 2.18 m; 2.00 m | ||||||||
| NMe/OMe | 26.8 CH3 | 2.89 s | 52.6 CH3 | 3.69 s | 52.7 CH3 | 3.70 s | 52.1 CH3 | 3.65 s | ||
| No. | 2 a | 5 a | 6 a | 7 b | 10 a | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | δC | δH | |
| 1 | 167.7 C | 167.1 C | 167.9 C | 175.9 C | ||||||
| 2 | 111.1 C | 109.1 C | 170.6 C | 42.8 CH2 | 2.41 (d, 6.5) | |||||
| 3 | 126.3 C | 171.2 C | 168.3 C | 120.3 C | 75.8 CH | 3.76, m | ||||
| 4 | 147.0 C | 113.4 C | 80.4 C | 157.7 C | 32.2 CH2 | 1.64, m; 1.21, m | ||||
| 5 | 160.8 C | 109.1 C | 96.5 CH | 5.77 (d, 6.8) | 24.6 CH2 | 1.84, m; 1.59, m | ||||
| 6 | 121.1 C | 75.2 C | 82.1 C | 127.8 C | 32.8 CH2 | 1.52, m; 1.21, m | ||||
| 7 | 128.4 CH | 7.64 (d, 8.1) | 147.7 CH | 6.38 (d, 1.4) | 90.5 CH | 4.09 s | 116.1 CH | 6.92 (d, 1.8) | 76.2 CH | 3.57, m |
| 8 | 136.0 CH | 7.77 (td, 8.4, 1.4) | 137.3 C | 134.1 C | 145.1 C | 46.5 CH2 | 1.48, m | |||
| 9 | 128.0 CH | 7.46 (t, 7.6) | 202.8 C | 133.3 CH | 5.84 s | 146.1 C | 65.3 CH | 3.93, m | ||
| 10 | 127.6 CH | 8.14 (dd, 8, 1.1) | 139.3 C | 136.6 C | 115.6 CH | 6.79 (d, 8.2) | 23.9 CH3 | 1.13 (d, 6.3) | ||
| 11 | 148.7 C | 143.7 CH | 6.28 (d, 1.4) | 133.2 CH | 5.53 s | 120.2 CH | 6.75 (dd, 8.2, 1.8) | |||
| 12 | 162.1 C | 81.3 C | 81.6 C | 31.0 CH2 | 3.85 (d, 15.1); 3.57 (d, 15.1) | |||||
| 13 | 68.0 CH | 3.64 s | 68.7 CH | 3.55 s | 126.5 C | |||||
| 14 | 56.2 CH | 5.34 (t, 6.6) | 68.7 C | 68.7 C | 129.6 CH | 6.99 (d, 8.4) | ||||
| 15 | 28.7 CH2 | 2.65 m; 2.15 m | 78.5 CH | 4.08 (dt, 6.8, 6.8) | 78.3 CH | 4.05 (d, 6.8) | 115.5 CH | 6.69 (d, 8.4) | ||
| 16 | 30.6 CH2 | 2.44 m | 14.6 CH3 | 2.03 s | 10.2 CH3 | 1.84 s | 156.1 C | |||
| 17 | 173.9 C | 11.1 CH3 | 2.09 s | 20.8 CH3 | 1.61 s | 115.5 CH | 6.69 (d, 8.4) | |||
| 18 | 129.6 CH | 6.33 (d, 10.4) | 27.0 CH3 | 1.68 (d, 0.8) | 19.7 CH3 | 1.28 s | 129.6 CH | 6.99 (d, 8.4) | ||
| 19 | 27.1 CH | 2.87 m | 13.4 CH3 | 1.67 s | 15.8 CH3 | 1.86 (d, 0.9) | ||||
| 20 | 22.5 CH3 | 1.13 (d, 6.6) | 10.3 CH3 | 2.02 (d, 1.4) | 19.1 CH3 | 1.93 s | ||||
| 21 | 22.6 CH3 | 1.16 (d, 6.6) | 21.1 CH3 | 1.38 s | 22.1 CH3 | 1.37 s | ||||
| 22 | 13.7 CH3 | 1.45 s | 13.8 CH3 | 1.45 s | ||||||
| 23 | 19.3 CH3 | 1.17 (d, 6.8) | 19.2 CH3 | 1.20 (d, 6.8) | ||||||
| OMe | 52.2 CH3 | 3.46 s | 61.3 CH3 | 3.87 s | 61.1 CH3 | 3.92 s | ||||
| Compound | IC50 (μM) |
|---|---|
| 13 | 60.3 |
| 14 | 167.0 |
| 29 | 134.0 |
| Others a | ≥ 200 |
| Loratadine b | 91.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, C.-P.; Chen, D.; Xie, C.-L.; Liu, Q.; Zhong, T.-H.; Shao, Z.; Liu, G.; Luo, L.-Z.; Yang, X.-W. Anti-Food Allergic Compounds from Penicillium griseofulvum MCCC 3A00225, a Deep-Sea-Derived Fungus. Mar. Drugs 2021, 19, 224. https://doi.org/10.3390/md19040224
Xing C-P, Chen D, Xie C-L, Liu Q, Zhong T-H, Shao Z, Liu G, Luo L-Z, Yang X-W. Anti-Food Allergic Compounds from Penicillium griseofulvum MCCC 3A00225, a Deep-Sea-Derived Fungus. Marine Drugs. 2021; 19(4):224. https://doi.org/10.3390/md19040224
Chicago/Turabian StyleXing, Cui-Ping, Dan Chen, Chun-Lan Xie, Qingmei Liu, Tian-Hua Zhong, Zongze Shao, Guangming Liu, Lian-Zhong Luo, and Xian-Wen Yang. 2021. "Anti-Food Allergic Compounds from Penicillium griseofulvum MCCC 3A00225, a Deep-Sea-Derived Fungus" Marine Drugs 19, no. 4: 224. https://doi.org/10.3390/md19040224
APA StyleXing, C.-P., Chen, D., Xie, C.-L., Liu, Q., Zhong, T.-H., Shao, Z., Liu, G., Luo, L.-Z., & Yang, X.-W. (2021). Anti-Food Allergic Compounds from Penicillium griseofulvum MCCC 3A00225, a Deep-Sea-Derived Fungus. Marine Drugs, 19(4), 224. https://doi.org/10.3390/md19040224