Rapid Sorting of Fucoxanthin-Producing Phaeodactylum tricornutum Mutants by Flow Cytometry
Abstract
1. Introduction
2. Results and Discussion
2.1. P. tricornutum Has a Specific Emission Wavelength
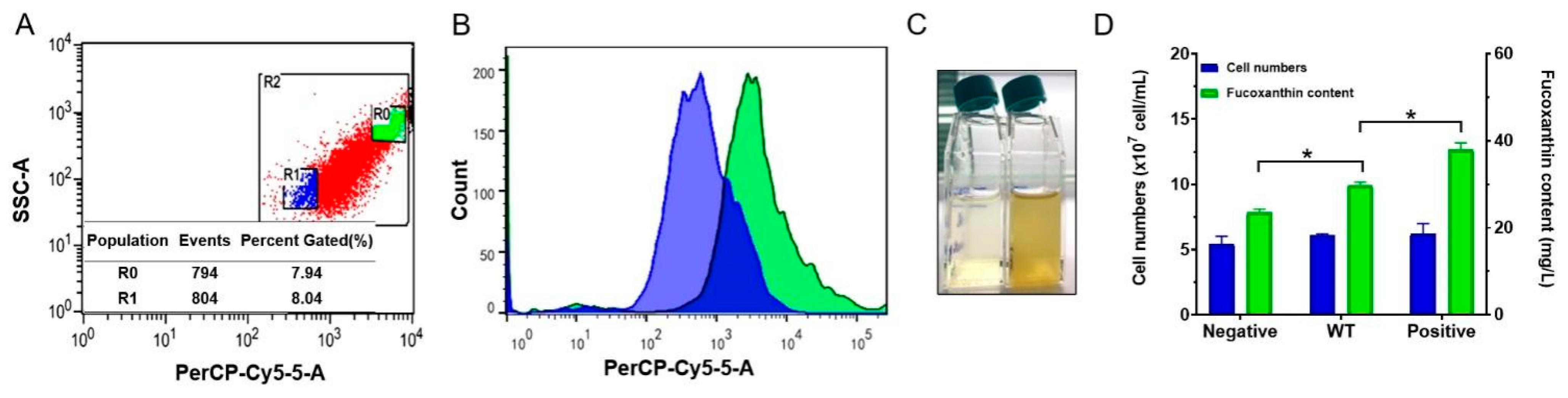
2.2. Flow Cytometry Analysis and Sorting
2.2.1. Flow Cytometry Can Be Efficiently Used to Analyze the Content of Fucoxanthin
2.2.2. Flow Cytometry Can Be Used for High-Throughput Separation of Cells with Different Fucoxanthin Contents
3. Materials and Methods
3.1. Strains and Culture Conditions
3.2. Fluorescence Spectrophotometer and Flow Cytometry Analysis
3.3. Fucoxanthin Assay
3.4. Screening Mutant Strains with High Fucoxanthin Content
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, S.; Wang, K.; Wan, L.L.; Li, A.F.; Hu, Q.; Zhang, C.W. Production, Characterization, and Antioxidant Activity of Fucoxanthin from the Marine Diatom. Odontella Aurita. Mar. Drugs 2013, 11, 2667–2681. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Yuan, J.P.; Wu, C.F.; Wang, J.H. Fucoxanthin, a Marine Carotenoid Present in Brown Seaweeds and Diatoms: Metabolism and Bioactivities Relevant to Human Health. Mar. Drugs 2011, 9, 1806–1828. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Sun, H.; Wu, T.; Fu, Y.L.; He, Y.J.; Mao, X.M.; Chen, F. Storage Carbon Metabolism of Isochrysis zhangjiangensis under Different Light Intensities and Its Application for Co-Production of Fucoxanthin and Stearidonic Acid. Bioresour. Technol. 2019, 282, 94–102. [Google Scholar] [CrossRef]
- Falciatore, A.; Jaubert, M.; Bouly, J.P.; Bailleul, B.; Mock, T. Diatom Molecular Research Comes of Age: Model Species for Studying Phytoplankton Biology and Diversity([OPEN]). Plant Cell 2020, 32, 547–572. [Google Scholar] [CrossRef]
- Gammone, M.A.; D’Orazio, N. Anti-Obesity Activity of the Marine Carotenoid Fucoxanthin. Mar. Drugs 2015, 13, 2196–2214. [Google Scholar] [CrossRef]
- Kang, S.I.; Ko, H.C.; Shin, H.S.; Kim, H.M.; Hong, Y.S.; Lee, N.H.; Kim, S.J. Fucoxanthin Exerts Differing Effects on 3t3-l1 Cells According to Differentiation Stage and Inhibits Glucose Uptake in Mature Adipocytes. Biochem. Bioph. Res. Co. 2011, 409, 769–774. [Google Scholar] [CrossRef]
- Kim, K.N.; Heo, S.J.; Yoon, W.J.; Kang, S.M.; Ahn, G.; Yi, T.H.; Jeon, Y.J. Fucoxanthin Inhibits the Inflammatory Response by Suppressing the Activation of Nf-Kappa b and Mapks in Lipopolysaccharide-Induced Raw 264 7 Macrophages. Eur. J. Pharmacol. 2010, 649, 369–375. [Google Scholar] [CrossRef]
- Liu, C.L.; Huang, Y.S.; Hosokawa, M.; Miyashita, K.; Hu, M.L. Inhibition of Proliferation of a Hepatoma Cell Line by Fucoxanthin in Relation to Cell Cycle Arrest and Enhanced Gap Junctional Intercellular Communication. Chem. Biol. Interact. 2009, 182, 165–172. [Google Scholar] [CrossRef]
- Wang, H.M.D.; Li, X.C.; Lee, D.J.; Chang, J.S. Potential Biomedical Applications of Marine Algae. Bioresour. Technol. 2017, 244, 1407–1415. [Google Scholar] [CrossRef]
- Mori, K.; Ooi, T.; Hiraoka, M.; Oka, N.; Hamada, H.; Tamura, M.; Kusumi, T. Fucoxanthin and Its Metabolites in Edible Brown Algae Cultivated in Deep Seawater. Mar. Drugs 2004, 2, 63–72. [Google Scholar] [CrossRef]
- Burch, A.R.; Franz, A.K. Combined Nitrogen Limitation and Hydrogen Peroxide Treatment Enhances Neutral Lipid Accumulation in the Marine Diatom Phaeodactylum Tricornutum. Bioresour. Technol. 2016, 219, 559–565. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- D’Adamo, S.; di Visconte, G.S.; Lowe, G.; Szaub-Newton, J.; Beacham, T.; Landels, A.; Allen, M.J.; Spicer, A.; Matthijs, M. Engineering the Unicellular Alga Phaeodactylum tricornutum for High-Value Plant Triterpenoid Production. Plant Biotechnol. J. 2019, 17, 75–87. [Google Scholar] [CrossRef]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular Toolbox for Studying Diatom Biology in Phaeodactylum tricornutum. Gene 2007, 406, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Eilers, U.; Dietzel, L.; Breitenbach, J.; Buchel, C.; Sandmann, G. Identification of Genes Coding for Functional Zeaxanthin Epoxidases in the Diatom Phaeodactylum Tricornutum. J. Plant Physiol. 2016, 192, 64–70. [Google Scholar] [CrossRef]
- Wang, W.D.; Yu, L.J.; Xu, C.Z.; Tomizaki, T.; Zhao, S.H.; Umena, Y.; Chen, X.B.; Qin, X.C.; Xin, Y.Y.; Suga, M.; et al. Structural Basis for Blue-Green Light Harvesting and Energy Dissipation in Diatoms. Science 2019, 363, 598. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.R.; Fan, Y.; Zhang, L.; Yuan, C.; Wang, J.F.; Li, W.J.; Hu, Q.; Li, F.L. Enhanced Lipid Productivity and Photosynthesis Efficiency in a Desmodesmus sp Mutant Induced by Heavy Carbon Ions. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Hada, M.; Georgakilas, A.G. Formation of Clustered DNA Camage after High-LET Irradiation: A Review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef]
- Hamada, N. Recent Insights into the Biological Action of Heavy-Ion Radiation. J. Radiat. Res. 2009, 50, 1–9. [Google Scholar] [CrossRef]
- Braumann, T.; Grimme, L.H. Reversed-Phase High-Performance Liquid-Chromatography of Chlorophylls and Carotenoids. Biochim. Biophys. Acta 1981, 637, 8–17. [Google Scholar] [CrossRef]
- Schwartz, S.J.; Woo, S.L.; Vonelbe, J.H. High-Performance Liquid-Chromatography of Chlorophylls and Their Derivatives in Fresh and Processed Spinach. J. Agr. Food Chem. 1981, 29, 533–535. [Google Scholar] [CrossRef]
- Di Bucchianico, S.; Cappellini, F.; Le Bihanic, F.; Zhang, Y.N.; Dreij, K.; Karlsson, H.L. Genotoxicity of TiO2 Nanoparticles Assessed by Mini-Gel Comet Assay and Micronucleus Scoring with Flow Cytometry. Mutagenesis 2017, 32, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, N.; Sung, M.; Yim, S.S.; Park, M.S.; Yang, J.W.; Jeong, K.J. Evaluation of Intracellular Lipid Bodies in Chlamydomonas reinhardtii Strains by Flow Cytometry. Bioresour. Technol. 2013, 138, 30–37. [Google Scholar] [CrossRef]
- Vives-Rego, J.; Lebaron, P.; Nebe-von Caron, G. Current and Future Applications of Flow Cytometry in Aquatic Microbiology. Fems. Microbiol. Rev. 2000, 24, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Ukibe, K.; Katsuragi, T.; Tani, Y.; Takagi, H. Efficient Screening for Astaxanthin-Overproducing Mutants of the Yeast Xanthophyllomyces dendrorhous by Flow Cytometry. Fems. Microbiol. Lett. 2008, 286, 241–248. [Google Scholar] [CrossRef]
- Chen, J.H.; Wei, D.; Pohnert, G. Rapid Estimation of Astaxanthin and the Carotenoid-to-Chlorophyll Ratio in the Green Microalga Chromochloris zofingiensis Using Flow Cytometry. Mar. Drugs 2017, 15, 231. [Google Scholar] [CrossRef]
- Xu, C.Z.; Pi, X.; Huang, Y.W.; Han, G.Y.; Chen, X.B.; Qin, X.C.; Huang, G.Q.; Zhao, S.H.; Yang, Y.Y.; Kuang, T.Y.; et al. Structural Basis for Energy Transfer in a Huge Diatom PSI-FCPI Supercomplex. Nat. Commun 2020, 11, 5081. [Google Scholar] [CrossRef]
- Nagao, R.; Kato, K.; Suzuki, T.; Ifuku, K.; Akita, F. Structural Basis for Energy Harvesting and Dissipation in a Diatom PSII–FCPII Supercomplex. Nat. Plants 2019, 5, 890–905. [Google Scholar] [CrossRef]
- Pi, X.; Zhao, S.; Wang, W.; Liu, D.; Shen, J.R. The Pigment-Protein Network of a Diatom Photosystem II-Light-Harvesting Antenna Supercomplex. Science 2019, 365. [Google Scholar] [CrossRef]
- Chumpolkulwong, N.; Kakizono, T.; Nagai, S.; Nishio, N. Increased Astaxanthin Production by Phaffia rhodozyma Mutants Isolated as Resistant to Diphenylamine. J. Ferment. Bioeng. 1997, 83, 429–434. [Google Scholar] [CrossRef]
- Goodwin, T.W. Biosynthesis of Carotenoids and Plant Triterpenes-Fifth Ciba Medal Lecture. Biochem. J. 1971, 123, 293. [Google Scholar] [CrossRef]
- Dong, X.; Li, W.; Di, C. Plant Mutation Breeding with Heavy Ion Irradiation at IMP. Eur. Biophys. J. Biophy. 2017, 46, 329. [Google Scholar] [CrossRef]
- Hu, W.; Li, W.; Chen, J. Recent Advances of Microbial Breeding via Heavy-Ion Mutagenesis at IMP. Lett. Appl. Microbiol. 2017, 65, 274–280. [Google Scholar] [CrossRef]
- Wang, L.J.; Fan, Y.; Parsons, R.L.; Hu, G.R.; Zhang, P.Y.; Li, F.L. A Rapid Method for the Determination of Fucoxanthin in Diatom. Mar. Drugs 2018, 16, 33. [Google Scholar] [CrossRef]
- Fan, Y.; Yuan, C.; Jin, Y.; Hu, G.R.; Li, F.L. Characterization of 3-Ketoacyl-CoA Synthase in a Nervonic Acid Producing Oleaginous Microalgae Mychonastes afer. Algal. Res. 2018, 31, 225–231. [Google Scholar] [CrossRef]
- Xu, F.; Fan, Y.; Miao, F.H.; Hu, G.R.; Sun, J.; Yang, G.F.; Li, F.L. Naphthylacetic Acid and Tea Polyphenol Application Promote Biomass and Lipid Production of Nervonic Acid-Producing Microalgae. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Yuan, C.; Liu, J.H.; Fan, Y.; Ren, X.H.; Hu, G.R.; Li, F.L. Mychonastes afer HSO-3-1 as a Potential New Source Of Biodiesel. Biotechnol. Biofuels. 2011, 4. [Google Scholar] [CrossRef]
- Yuan, C.; Zheng, Y.L.; Zhang, W.L.; He, R.; Fan, Y.; Hu, G.R.; Li, F. Lipid Accumulation and Anti-Rotifer Robustness of Microalgal Strains Isolated from Eastern China. J. Appl. Phycol. 2017, 29, 2789–2800. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Hu, H.H. High-Efficiency Nuclear Transformation of the Diatom Phaeodactylum tricornutum by Electroporation. Mar. Genom. 2014, 16, 63–66. [Google Scholar] [CrossRef]
- Xia, J.W.; Zhan, W.L.; Wei, B.W.; Yuan, Y.J.; Yuan, P.; Qiao, W.M.; Gao, D.Q.; Yang, X.T.; Man, K.D.; Xiao, G.Q.; et al. HIRFL-CSR complex. Chin. Phys. C 2009, 33, 804–810. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Ding, X.-T.; Wang, L.-J.; Jiang, E.-Y.; Van, P.N.; Li, F.-L. Rapid Sorting of Fucoxanthin-Producing Phaeodactylum tricornutum Mutants by Flow Cytometry. Mar. Drugs 2021, 19, 228. https://doi.org/10.3390/md19040228
Fan Y, Ding X-T, Wang L-J, Jiang E-Y, Van PN, Li F-L. Rapid Sorting of Fucoxanthin-Producing Phaeodactylum tricornutum Mutants by Flow Cytometry. Marine Drugs. 2021; 19(4):228. https://doi.org/10.3390/md19040228
Chicago/Turabian StyleFan, Yong, Xiao-Ting Ding, Li-Juan Wang, Er-Ying Jiang, Phung Nghi Van, and Fu-Li Li. 2021. "Rapid Sorting of Fucoxanthin-Producing Phaeodactylum tricornutum Mutants by Flow Cytometry" Marine Drugs 19, no. 4: 228. https://doi.org/10.3390/md19040228
APA StyleFan, Y., Ding, X.-T., Wang, L.-J., Jiang, E.-Y., Van, P. N., & Li, F.-L. (2021). Rapid Sorting of Fucoxanthin-Producing Phaeodactylum tricornutum Mutants by Flow Cytometry. Marine Drugs, 19(4), 228. https://doi.org/10.3390/md19040228





