Bacteria Associated with Benthic Invertebrates from Extreme Marine Environments: Promising but Underexplored Sources of Biotechnologically Relevant Molecules
Abstract
:1. Introduction
2. Extremophilic Microorganisms
3. Extreme Marine Environments and Their Benthic Fauna
3.1. The Deep-Sea
3.1.1. Deep-Sea Hydrothermal Vents (DSHVs)
3.1.2. Cold Seeps
3.2. Shallow Hydrothermal Vents (SHVs)
3.3. Cold Water Habitats
3.3.1. Polar Marine Environments
3.3.2. Deep-Sea and Submarine Canyons
3.4. Other Extreme Environments
3.4.1. Hypersaline Habitats
3.4.2. Marine Caves
3.4.3. Hypoxic and Anoxic Environments
4. Invertebrates from Extreme Benthic Habitats as Hosts of Microbial Communities
4.1. Deep-Sea Hot-Spots of Biodiversity
4.1.1. Deep-Sea Hydrothermal Vents
4.1.2. Cold Seeps
4.2. Shallow Hydrothermal Vents
4.3. Cold Marine Habitats
4.3.1. Antarctic Marine Environments
4.3.2. Arctic Marine Environments
4.3.3. Deep-Sea and Submarine Canyons
4.3.4. Marine Caves
5. Molecules Involved in the Microbe–Invertebrate Associations Become Biotechnologically Relevant
5.1. Bioactive Molecules from Invertebrate-Associated Bacteria
5.1.1. Antibiotic and Antitumor Compounds
5.1.2. Antibiofilm Capabilities and Quorum Sensing
5.2. Extracellular Polymeric Substances
5.2.1. EPS from Deep-Sea Hydrothermal Vents
5.2.2. EPSs from Cold Marine Systems
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Zeppilli, D.; Leduc, D.; Fontanier, C.; Fontaneto, D.; Fuchs, S.; Gooday, A.J.; Goineau, A.; Ingels, J.; Ivanenko, V.N.; Kristensen, R.M.; et al. Characteristics of meiofauna in extreme marine ecosystems: A review. Mar. Biodiver. 2018, 48, 35–71. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Chemical diversity: A function of biodiversity. Trends Pharmacol. Sci. 2002, 23, 404–405. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.R.; Nowak, M.; Austin, B.; Colwell, R.R. Specificity of bacterial symbionts in Mediterranean and Great Barrier Reef sponges. Microb. Ecol. 1981, 7, 1321. [Google Scholar] [CrossRef] [PubMed]
- Regoli, F.; Cerrano, C.; Chierici, E.; Bompadre, S.; Bavestrello, G. Susceptibility to the oxidative stress of the Mediterranean demosponge Petrosia ficiformis: Role of endosymbionts and solar irradiance. Mar. Biol. 2000, 137, 453461. [Google Scholar] [CrossRef]
- Usher, K.M. The ecology and phylogeny of cyanobacterial symbionts in sponges. Mar. Ecol. 2008, 29, 178192. [Google Scholar] [CrossRef]
- De Goeij, J.M.; Van Oevelen, D.; Vermeij, M.J.A.; Osinga, R.; Middelburg, J.J.; De Goeij, A.F.P.M.; Admiraal, W. Surviving in a marine desert: The sponge loop retains resources within coral reefs. Science 2013, 342, 108110. [Google Scholar] [CrossRef]
- Hoer, D.R.; Gibson, P.J.; Tommerdahl, J.P.; Lindquist, N.L.; Martens, C.S. Consumption of dissolved organic carbon by Caribbean reef sponges. Limnonol. Oceanogr. 2018, 63, 337351. [Google Scholar] [CrossRef]
- Maldonado, M.; Ribes, M.; Van Duyl, F.C. Nutrient fluxes through sponges: Biology, budgets, and ecological implications. Adv. Mar. Biol. 2012, 62, 113182. [Google Scholar]
- McMurray, S.E.; Stubler, A.D.; Erwin, P.M.; Finelli, C.M.; Pawlik, J.R. A test of the spongeloop hypothesis for emergent Caribbean reef sponges. Mar. Ecol. Progr. Ser. 2018, 588, 114. [Google Scholar] [CrossRef] [Green Version]
- Beinart, R.A.; Sanders, J.G.; Faure, B.; Sylva, S.P.; Lee, R.W.; Becker, E.L.; Gartman, A.; Luther, G.W.; Seewald, J.S.; Fisher, C.R.; et al. Evidence for the role of endosymbionts in regional-scale habitat partitioning by hydrothermal vent symbioses. Proc. Natl. Acad. Sci. USA 2012, 109, E3241–E3250. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. Microbes drive evolution of animals and plants: The hologenome concept. mBio 2016, 7, e01395-15. [Google Scholar] [CrossRef]
- Duperron, S.; de Beer, D.; Zbinden, M.; Boetius, A.; Schipani, V.; Kahil, N.; Gaill, F. Molecular characterization of bacteria associated with the trophosome and the tube of Lamellibrachia sp., a siboglinid annelid from cold seeps in the eastern Mediterranean. FEMS Microbiol. Ecol. 2009, 69, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.; Indraningrat, A.A.G.; Erngren, I.; Haglöf, J.; Becking, L.E.; Smidt, H.; Yashayaev, I.; Kenchington, E.; Pettersson, C.; Cárdenas, P.; et al. Oceanographic setting influences the prokaryotic community and metabolome in deep-sea sponges. Sci. Rep. 2022, 12, 3356. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Zhang, L.; Bazylinski, D.A. Deep-sea piezosphere and piezophiles: Geomicrobiology and biogeochemistry. Trends Microbiol. 2010, 18, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Somayaji, A.; Dhanjal, C.R.; Lingamsetty, R.; Vinayagam, R.; Selvaraj, R.; Varadavenkatesan, T.; Govarthanan, M. An insight into the mechanisms of homeostasis in extremophiles. Microbiol. Res. 2022, 263, 127115. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Llodra, E.; Brandt, A.; Danovaro, R.; De Mol, B.; Escobar, E.; German, C.; Levin, L.; Arbizu, P.; Menot, L.; Buhl-Mortensen, P. Deep, diverse and definitely different: Unique attributes of the world’s largest ecosystem. Biogeosciences 2010, 7, 2851–2899. [Google Scholar] [CrossRef]
- Van Dover, C.L.; German, C.R.; Speer, K.G.; Parson, L.M.; Vrijenhoek, R.C. Evolution and biogeography of deep-sea vent and seep invertebrates. Science 2002, 295, 1253–1257. [Google Scholar] [CrossRef]
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef]
- Petersen, J.M.; Dubilier, N. Methanotrophic symbioses in marine invertebrates. Environ. Microbiol. Rep. 2009, 1, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, S.E.; Baker, E.T.; German, C.R.; Maffei, A. An authoritative global database for active submarine hydrothermal vent fields: Global vents database. Geochem. Geophys. 2013, 14, 4892–4905. [Google Scholar] [CrossRef]
- Burgess, E.A.; Wagner, I.D.; Wiegel, J. Thermal Environments and Biodiversity. In Physiology and Biochemistry of Extremophiles; Gerday, C., Glansdorff, N., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 13–29. [Google Scholar]
- Le Bris, N.; Yücel, M.; Das, A.; Sievert, S.M.; LokaBharathi, P.; Girguis, P.R. Hydrothermal energy transfer and organic carbon production at the deep seafloor. Front. Mar. Sci. 2019, 5, 531. [Google Scholar] [CrossRef]
- Rizzo, C.; Arcadi, E.; Calogero, R.; Sciutteri, V.; Consoli, P.; Esposito, V.; Canese, S.; Andaloro, F.; Romeo, T. Ecological and Biotechnological Relevance of Mediterranean Hydrothermal Vent Systems. Minerals 2022, 12, 251. [Google Scholar] [CrossRef]
- Kennedy, J.; Flemer, B.; Jackson, S.A.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D.W. Evidence of a putative deep sea specific microbiome in marine sponges. PLoS ONE 2014, 9, e91092. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, D.J.; Fielman, K.T.; Santos, S.R.; Halanych, K.M. Siboglinid-bacteria endosymbiosis: A model system for studying symbiotic mechanisms. Commun. Integr. Biol. 2008, 1, 163–166. [Google Scholar] [CrossRef]
- Zwirglmaier, K.; Reid, W.D.K.; Heywood, J.; Sweeting, C.J.; Wigham, B.D.; Polunin, N.V.C.; Hawkes, J.A.; Connelly, D.P.; Pearce, D.; Linse, K. Linking regional variation of epibiotic bacterial diversity and trophic ecology in a new species of Kiwaidae (Decapoda, Anomura) from East Scotia Ridge (Antarctica) hydrothermal vents. Microbiol. Open 2015, 4, 136–150. [Google Scholar] [CrossRef]
- Hestetun, J.T.; Dahle, H.; Jørgensen, S.L.; Olsen, B.R.; Rapp, H.T. The microbiome and occurrence of methanotrophy in carnivorous sponges. Front. Microbiol. 2016, 7, 1781. [Google Scholar] [CrossRef]
- Rubin-Blum, M.; Antony, C.P.; Sayavedra, L.; Martínez-Pérez, C.; Birgel, D.; Peckmann, J.; Wu, Y.C.; Cardenas, P.; MacDonald, I.; Marcon, Y.; et al. Fueled by methane: Deep-sea sponges from asphalt seeps gain their nutrition from methane-oxidizing symbionts. ISME J. 2019, 13, 1209–1225. [Google Scholar] [CrossRef]
- Arellano, S.M.; Lee, O.O.; Lafi, F.F.; Yang, J.; Wang, Y.; Young, C.M.; Qian, P.-Y. Deep sequencing of Myxilla (Ectyomyxilla) methanophila, an epibiotic sponge on cold-seep tubeworms, reveals methylotrophic, thiotrophic, and putative hydrocarbon-degrading microbial associations. Microb. Ecol. 2013, 65, 450–461. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, R.; Sun, J.; Zhang, W.; Tian, R.-M.; Chen, C.; Kawagucci, S.; Xu, Y. Potential SUP05-phage interactions in hydrothermal vent sponges. Appl. Environ. Microbiol. 2019, 85, e992-19. [Google Scholar] [CrossRef] [PubMed]
- Chapman, A.S.A.; Beaulieu, S.E.; Colaço, A.; Gebruk, A.V.; Hilario, A.; Kihara, T.C.; Ramirez-Llodra, E.; Sarrazin, J.; Tunnicliffe, V.; Amon, D.J.; et al. sFDvent: A global trait database for deep-sea hydrothermal-vent fauna. Glob. Ecol. Biogeogr. 2019, 28, 1538–1551. [Google Scholar] [CrossRef]
- Paull, C.K.; Hecker, B.; Commeau, R.; Freeman-Lynde, R.P.; Neumann, C.; Corso, W.P.; Golubic, S.; Hook, J.E.; Sikes, E.; Curray, J. Biological communities at the Florida Escarpment resemble hydrothermal vent taxa. Science 1984, 226, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Levin, L.A. Ecology of cold seep sediments: Interactions of fauna with flow, chemistry and microbes. In Oceanography and Marine Biology, 1st ed.; CRC Press: Boca Raton, FL, USA, 2005; 46p. [Google Scholar]
- Muyzer, G.; Van Der Kraan, G.M. Bacteria from hydrocarbon seep areas growing on short-chain alkanes. Trends Microbiol. 2008, 16, 138–141. [Google Scholar] [CrossRef] [PubMed]
- Baco, A.R.; Rowden, A.A.; Levin, L.A.; Smith, C.R.; Bowden, D.A. Initial characterization of cold seep faunal communities on the New Zealand Hikurangi margin. Mar. Geol. 2010, 272, 251–259. [Google Scholar] [CrossRef]
- Tunniclife, V.; Juniper, S.K.; Sibuet, M. Reducing environments of the deep-sea foor. In Ecosystems of the Deep Oceans. Ecosystems of the World, 1st ed.; Tyler, P.A., Ed.; Elsevier Science: Amsterdam, The Netherlands, 2003; pp. 81–110. [Google Scholar]
- Imhof, J.F.; Sahling, H.; Suling, J.; Kath, T. 16S rDNA-based phylogeny of sulfur oxidizing bacterial endosymbionts in marine bivalves from cold-seeps environments. Mar. Ecol. Prog. Ser. 2003, 249, 39–51. [Google Scholar] [CrossRef]
- Åström, E.K.L.; Carroll, M.L.; Sen, A.; Niemann, H.; Ambrose, W.G., Jr.; Lehmann, M.F.; Carroll, J. Chemosynthesis influences food web and community structure in high-Arctic benthos. Mar. Ecol. Progr. Ser. 2019, 629, 19–42. [Google Scholar] [CrossRef]
- Hilário, A.; Capa, M.; Dahlgren, T.G.; Halanych, K.M.; Little, C.T.S.; Thornhill, D.J.; Verna, C.; Glover, A.G. New perspectives on the ecology and evolution of siboglinid tubeworms. PLoS ONE 2011, 6, e16309. [Google Scholar] [CrossRef]
- Mazumdar, A.; Dewangan, P.; Peketi, A.; Gullapalli, S.; Kalpana, M.S.; Naik, G.P.; Shetty, D.; Pujari, S.; Pillutla, S.P.K.; Gaikwad, V.V.; et al. The first record of active methane (cold) seep ecosystem associated with shallow methane hydrate from the Indian EEZ. J. Earth Syst. Sci. 2019, 128, 18. [Google Scholar] [CrossRef]
- Sibuet, M.; Olu, K. Biogeography, biodiversity and fluid dependence of deep-sea cold-seep communities at active and passive margins. Deep Sea Res. Part II 1998, 45, 517–567. [Google Scholar] [CrossRef]
- Sangodkar, N.; Gonsalves, M.J.; Nazareth, D.R. Macrofaunal distribution, diversity, and its ecological interaction at the cold seep site of Krishna-Godavari Basin, East Coast of India. Microb. Ecol. 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, V.G.; Gebruk, A.V.; Mironov, A.N.; Moskalev, L.I. Deep-sea and shallow-water hydrothermal vent communities: Two different phenomena? Chem. Geol. 2005, 224, 5–39. [Google Scholar] [CrossRef]
- Dando, P.R.; Stüben, D.; Varnavas, S.P. Hydrothermalism in the Mediterranean Sea. Prog. Oceanogr. 1999, 44, 333–367. [Google Scholar] [CrossRef]
- Donnarumma, L.; Appolloni, L.; Chianese, E.; Bruno, R.; Baldrighi, E.; Guglielmo, R.; Russo, G.F.; Zeppilli, D.; Sandulli, R. Environmental and benthic community patterns of the shallow hydrothermal area of Secca delle Fumose (Baia, Naples, Italy). Front. Mar. Sci. 2019, 6, 685. [Google Scholar] [CrossRef]
- Garilli, V.; Rodolfo-Metalpa, R.; Scuderi, D.; Brusca, L.; Parrinello, D.; Rastrick, S.P.S.; Foggo, A.; Twitchett, R.J.; Hall-Spencer, J.M.; Milazzo, M. Physiological advantages of dwarfing in surviving extinctions in high-CO2 oceans. Nat. Clim. Change 2015, 5, 678–682. [Google Scholar] [CrossRef]
- Harvey, B.P.; McKeown, N.J.; Rastrick, S.P.S.; Bertolini, C.; Foggo, A.; Graham, H.; Hall-Spencer, J.M.; Milazzo, M.; Shaw, P.W.; Small, D.P.; et al. Individual and population-level responses to ocean acidification. Sci. Rep. 2016, 6, 20194. [Google Scholar] [CrossRef]
- Wäge, J.; Valvassori, G.; Hardege, J.D.; Shulze, A.; Gambi, M.C. The sibling polychaetes Platynereis dumerilii and Platynereis massiliensis in the Mediterranean Sea: Are phylogeographic patterns related to exposure to ocean acidification? Mar. Biol. 2017, 164, 199. [Google Scholar] [CrossRef]
- Valvassori, G.; Massa-Gallucci, A.; Gambi, M.C. Reappraisal of Platynereis massiliensis (Moquin-Tandon) (Annelida, Nereididae), a neglected sibling species of Platynereis dumerilii (Audouin & Milne Edwards). Biol. Mar. Mediterr. 2015, 22, 113–116. [Google Scholar]
- Scipione, M.B. On the presence of the Mediterranean endemic Microdeutopus sporadhi Myers, 1969 (Crustacea: Amphipoda: Aoridae) in the Gulf of Naples (Italy) with a review on its distribution and ecology. Mediterr. Mar. Sci. 2013, 4, 56–63. [Google Scholar] [CrossRef]
- Esposito, V.; Giacobbe, S.; Cosentino, A.; Minerva, C.S.; Romeo, T.; Canese, S.; Andaloro, F. Distribution and ecology of the tube-dweller Ampelisca ledoyeri (Amphipoda: Ampeliscidae) associated to the hydrothermal field off Panarea Island (Tyrrhenian Sea, Mediterranean). Mar. Biodivers. 2015, 45, 763–768. [Google Scholar] [CrossRef]
- Dubilier, N.; Bergin, C.; Lott, C. Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nat. Rev. Microbiol. 2008, 6, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.M.; Wentrup, C.; Verna, C.; Knittel, K.; Dubilier, N. Origins and evolutionary flexibility of chemosynthetic symbionts from deep-sea animals. Biol. Bull. 2012, 223, 123–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalli, C.; Parsons, T. Biological Oceanography: An Introduction; Butterworth–Heinemann: Oxford, UK, 1997; p. 314. [Google Scholar]
- Lebar, M.D.; Heimbegner, J.L.; Baker, B.J. Cold-water marine natural products. Nat. Prod. Rep. 2007, 24, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Clark, G.F.; Raymond, B.; Riddle, M.J.; Stark, J.S.; Johnston, E.L. Vulnerability of Antarctic shallow invertebrate-dominated ecosystems. Austral Ecol. 2015, 40, 482–491. [Google Scholar] [CrossRef]
- Gili, J.-M.; Arntz, W.E.; Palanques, A.; Orejas, C.; Clarke, A.; Dayton, P.K.; Isla, E.; Teixidó, N.; Rossi, S.; López-González, P.J. A unique assemblage of epibenthic sessile suspension feeders with archaic features in the high-Antarctic. Deep Sea Res. Part II 2006, 53, 1029–1052. [Google Scholar] [CrossRef]
- Chiantore, M.; Guidetti, M.; Cavallero, M.; Albertelli, G.; Cattaneo-Vietti, R. Sea urchins, sea stars and brittle stars from Terra Nova Bay (Ross Sea, Antarctica). Polar Biol. 2006, 29, 467–475. [Google Scholar] [CrossRef]
- Oleszczuk, B.; Grzelak, K.; Kędra, M. Community structure and productivity of Arctic benthic fauna across depth gradients during springtime. Deep Sea Res. Part I 2021, 170, 103457. [Google Scholar] [CrossRef]
- Roberts, J.M.; Wheeler, A.J.; Freiwald, A. Reefs of the deep: The biology and geology of cold-water coral ecosystems. Science 2006, 312, 543–547. [Google Scholar] [CrossRef]
- Roberts, J.M.; Wheeler, A.J.; Freiwald, A.; Cairns, S.D. Cold-Water Corals: The Biology and Geology of Deep-Sea Coral Habitats; Cambridge University Press: Cambridge, UK, 2009; p. 334. [Google Scholar]
- Harris, P.; Macmillan-Lawler, M.; Rupp, J.; Baker, E. Geomorphology of the oceans. Mar. Geol. 2014, 352, 4–24. [Google Scholar] [CrossRef]
- De Leo, F.C.; Vetter, E.W.; Smith, C.R.; Rowden, A.A.; McGranaghan, M. Spatial scale-dependent habitat heterogeneity influences submarine canyon macrofaunal abundance and diversity off the Main and Northwest Hawaiian Islands. Deep Sea Res. Part II Top. Stud. Oceanogr. 2014, 104, 267–290. [Google Scholar] [CrossRef]
- Puig, P.; Canals, M.; Company, J.B.; Martin, J.; Amblas, D.; Lastras, G.; Palanques, A.; Calafat, A.M. Ploughing the deep sea floor. Nature 2012, 489, 286–289. [Google Scholar] [CrossRef] [PubMed]
- Huvenne, V.A.I.; Tyler, P.A.; Masson, D.G.; Fisher, E.H.; Hauton, C.; Hühnerbach, V.; Le Bas, T.P.; Wolff, G.A. A Picture on the wall: Innovative mapping reveals cold-water coral refuge in submarine canyon. PLoS ONE 2011, 6, e28755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Arcaya, U.; Ramirez-Llodra, E.; Aguzzi, J.; Allcock, A.L.; Davies, J.S.; Dissanayake, A.; Harris, P.; Howell, K.; Huvenne, V.A.I.; Macmillan-Lawler, M.; et al. Ecological role of submarine canyons and need for canyon conservation: A review. Front. Mar. Sci. 2017, 4, 5. [Google Scholar] [CrossRef]
- Jobstvogt, N.; Townsend, M.; Witte, U.; Hanley, N. How can we identify and communicate the ecological value of deep-sea ecosystem services? PLoS ONE 2014, 9, e100646. [Google Scholar] [CrossRef] [PubMed]
- Cunha, M.R.; Paterson, G.L.J.; Amaro, T.; Blackbird, S.; de Stigter, H.C.; Ferreira, C.; Glover, A.; Hilário, A.; Kiriakoulakis, K.; Neal, L.; et al. Biodiversity of macrofaunal assemblages from three Portuguese submarine canyons (NE Atlantic). Deep Sea Res. Part II 2011, 58, 2433–2447. [Google Scholar] [CrossRef]
- Louzao, M.; Anadón, N.; Arrontes, J.; Álvarez-Claudio, C.; Fuente, D.C.; Ocharan, F.; Anadón, A.; Acuña, J.L. Historical macrobenthic community assemblages in the Avilés Canyon, N Iberian Shelf: Baseline biodiversity information for a marine protected area. J. Mar. Syst. 2010, 80, 47–56. [Google Scholar] [CrossRef]
- Kaiser, M.J.; Attrill, M.J.; Jennings, S.; Thomas, D.N.; Barnes, D.K.A.; Brierley, A.S.; Polunin, N.V.C.; Raffaelli, D.G.; Williams, P.J.L.B. Marine Ecology: Processes, Systems, and Impacts; Oxford University Press: Oxford, UK, 2005. [Google Scholar]
- Hugoni, M.; Escalas, A.; Bernard, C.; Nicolas, S.; Jézéquel, D.; Vazzoler, F.; Sarazin, G.; Leboulanger, C.; Bouvy, M.; Got, P.; et al. Spatiotemporal variations in microbial diversity across the three domains of life in a tropical thalassohaline lake (Dziani Dzaha, Mayotte Island). Mol. Ecol. 2018, 27, 4775–4786. [Google Scholar] [CrossRef]
- Paul, V.G.; Mormile, M.R. A case for the protection of saline and hypersaline environments: A microbiological perspective. FEMS Microbiol. Ecol. 2017, 93, fix091. [Google Scholar] [CrossRef]
- Ólafsson, E.; Carlström, S.; Ndaro, S.G. Meiobenthos of hypersaline tropical mangrove sediment in relation to spring tide inundation. Hydrobiologia 2000, 426, 57–64. [Google Scholar] [CrossRef]
- Carrasco, N.K.; Perissinotto, R. Development of a halotolerant community in the St. Lucia Estuary (South Africa) during a hypersaline phase. PLoS ONE 2012, 7, e29927. [Google Scholar] [CrossRef]
- Bassler-Veit, B.; Barut, I.F.; Meric, E.; Avsar, N.; Nazik, A.; Kapan-Yesilyurt, S.; Yildiz, A. Distribution of microflora, meiofauna, and macrofauna assemblages in the hypersaline environment of northeastern Aegean Sea coasts. J. Coast. Res. 2013, 29, 883–898. [Google Scholar] [CrossRef]
- Abu-Zied, R.H.; Bantan, R.A. Hypersaline benthic foraminifera from the Shuaiba Lagoon, eastern Red Sea, Saudi Arabia: Their environmental controls and usefulness in sea-level reconstruction. Mar. Micropaleontol. 2013, 103, 51–67. [Google Scholar] [CrossRef]
- Canganella, F.; Bianconi, G.; Kato, C.; Gonzalez, J. Microbial ecology of submerged marine caves and holes characterised by high levels of hydrogen sulphide. Rev. Environ. Sci. Biotechnol. 2007, 6, 61–70. [Google Scholar] [CrossRef]
- Sket, B. The ecology of anchihaline caves. Trends Ecol. Evol. 1996, 11, 221–225. [Google Scholar] [CrossRef]
- Riesch, R.; Plath, M.; Schlupp, I. Toxic hydrogen sulfide and dark caves: Life-history adaptations in a livebearing fish (Poecilia mexicana, Poeciliidae). Ecology 2010, 91, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Bakran-Petricioli, T.; Vacelet, J.; Zibrowius, H.; Petricioli, D.; Chevaldonné, P.; Rada, T. New data on the distribution of the ‘deep-sea’ sponges Asbestopluma hypogea and Oopsacas minuta in the Mediterranean Sea. Mar. Ecol. 2007, 28, 10–23. [Google Scholar] [CrossRef]
- Janssen, A.; Chevaldonné, P.; Martinez-Arbizu, P. The meiobenthic copepod fauna of a marine cave (3PP cave, NW Mediterranean) closely resembles that of deep-sea communities. Mar. Ecol. Prog. Ser. 2013, 479, 99113. [Google Scholar] [CrossRef]
- Harmelin, J.G. Diversity of bryozoans in a Mediterranean sublittoral cave with bathyal-like conditions: Role of dispersal processes and local factors. Mar. Ecol. Prog. Ser. 1997, 153, 139152. [Google Scholar] [CrossRef]
- Grenier, M.; Ruiz, C.; Fourt, M.; Santonja, M.; Dubois, M.; Klautau, M.; Vacelet, J.; Boury-Esnault, N.; Pérez, T. Sponge inventory of the French Mediterranean waters, with an emphasis on cave-dwelling species. Zootaxa 2018, 4466, 205228. [Google Scholar] [CrossRef]
- Rabalais, N.N.; Diaz, R.J.; Levin, L.A.; Turner, R.E.; Gilbert, D.; Zhang, J. Dynamics and distribution of natural and human-caused hypoxia. Biogeosciences 2010, 7, 585–619. [Google Scholar] [CrossRef]
- Helly, J.J.; Levin, L.A. Global distribution of naturally occurring marine hypoxia on continental margins. Deep Sea Res. Part I 2004, 51, 1159–1168. [Google Scholar] [CrossRef]
- Diaz, R.J. Overview of hypoxia around the world. J. Environ. Qual. 2001, 30, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, R.J.; Rosenberg, R. Spreading dead zones and consequences for marine ecosystems. Science 2008, 321, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Sagasti, A.; Schaffner, L.C.; Duffy, J.E. Effects of periodic hypoxia on mortality, feeding and predation in an estuarine epifaunal community. J. Exp. Mar. Biol. Ecol. 2001, 258, 257–283. [Google Scholar] [CrossRef]
- Riedel, B.; Zuschin, M.; Heselmair, A.; Stachowitsch, M. Oxygen depletion under glass: Behavioural responses of benthic macrofauna to induced anoxia in the Northern Adriatic. J. Exp. Mar. Biol. Ecol. 2008, 367, 17–27. [Google Scholar] [CrossRef]
- Nakagawa, S.; Takai, K. Deep-sea vent chemoautotrophs: Diversity, biochemistry and ecological significance. FEMS Microbiol. Ecol. 2008, 65, 1–14. [Google Scholar] [CrossRef]
- Durand, L.; Zbinden, M.; Cueff-Gauchard, V.; Duperron, S.; Roussel, E.G.; Shillito, B.; Cambon-Bonavita, M.A. Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. FEMS Microbiol. Ecol. 2010, 71, 291–303. [Google Scholar] [CrossRef]
- Jan, C.; Petersen, J.M.; Werner, J.; Teeling, H.; Huang, S.; Glöckner, F.O.; Golyshina, O.V.; Dubilier, N.; Golyshin, P.N.; Jebbar, M.; et al. The gill chamber epibiosis of deep-sea shrimp Rimicaris exoculata: An in-depth metagenomic investigation and discovery of Zetaproteobacteria. Environ. Microbiol. 2014, 16, 2723–2738. [Google Scholar] [CrossRef]
- Zbinden, M.I.; Shillito, B.; Le Bris, N.; de Villardi de Montlaur, C.; Roussel, E.; Guyot, F.; Gaill, F.; Cambon-Bonavita, M.A. New insights on the metabolic diversity among the epibiotic microbial community of the hydrothermal shrimp Rimicaris exoculata. J. Exp. Mar. Bio. Ecol. 2008, 359, 131–140. [Google Scholar] [CrossRef]
- Tsuchida, S.; Suzuki, Y.; Fujiwara, Y.; Kawato, M.; Uematsu, K.; Yamanaka, T.; Mizota, C.; Yamamoto, H. Epibiotic association between filamentous bacteria and the vent-associated galatheid crab, Shinkaia crosnieri (Decapoda: Anomura). J. Mar. Biol. Assoc. U. K. 2011, 91, 23–32. [Google Scholar] [CrossRef]
- Ponsard, J.; Cambon-Bonavita, M.A.; Zbinden, M.; Lepoint, G.; Joassin, A.; Corbari, L.; Shillito, B.; Durand, L.; Cueff-Gauchard, V.; Compère, P. Inorganic carbon fixation by chemosynthetic ectosymbionts and nutritional transfers to the hydrothermal vent host-shrimp Rimicaris exoculata. ISME J. 2013, 7, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.J.; Cary, S.C. Characterization of a novel spirochete associated with the hydrothermal vent polychaete annelid, Alvinella pompejana. Appl. Environ. Microbiol. 2001, 67, 110–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cary, S.C.; Cottrell, M.T.; Stein, J.L.; Camacho, F.; Desbruyeres, D. Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent Annelid Alvinella pompejana. Appl. Environ. Microbiol. 1997, 63, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.J.; Jeanthon, C.; Kostka, J.E.; Luther, G.W., 3rd; Cary, S.C. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 2001, 67, 4566–4572. [Google Scholar] [CrossRef] [PubMed]
- Haddad, A.; Camacho, F.; Durand, P.; Cary, S.C. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana. Appl. Environ. Microbiol. 1995, 61, 1679–1687. [Google Scholar] [CrossRef]
- Gaill, F.; Desbruyeres, D.; Laubier, L. Relationships between the “Pompeii worms” and their epibiotic bacteria. Oceanol. Acta 1988, 8, 147–155. [Google Scholar]
- Grzymski, J.J.; Murray, A.E.; Campbell, B.J.; Kaplarevic, M.; Gao, G.R.; Lee, C.; Daniel, R.; Ghadiri, A.; Feldman, R.A.; Cary, S.C. Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. Proc. Natl. Acad. Sci. USA 2008, 105, 17516–17521. [Google Scholar] [CrossRef]
- Le Bris, N.; Gaill, F. How does the annelid Alvinella pompejana deal with an extreme hydrothermal environment? Rev. Environ. Sci. Biotechnol. 2007, 6, 197–221. [Google Scholar] [CrossRef]
- Tasiemski, A.; Jung, S.; Boidin-Wichlacz, C.; Jollivet, D.; Cuvillier-Hot, V.; Pradillon, F.; Vetriani, C.; Hecht, O.; Sönnichsen, F.D.; Gelhaus, C.; et al. Characterization and function of the first antibiotic isolated from a vent organism: The extremophile metazoan Alvinella pompejana. PLoS ONE 2014, 9, e95737. [Google Scholar] [CrossRef]
- Minic, Z.; Hervé, G. Biochemical and enzymological aspects of the symbiosis between the deep-sea tubeworm Riftia pachyptila and its bacterial endosymbiont. Eur. J. Biochem. 2004, 271, 3093–3102. [Google Scholar] [CrossRef]
- Polzin, J.; Arevalo, P.; Nussbaumer, T.; Polz, M.F.; Bright, M. Polyclonal symbiont populations in hydrothermal vent tubeworms and the environment. Proc. R. Soc. B 2019, 286, 20181281. [Google Scholar] [CrossRef] [PubMed]
- Newton, I.L.G.; Woyke, T.; Auchtung, T.A.; Dilly, G.F.; Dutton, R.J.; Fisher, M.C.; Fontanez, K.M.; Lau, E.; Stewart, F.J.; Richardson, P.M.; et al. The Calyptogena magnifica chemoautotrophic symbiont genome. Science 2007, 315, 998–1000. [Google Scholar] [CrossRef] [PubMed]
- Katz, S.; Cavanaugh, C.M.; Bright, M. Symbiosis of epi- and endocuticular bacteria with Helicoradomenia spp. (Mollusca, Aplacophora, Solenogastres) from deep-sea hydrothermal vents. Mar. Ecol. Progr. Ser. 2006, 320, 89–99. [Google Scholar] [CrossRef] [Green Version]
- Duperron, S.; Bergin, C.; Zielinski, F.; Blazejak, A.; Pernthaler, A.; McKiness, Z.P.; DeChaine, E.; Cavanaugh, C.M.; Dubilier, N. A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ. Microbiol. 2006, 8, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, M.; Lindsay, D.J.; Hata, J.; Nakamura, A.; Kasai, H.; Ise, Y.; Fisher, C.R.; Fujiwara, Y.; Kawato, M.; Maruyama, T. Association of thioautotrophic bacteria with deep-sea sponges. Mar. Biotechnol. 2010, 12, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Thiel, V.; Hügler, M.; Blümel, M.; Baumann, H.I.; Gärtner, A.; Schmaljohann, R.; Strauss, H.; Garbe-Schönberg, D.; Petersen, S.; Cowart, D.A.; et al. Widespread occurrence of two carbon fixation pathways in tubeworm endosymbionts: Lessons from hydrothermal vent associated tubeworms from the Mediterranean Sea. Front. Microbiol. 2012, 3, 423. [Google Scholar] [CrossRef] [PubMed]
- Goffredi, S.K.; Warén, A.; Orphan, V.J.; Van Dover, C.L.; Vrijenhoek, R.C. Novel forms of structural integration between microbes and a hydrothermal vent gastropod from the Indian Ocean. Appl. Environ. Microbiol. 2004, 70, 3082–3090. [Google Scholar] [CrossRef]
- Vacelet, J.; Boury-Esnault, N.; Fiala-Medioni, A.; Fisher, C.R. A methanotrophic carnivorous sponge. Nature 1995, 377, 296. [Google Scholar] [CrossRef]
- Vacelet, J.; Boury-Esnault, N. A new species of carnivorous deep-sea sponge (Demospongiae, Cladorhizidae) associated with methanotrophic bacteria. Cah. Biol. Mar. 2002, 43, 141–148. [Google Scholar]
- Medina-Silva, R.; Oliveira, R.R.; Trindade, F.J.; Borges, L.G.A.; Lopes Simão, T.L.; Augustin, A.H.; Valdez, F.P.; Constant, M.J.; Simundi, C.L.; Eizirik, E.; et al. Microbiota associated with tubes of Escarpia sp. from cold seeps in the southwestern Atlantic Ocean constitutes a community distinct from that of surrounding marine sediment and water. Antonie Van Leeuwenhoek 2018, 111, 533–550. [Google Scholar] [CrossRef]
- Mohamed, N.M.; Saito, K.; Tal, Y.; Hill, R.T. Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J. 2010, 4, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Lòpez-Garcìa, P.; Gaill, F.; Moreira, D. Wide bacterial diversity associated with tubes of the vent worm Riftia pachyptila. Environ. Microbiol. 2002, 4, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, J.; Sun, Y.; Kwan, Y.H.; Wong, W.C.; Zhang, Y.; Xu, T.; Feng, D.; Zhang, Y.; Qiu, J.W.; et al. Genomic, transcriptomic, and proteomic insights into the symbiosis of deep-sea tubeworm holobionts. ISME J. 2020, 14, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Sahling, H.; Borowski, C.; Escobar-Briones, E.; Gaytán-Caballero, A.; Hsu, C.-W.; Loher, M.; MacDonald, I.; Marcon, Y.; Pape, T.; Römer, M.; et al. Massive asphalt deposits, oil seepage, and gas venting support abundant chemosynthetic communities at the Campeche Knolls, southern Gulf of Mexico. Biogeosciences 2016, 13, 4491–4512. [Google Scholar] [CrossRef] [Green Version]
- Dando, P.R.; Hughes, J.A.; Thiermann, F. Preliminary observations on biological communities at shallow hydrothermal vents in the Aegean Sea. In Hydrothermal Vents and Processes; Parson, L.M., Walker, C.L., Dixon, D.R., Eds.; Special Publications: London, UK, 1995; Volume 87, pp. 303–317. [Google Scholar]
- Thiermann, F.; Akoumianaki, I.; Hugjes, A.J.; Giere, O. Benthic fauna of a shallow-water gaseohydrothermal vent area in the Aegean Sea (Milos, Greece). Mar. Biol. 1997, 128, 149–159. [Google Scholar] [CrossRef]
- Zeppilli, D.; Danovaro, R. Meiofaunal diversity and assemblage structure in a shallow-water hydrothermal vent in the Pacific Ocean. Aquat. Biol. 2009, 5, 75–84. [Google Scholar] [CrossRef]
- Bellec, L.; Cambon-Bonavita, M.-A.; Durand, L.; Aube, J.; Gayet, N.; Sandulli, R.; Brandily, C.; Zeppilli, D. Microbial communities of the shallow-water hydrothermal vent near Naples, Italy, and chemosynthetic symbionts associated with a free-living marine nematode. Front. Microbiol. 2020, 11, 2023. [Google Scholar] [CrossRef]
- Waite, D.W.; Vanwonterghem, I.; Rinke, C.; Parks, D.H.; Zhang, Y.; Takai, K.; Sievert, S.M.; Simon, J.; Campbell, B.J.; Hanson, T.E.; et al. Comparative genomic analysis of the class epsilonproteobacteria and proposed reclassification to epsilonbacteraeota (phyl. nov.). Front. Microbiol. 2017, 8, 682. [Google Scholar] [CrossRef]
- Yang, S.-H.; Chiang, P.-W.; Hsu, T.-C.; Kao, S.-J.; Tang, S.-L. Bacterial community associated with organs of shallow hydrothermal vent crab Xenograpsus testudinatus near Kuishan Island, Taiwan. PLoS ONE 2016, 11, e0150597. [Google Scholar] [CrossRef]
- Kuo, F.W. Preliminary Investigation of the Hydrothermal Activities of Kueishantao Island. Ph.D. Thesis, National Sun Yat-Sen University, Kaohsiung, Taiwan, 2001. [Google Scholar]
- Hu, M.Y.A.; Hagen, W.; Jeng, M.S.; Saborowski, E. Metabolic energy demand and food utilization of the hydrothermal vent crab Xenograpsus testudinatus (Crustacea: Brachyura). Aquat. Biol. 2012, 15, 11–25. [Google Scholar] [CrossRef]
- Chen, C.T.A.; Zeng, Z.; Kuo, F.W.; Yang, T.F.; Wang, B.J.; Tu, Y.Y. Tide-influenced acidic hydrothermal system offshore NE Taiwan. Chem. Geol. 2005, 224, 69–81. [Google Scholar] [CrossRef]
- Coelho, F.J.R.C.; Cleary, D.F.R.; Gomes, N.C.M.; Pólonia, A.R.M.; Huang, Y.M.; Liu, L.-L.; de Voogd, N.J. Sponge prokaryote communities in Taiwanese Coral Reef and shallow hydrothermal vent ecosystems. Microb. Ecol. 2018, 75, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Eythorsdottir, A.; Omarsdottir, S.; Einarsson, H. Antimicrobial activity of marine bacterial symbionts retrieved from shallow water hydrothermal vents. Mar. Biotechnol. 2016, 18, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.; Fine, M.; Ritchie, K. The stable microbiome of inter and sub-tidal anemone species under increasing pCO2. Sci. Rep. 2016, 6, 37387. [Google Scholar] [CrossRef] [Green Version]
- Meron, D.; Buia, M.C.; Fine, M.; Banin, E. Changes in microbial communities associated with the sea anemone Anemonia viridis in a natural pH gradient. Microb. Ecol. 2013, 65, 269–276. [Google Scholar] [CrossRef]
- Biagi, E.; Caroselli, E.; Barone, M.; Pezzimenti, M.; Teixido, N.; Soverini, M.; Rampelli, S.; Turroni, S.; Gambi, M.C.; Brigidi, P.; et al. Patterns in microbiome composition differ with ocean acidification in anatomic compartments of the Mediterranean coral Astroides calycularis living at CO2 vents. Sci. Total Environ. 2020, 724, 138048. [Google Scholar] [CrossRef]
- Webster, N.S.; Negri, A.P.; Munro, M.M.H.G.; Battershill, C.N. Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 2004, 6, 288–300. [Google Scholar] [CrossRef]
- Rodríguez-Marconi, S.; De la Iglesia, R.; Díez, B.; Fonseca, C.A.; Hajdu, E.; Trefault, N. Characterization of bacterial, archaeal and eukaryote symbionts from Antarctic sponges reveals a high diversity at a three-domain level and a particular signature for this ecosystem. PLoS ONE 2015, 10, e0138837. [Google Scholar] [CrossRef]
- Steinert, G.; Wemheuer, B.; Janussen, D.; Erpenbeck, D.; Daniel, R.; Simon, M.; Brinkhoff, T.; Schupp, P.J. Prokaryotic diversity and community patterns in Antarctic continental shelf sponges. Front. Mar. Sci. 2019, 6, 297. [Google Scholar] [CrossRef]
- Cárdenas, C.A.; González-Aravena, M.; Font, A.; Hestetun, J.T.; Hajdu, E.; Trefault, N.; Malmberg, M.; Bongcam-Rudloff, E. High similarity in the microbiota of cold-water sponges of the genus Mycale from two different geographical areas. PeerJ 2018, 6, e4935. [Google Scholar] [CrossRef]
- Cárdenas, C.A.; Font, A.; Steinert, G.; Rondon, R.; González-Aravena, M. Temporal stability of bacterial communities in Antarctic sponges. Front. Microbiol. 2019, 10, 2699. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pino, M.; Cristi, A.; Gillooly, J.F.; Trefault, N. Characterizing the microbiomes of Antarctic sponges: A functional metagenomic approach. Sci. Rep. 2020, 10, 645. [Google Scholar] [CrossRef] [PubMed]
- Papale, M.; Rizzo, C.; Fani, R.; Bertolino, M.; Costa, G.; Paytuví-Gallart, A.; Schiaparelli, S.; Michaud, L.; Azzaro, M.; Lo Giudice, A. Exploring the Diversity and Metabolic Profiles of Bacterial Communities Associated With Antarctic Sponges (Terra Nova Bay, Ross Sea). Front. Ecol. Evol. 2020, 8, 268. [Google Scholar] [CrossRef]
- Sacristán-Soriano, O.; Pérez Criado, N.; Avila, C. Host species determines symbiotic community composition in Antarctic sponges (Porifera: Demospongiae). Front. Mar. Sci. 2020, 7, 474. [Google Scholar] [CrossRef]
- Happel, L.; Rondon, R.; Font, A.; González-Aravena, M.; Cárdenas, C.A. Stability of the microbiome of the sponge Mycale (Oxymycale) acerata in the Western Antarctic Peninsula. Front. Microbiol. 2022, 13, 827863. [Google Scholar] [CrossRef]
- Moreno-Pino, M.; Ugalde, J.A.; Valdés, J.H.; Rodríguez-Marconi, S.; Parada-Pozo, G.; Trefault, N. Bacteria isolated from the Antarctic sponge Iophon sp. reveals mechanisms of symbiosis in Sporosarcina, Cellulophaga, and Nesterenkonia. Front. Microbiol. 2021, 12, 660779. [Google Scholar] [CrossRef]
- Cristi, A.; Parada-Pozo, G.; Morales-Vicencio, F.; Cárdenas, C.A.; Trefault, N. Variability in host specificity and functional potential of Antarctic sponge-associated bacterial communities. Front. Microbiol. 2022, 12, 771589. [Google Scholar] [CrossRef]
- Ruocco, N.; Esposito, R.; Bertolino, M.; Zazo, G.; Sonnessa, M.; Andreani, F.; Coppola, D.; Giordano, D.; Nuzzo, G.; Lauritano, C.; et al. A metataxonomic approach reveals diversified bacterial communities in antarctic sponges. Mar. Drugs 2021, 19, 173. [Google Scholar] [CrossRef]
- Papaleo, M.C.; Fondi, M.; Maida, I.; Perrin, E.; Giudice, A.L.; Michaud, L.; Mangano, S.; Bartolucci, G.; Romoli, R.; Fani, R. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol. Adv. 2012, 30, 272–293. [Google Scholar] [CrossRef]
- Mangano, S.; Michaud, L.; Caruso, C.; Brilli, M.; Bruni, V.; Fani, R.; Giudice, A.L. Antagonistic interactions between psychrotrophic cultivable bacteria isolated from Antarctic sponges: A preliminary analysis. Res. Microbiol. 2009, 160, 27–37. [Google Scholar] [CrossRef]
- Mangano, S.; Michaud, L.; Caruso, C.; Lo Giudice, A. Metal and antibiotic-resistance in psychrotrophic bacteria associated with the Antarctic sponge Hemigellius pilosus (Kirkpatrick, 1907). Polar Biol. 2014, 37, 227–235. [Google Scholar] [CrossRef]
- Xin, Y.; Kanagasabhapathy, M.; Janussen, D.; Xue, S.; Zhang, W. Phylogenetic diversity of Gram-positive bacteria cultured from Antarctic deep-sea sponges. Polar Biol. 2011, 34, 1501–1512. [Google Scholar] [CrossRef]
- Savoca, S.; Lo Giudice, A.; Papale, M.; Mangano, S.; Caruso, C.; Spanò, N.; Michaud, L.; Rizzo, C. Antarctic sponges from the Terra Nova Bay (Ross Sea) host a diversified bacterial community. Sci. Rep. 2019, 9, 16135. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Bourne, D. Bacterial community structure associated with the Antarctic soft coral, Alcyonium antarcticum. FEMS Microbiol. Ecol. 2007, 59, 81–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, A.E.; Rack, F.R.; Zook, R.; Williams, M.J.M.; Higham, M.L.; Broe, M.; Kaufmann, R.S.; Daly, M. Microbiome composition and diversity of the ice-dwelling sea anemone, Edwardsiella andrillae. Integr. Comp. Biol. 2016, 56, 542–555. [Google Scholar] [CrossRef]
- González-Aravena, M.; Urtubia, R.; Del Campo, K.; Lavín, P.; Wong, C.M.V.L.; Cárdenas, C.A.; González-Rocha, G. Antibiotic and metal resistance of cultivable bacteria in the Antarctic sea urchin. Antarct. Sci. 2016, 28, 261–268. [Google Scholar] [CrossRef]
- Schwob, G.; Cabrol, L.; Poulin, E.; Orlando, J. Characterization of the gut microbiota of the Antarctic heart urchin (Spatangoida) Abatus agassizii. Front. Microbiol. 2020, 11, 308. [Google Scholar] [CrossRef] [PubMed]
- Herrera, L.M.; García-Laviña, C.X.; Marizcurrena, J.J.; Volonterio, O.; Ponce de León, R.; Castro-Sowinski, S. Hydrolytic enzyme-producing microbes in the Antarctic oligochaete Grania sp. (Annelida). Polar Biol. 2017, 40, 947–953. [Google Scholar] [CrossRef]
- Riesenfeld, C.S.; Murray, A.E.; Baker, B.J. Characterization of the microbial community and polyketide biosynthetic potential in the palmerolide-producing tunicate Synoicum adareanum. J. Nat. Prod. 2008, 71, 1812–1818. [Google Scholar] [CrossRef]
- Murray, A.E.; Avalon, N.E.; Bishop, L.; Davenport, K.W.; Delage, E.; Dichosa, A.E.K.; Eveillard, D.; Higham, M.L.; Kokkaliari, S.; Lo, C.C.; et al. Uncovering the core microbiome and distribution of palmerolide in Synoicum adareanum across the Anvers Island Archipelago, Antarctica. Mar. Drugs 2020, 18, 298. [Google Scholar] [CrossRef]
- Verhoeven, J.T.P.; Dufour, S.C. Microbiomes of the Arctic carnivorous sponges Chondrocladia grandis and Cladorhiza oxeata suggest a specific, but differential involvement of bacterial associates. Arct. Sci. 2017, 4, 186–204. [Google Scholar] [CrossRef]
- Gloeckner, V.; Wehrl, M.; Moitinho-Silva, L.; Gernert, C.; Schupp, P.; Pawlik, J.R.; Lindquist, N.L.; Erpenbeck, D.; Wörheide, G.; Hentschel, U. The HMA-LMA dichotomy revisited: An electron microscopical survey of 56 sponge species. Biol. Bull. 2014, 227, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Morganti, T.M.; Slaby, B.M.; de Kluijver, A.; Busch, K.; Hentschel, U.; Middelburg, J.J.; Grotheer, H.; Mollenhauer, G.; Dannheim, J.; Rapp, H.T.; et al. Giant sponge grounds of Central Arctic seamounts are associated with extinct seep life. Nat. Commun. 2022, 13, 638. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, S.; Jóhannsson, R.; Marteinsson, V. Bacterial diversity in the marine sponge Halichondria panicea from Icelandic waters and host-specificity of its dominant symbiont “Candidatus Halichondribacter symbioticus”. FEMS Microbiol. Ecol. 2019, 95, fiy220. [Google Scholar] [CrossRef] [PubMed]
- Borchert, E.; Jackson, S.A.; O’Gara, F.; Dobson, A.D.W. Diversity of natural product biosynthetic genes in the microbiome of the deep sea sponges Inflatella pellicula, Poecillastra compressa, and Stelletta normani. Front. Microbiol. 2016, 7, 1027. [Google Scholar] [CrossRef]
- Brück, W.M.; Sennett, S.H.; Pomponi, S.A.; Willenz, P.; McCarthy, P.J. Identification of the bacterial symbiont Entotheonella sp. in the mesohyl of the marine sponge Discodermia sp. ISME J. 2008, 2, 335–339. [Google Scholar] [CrossRef]
- Brück, W.M.; Brück, T.B.; Self, W.T.; Reed, J.K.; Nitecki, S.S.; Mc Carthy, P.J. Comparison of the anaerobic microbiota of deep-water Geodia spp. and sandy sediments in the Straits of Florida. ISME J. 2010, 4, 686–699. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Uchino, M.; Falsen, E.; Frolova, G.M.; Zhukova, N.V.; Mikhailov, V.V. Pseudomonas pachastrellae sp. nov., isolated from a marine sponge. Int. J. Syst. Evol. Microbiol. 2005, 55, 919–924. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Uchino, M.; Tanaka, N.; Frolova, G.M.; Mikhailov, V.V. Lysobacter spongiicola sp. nov., isolated from a deep-sea sponge. Int. J. Syst. Evol. Microbiol. 2008, 58, 370–374. [Google Scholar] [CrossRef]
- Jackson, S.A.; Flemer, B.; McCann, A.; Kennedy, J.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D. Archaea appear to dominate the microbiome of Inflatella pellicula deep sea sponges. PLoS ONE 2013, 8, e84438. [Google Scholar] [CrossRef]
- Li, Z.Y.; Wang, Y.Z.; He, L.M.; Zheng, H.J. Metabolic profiles of prokaryotic and eukaryotic communities in deep-sea sponge Neamphius huxleyi indicated by metagenomics. Sci. Rep. 2014, 4, 3895. [Google Scholar] [CrossRef] [PubMed]
- Penn, K.; Wu, D.; Eisen, J.A.; Ward, N. Characterization of bacterial communities associated with deep-sea corals on Gulf of Alaska Seamounts. Appl. Environ. Microbiol. 2006, 72, 1680–1683. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Cappello, S.; Crisafi, E.; Tursi, A.; Savini, A.; Corselli, C.; Scarfi, S.; Giuliano, L. Phylogenetic survey of metabolically active microbial communities associated with the deep-sea coral Lophelia pertusa from the Apulian plateau, Central Mediterranean Sea. Deep Sea Res. Part I 2006, 53, 62–75. [Google Scholar] [CrossRef]
- Neulinger, S.C.; Jarnegren, J.; Ludvigsen, M.; Lochte, K.; Dullo, W.-C. Phenotype-specific bacterial communities in the cold-water coral Lophelia pertusa (Scleractinia) and their implications for the coral’s nutrition, health, and distribution. Appl. Environ. Microbiol. 2008, 74, 7272–7285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellogg, C.A.; Lisle, J.T.; Galkiewicz, J.P. Culture-independent characterization of bacterial communities associated with the cold-water coral Lophelia pertusa in the northeastern Gulf of Mexico. Appl. Environ. Microbiol. 2009, 75, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Schottner, S.; Hoffmann, F.; Wild, C.; Rapp, H.T.; Boetius, A.; Ramette, A. Inter- and intra-habitat bacterial diversity associated with cold-water corals. ISME J. 2009, 3, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Galkiewicz, J.P.; Pratte, Z.A.; Gray, M.A.; Kellogg, C.A. Characterization of culturable bacteria isolated from the cold-water coral Lophelia pertusa. FEMS Microbiol. Ecol. 2011, 77, 333–346. [Google Scholar] [CrossRef]
- Hansson, L.; Agis, M.; Maier, C.; Weinbauer, M.G. Community composition of bacteria associated with cold-water coral Madrepora oculata: Within and between colony variability. Mar. Ecol. Prog. Ser. 2009, 397, 89–102. [Google Scholar] [CrossRef]
- Gray, M.A.; Stone, R.P.; McLaughlin, M.R.; Kellogg, C.A. Microbial consortia of gorgonian corals from the Aleutian Islands. FEMS Microbiol. Ecol. 2009, 76, 109–120. [Google Scholar] [CrossRef]
- Lawler, S.N.; Kellogg, C.A.; France, S.C.; Clostio, R.W.; Brooke, S.D.; Ross, S.W. Coral-associated bacterial diversity is conserved across two deep-sea Anthothela species. Front. Microbiol. 2016, 7, 458. [Google Scholar] [CrossRef]
- Vortsepnev, E.; Chevaldonné, P.; Klyukina, A.; Naduvaeva1, E.; Todt, C.; Zhadan, A.; Tzetlin, A.; Kublanov, I. Microbial associations of shallow-water Mediterranean marine cave Solenogastres (Mollusca). PeerJ 2021, 9, e12655. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M.; Roeser-Mueller, K.; Koehler, S.; Peterson, A.; Nechitaylo, T.Y.; Stubblefield, J.W.; Herzner, G.; Seger, J.; Strohm, E. Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc. Natl. Acad. Sci. USA 2014, 111, 6359–6364. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Schmidt, E.W. Parallel lives of symbionts and hosts: Chemical mutualism in marine animals. Nat. Prod. Rep. 2018, 35, 357. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Xu, D.; Han, L.; Li, C.; Cao, Q.; Zhu, D.; Barrett, N.H.; Harmody, D.; Chen, J.; Zhu, H.; McCarthy, P.J.; et al. Bioprospecting deep-sea Actinobacteria for novel anti-infective natural products. Front. Microbiol. 2018, 9, 787. [Google Scholar] [CrossRef] [Green Version]
- Rayan, A.; Raiyn, J.; Falah, M. Nature is the best source of anticancer drugs: Indexing natural products for their anticancer bioactivity. PLoS ONE 2017, 12, e0187925. [Google Scholar] [CrossRef]
- Helfrich, E.J.; Reiter, S.; Piel, J. Recent advances in genome-based polyketide discovery. Curr. Opin. Biotechnol. 2014, 29, 107–115. [Google Scholar] [CrossRef]
- Borchert, E.; Knobloch, S.; Dwyer, E.; Flynn, S.; Jackson, S.A.; Jóhannsson, R.; Marteinsson, V.T.; O’Gara, F.; Dobson, A.D.W. Biotechnological potential of cold adapted Pseudoalteromonas spp. isolated from ‘Deep Sea’ sponges. Mar. Drugs 2017, 15, 184. [Google Scholar] [CrossRef]
- Murray, A.E.; Lo, C.C.; Daligault, H.E.; Avalon, N.E.; Read, R.W.; Davenport, K.W.; Higham, M.L.; Kunde, Y.; Dichosa, A.E.K.; Baker, B.J.; et al. Discovery of an Antarctic ascidian-associated uncultivated Verrucomicrobia with antimelanoma palmerolide biosynthetic potential. mSphere 2021, 6, e0075921. [Google Scholar] [CrossRef]
- Modolon, F.; Barno, A.; Villela, H.; Peixoto, R. Ecological and biotechnological importance of secondary metabolites produced by coral-associated bacteria. J. Appl. Microbiol. 2020, 129, 1441–1457. [Google Scholar] [CrossRef]
- Back, C.R.; Stennett, H.L.; Williams, S.E.; Wang, L.; Ojeda Gomez, J.; Abdulle, O.M.; Duffy, T.; Neal, C.; Mantell, J.; Jepson, M.A.; et al. A new Micromonospora strain with antibiotic activity isolated from the microbiome of a Mid-Atlantic deep-sea sponge. Mar. Drugs 2021, 19, 105. [Google Scholar] [CrossRef] [PubMed]
- Jayatilake, G.S.; Thornton, M.P.; Leonard, A.C.; Grimwade, J.E.; Baker, B.J. Metabolites from an Antarctic sponge-associated bacterium, Pseudomonas aeruginosa. J. Nat. Prod. 1996, 59, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Romoli, R.; Papaleo, M.C.; de Pascale, D.; Tutino, M.L.; Michaud, L.; Lo Giudice, A.; Fani, R.; Bartolucci, G. Characterization of the volatile profile of Antarctic bacteria by using solid-phase microextraction–gas chromatography–mass spectrometry. J. Mass Spectr. 2011, 46, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, M.C.; Romoli, R.; Bartolucci, G.; Maida, I.; Perrin, E.; Fondi, M.; Orlandini, V.; Mengoni, A.; Emiliani, G.; Tutino, M.L.; et al. Bioactive volatile organic compounds from Antarctic (sponges) bacteria. New Biotechnol. 2013, 30, 824–838. [Google Scholar] [CrossRef]
- Maida, I.; Bosi, E.; Fondi, M.; Perrin, E.; Orlandini, V.; Papaleo, M.C.; Mengoni, A.; de Pascale, D.; Tutino, M.L.; Michaud, L.; et al. Antimicrobial activity of Pseudoalteromonas strains isolated from the Ross Sea (Antarctica) vs. Cystic Fibrosis opportunistic pathogens. Hydrobiologia 2015, 761, 443–457. [Google Scholar] [CrossRef] [Green Version]
- Bosi, E.; Fondi, M.; Maida, I.; Perrin, E.; de Pascale, D.; Tutino, M.L.; Parrilli, E.; Lo Giudice, A.; Filloux, A.; Fani, R. Genome-scale phylogenetic and DNA composition analyses of Antarctic Pseudoalteromonas bacteria reveal inconsistencies in current taxonomic affiliation. Hydrobiologia 2015, 761, 85–95. [Google Scholar] [CrossRef]
- Orlandini, V.; Maida, I.; Fondi, M.; Perrin, E.; Papaleo, M.C.; Bosi, E.; de Pascale, D.; Tutino, M.L.; Michaud, L.; Lo Giudice, A.; et al. Genomic analysis of three sponge-associated Arthrobacter Antarctic strains, inhibiting the growth of Burkholderia cepacia complex bacteria by synthesizing volatile organic compounds. Microbiol. Res. 2014, 169, 593–601. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; De Pedro, N.; De la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K, a new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral Lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Vizcaíno, A.; González, V.; Braña, A.F.; Palacios, J.J.; Otero, L.; Fernández, J.; Molina, A.; Kulik, A.; Vázquez, F.; Acuña, J.L.; et al. Pharmacological potential of phylogenetically diverse Actinobacteria isolated from deep-sea coral ecosystems of the submarine Avilés Canyon in the Cantabrian Sea. Microb. Ecol. 2017, 73, 338–352. [Google Scholar] [CrossRef]
- Sarmiento Vizcaíno, A.; González Iglesias, V.; Fernández Braña, A.J.; Molina Ramírez, A.; Acuña Fernández, J.L.; García Díaz, L.A.; Blanco Blanco, M.G. Myceligenerans cantabricum sp. nov., a barotolerant actinobacterium isolated from a deep cold-water coral. Int. J. Syst. Evol. Microbiol. 2015, 65, 1328–1334. [Google Scholar] [CrossRef]
- Sarmiento-Vizcaíno, A.; Braña, A.F.; González, V.; Nava, H.; Molina, H.; Llera, E.; Fiedler, H.-P.; Rico, J.M.; García-Flórez, L.; Acuña, J.L.; et al. Atmospheric dispersal of bioactive Streptomyces albidoflavus strains among terrestrial and marine environments. Microb. Ecol. 2016, 71, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Vizcaíno, A.; Braña, A.F.; Pérez-Victoria, I.; Martín, J.; De Pedro, N.; Cruz, M.D.L.; Díaz, C.; Vicente, F.; Acuña, J.L.; Reyes, F.; et al. Paulomycin G, a new natural product with cytotoxic activity against tumor cell lines produced by deep-sea sediment derived Micromonospora matsumotoense M-412 from the Avilés Canyon in the Cantabrian Sea. Mar. Drugs 2017, 15, 271. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, E.; Yan, L.; Boyd, K.G.; Wright, P.-C.; Burgess, J.G. The symbiotic role of marine microbes on living surfaces. Hydrobiologia 2001, 461, 37–40. [Google Scholar] [CrossRef]
- Qian, P.Y.; Cheng, A.; Wang, R.; Zhang, R. Marine biofilms: Diversity, interactions and biofouling. Nat. Rev. Microbiol. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Little, B.J.; Lee, J.S.; Ray, R.I. The influence of marine biofilms on corrosion: A concise review. Electrochim. Acta 2008, 54, 2–7. [Google Scholar] [CrossRef]
- Slattery, M.; McClintock, J.B.; Heine, J.N. Chemical defenses in Antarctic soft corals: Evidence for antifouling compounds. J. Exp. Mar. Biol. Ecol. 1995, 190, 61–77. [Google Scholar] [CrossRef]
- Angulo-Preckler, C.; Cid, C.; Oliva, F.; Avila, C. Antifouling activity in some benthic Antarctic invertebrates by “in situ” experiments at Deception Island, Antarctica. Mar. Environ. Res. 2015, 105, 30–38. [Google Scholar] [CrossRef]
- Furrow, F.B.; Amsler, C.D.; McClintock, J.B.; Baker, B.J. Surface sequestration of chemical feeding deterrents in the Antarctic sponge Latrunculia apicalis as an optimal defense against sea star spongivory. Mar. Biol. 2003, 143, 443–449. [Google Scholar] [CrossRef]
- Hoiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.O. The clinical impact of bacterial biofilms. Int. J. Oral. Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef]
- Chen, J.; Rossman, M.L.; Pawar, D.M. Attachment of enterohemorrhagic Escherichia coli to the surface of beef and a culture medium. LWT—Food Sci. Technol. 2007, 40, 249–254. [Google Scholar] [CrossRef]
- Melo, W.; Perussi, J.R. Strategies to overcome biofilm resistance. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2013; Volume 1, pp. 179–187. [Google Scholar]
- Rizzo, C.; Zammuto, V.; Lo Giudice, A.; Rizzo, M.G.; Spanò, A.; Laganà, P.; Martinez, M.; Guglielmino, S.; Gugliandolo, C. Antibiofilm activity of Antarctic sponge-associated bacteria against Pseudomonas aeruginosa and Staphylococcus aureus. J. Mar. Sci. Eng. 2021, 9, 243. [Google Scholar] [CrossRef]
- Bassler, B.L.; Losick, R. Bacterially speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Mangano, S.; Caruso, C.; Michaud, L.; Lo Giudice, A. First evidence of quorum sensing activity in bacteria associated with Antarctic sponges. Polar Biol. 2018, 41, 1435–1445. [Google Scholar] [CrossRef]
- Decho, A.W.; Gutierrez, T. Microbial extracellular polymeric substances (EPSs) in ocean systems. Front. Microbiol. 2017, 8, 922. [Google Scholar] [CrossRef] [PubMed]
- Carillo, S.; Casillo, A.; Pieretti, G.; Parrilli, E.; Sannino, F.; Bayer-Giraldi, M.; Cosconati, S.; Novellino, E.; Ewert, M.; Deming, J.W.; et al. A unique capsular polysaccharide structure from the psychrophilic marine bacterium Colwellia psychrerythraea 34H that mimics antifreeze (glyco)proteins. J. Am. Chem. Soc. 2015, 137, 179–189. [Google Scholar] [CrossRef]
- Carrión, O.; Delgado, L.; Mercade, E. New emulsifying and cryoprotective exopolysaccharide from Antarctic Pseudomonas sp. ID1. Carbohydr. Polym. 2015, 117, 1028–1034. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, B.-G.; Park, H.J.; Yim, J.H. Cryoprotective Properties and Preliminary Characterization of Exopolysaccharide (P-ArcPo 15) Produced by the Arctic bacterium Pseudoalteromonas elyakovii ArcPo 15. Prep. Biochem. Biotechnol. 2016, 46, 261–266. [Google Scholar] [CrossRef]
- Kim, S.J.; Yim, J.H. Cryoprotective properties of exopolysaccharide (P-21653) produced by the Antarctic bacterium, Pseudoalteromonas arctica KOPRI 21653. J. Microbiol. 2007, 45, 510–514. [Google Scholar]
- Cambon-Bonavita, M.-A.; Raguénès, G.; Jean, J.; Vincent, P.; Guezennec, J. A novel polymer produced by a bacterium isolated from a deep-sea hydrothermal vent polychaete annelid. J. Appl. Microbiol. 2002, 93, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Lelchat, F.; Cozien, J.; Le Costaouec, T.; Brandilly, C.; Schmitt, S.; Baudoux, A.C.; Colliec-Jouault, S.; Boisset, C. Exopolysaccharide biosynthesis and biodegradation by a marine hydrothermal Alteromonas sp. strain. Appl. Microbiol. Biotechnol. 2015, 99, 2637–2647. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, A.; Takeoka, A. The applications and functions of new exopolysaccharide “Deepsane” from the deepest oceans. Fragr. J. 2006, 34, 61–68. [Google Scholar]
- Dubreucq, G.; Domon, B.; Fournet, B. Structure determination of a novel uronic acid residue isolated from the exopolysaccharide produced by a bacterium originating from deep sea hydrothermal vents. Carbohyd. Res. 1996, 290, 175–181. [Google Scholar] [CrossRef]
- Vincent, P.; Pignet, P.; Talmont, F.; Bozzi, L.; Fournet, B.; Guezennec, J.; Jeanthon, C.; Prieur, D. Production and characterization of an exopolysaccharide excreted by a deep-sea hydrothermal vent bacterium isolated from the polychaete Annelid Alvinella pompejana. Appl. Environ. Microbiol. 1994, 60, 4134–4141. [Google Scholar] [CrossRef] [PubMed]
- Raguénès, G.; Christen, R.; Guezennec, J.G.; Pignet, P.; Barbier, G. Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Bacteriol. 1997, 47, 989–995. [Google Scholar] [CrossRef]
- Rougeaux, H.; Pichon, R.; Kervarec, N.; Raguénès, G.H.C.; Guezennec, J.G. Novel bacterial exopolysaccharides from deep-sea hydrothermal vents. Carbohyd. Polym. 1996, 31, 237–242. [Google Scholar] [CrossRef]
- Rougeaux, H.; Kervarec, N.; Pichon, R.; Guezennec, J. Structure of the exopolysaccharide of Vibrio diabolicus isolated from a deep-sea hydrothermal vent. Carbohyd. Res. 1999, 322, 40–45. [Google Scholar] [CrossRef]
- Senni, K.; Gueniche, F.; Changotade, S.; Septier, D.; Sinquin, C.; Ratiskol, J.; Lutomski, D.; Godeau, G.; Guezennec, J.; Colliec-Jouault, S. Unusual glycosaminoglycans from a deep sea hydrothermal bacterium improve fibrillar collagen structuring and fibroblast activities in engineered connective tissues. Mar. Drugs 2013, 11, 1351–1369. [Google Scholar] [CrossRef]
- Goudenège, D.; Boursicot, V.; Versigny, T.; Bonnetot, S.; Ratiskol, J.; Sinquin, C.; LaPointe, G.; Le Rous, F.; Delbarre-Ladrat, C. Genome sequence of Vibrio diabolicus and identification of the exopolysaccharide HE800 biosynthesis locus. Appl. Microbiol. Biotechnol. 2014, 98, 10165–10176. [Google Scholar] [CrossRef]
- Raguénès, G.; Peres, A.; Ruimy, R.; Pignet, P.; Christen, R.; Loaec, M.; Rougeaux, H.; Barbier, G.; Guezennec, J. Alteromonas infernus sp. nov., a new polysaccharide-producing bacterium isolated from a deep-sea hydrothermal vent. J. Appl. Microbiol. 1997, 82, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Roger, O.; Kervarec, N.; Ratiskol, J.; Colliec-Jouault, S.; Chevolot, L. Structural studies of the main exopolysaccharide produced by the deep-sea bacterium Alteromonas infernus. Carbohyd. Res. 2004, 339, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Colliec Jouault, S.; Chevolot, L.; Helley, D.; Ratiskol, J.; Bros, A.; Sinquin, C.; Roger, O.; Fischer, A.-M. Characterization, chemical modifications and in vitro anticoagulant properties of an exopolysaccharide produced by Alteromonas infernus. Biochim. Biophys. Acta 2001, 1528, 141–151. [Google Scholar] [CrossRef]
- Heymann, D.; Ruiz-Velasco, C.; Chesneau, J.; Ratiskol, J.; Sinquin, C.; Colliec-Jouault, S. Anti-metastatic properties of a marine bacterial exopolysaccharide-based derivative designed to mimic glycosaminoglycans. Molecules 2016, 21, 309. [Google Scholar] [CrossRef]
- Rederstorff, E.; Rethore, G.; Weiss, P.; Sourice, S.; Beck-Cormier, S.; Mathieu, E.; Maillasson, M.; Jacques, Y.; Colliec-Jouault, S.; Fellah, B.H.; et al. Enriching a cellulose hydrogel with a biologically active marine exopolysaccharide for cell-based cartilage engineering. J. Tissue Eng. Regen. Med. 2017, 11, 1152–1164. [Google Scholar] [CrossRef] [PubMed]
- Caruso, C.; Rizzo, C.; Mangano, S.; Poli, A.; Di Donato, P.; Finore, I.; Nicolaus, B.; Di Marco, G.; Michaud, L.; Lo Giudice, A. Production and biotechnological potentialities of extracellular polymeric substances from sponge-associated Antarctic bacteria. Appl. Environ. Microbiol. 2018, 84, e01624-17. [Google Scholar] [CrossRef] [Green Version]

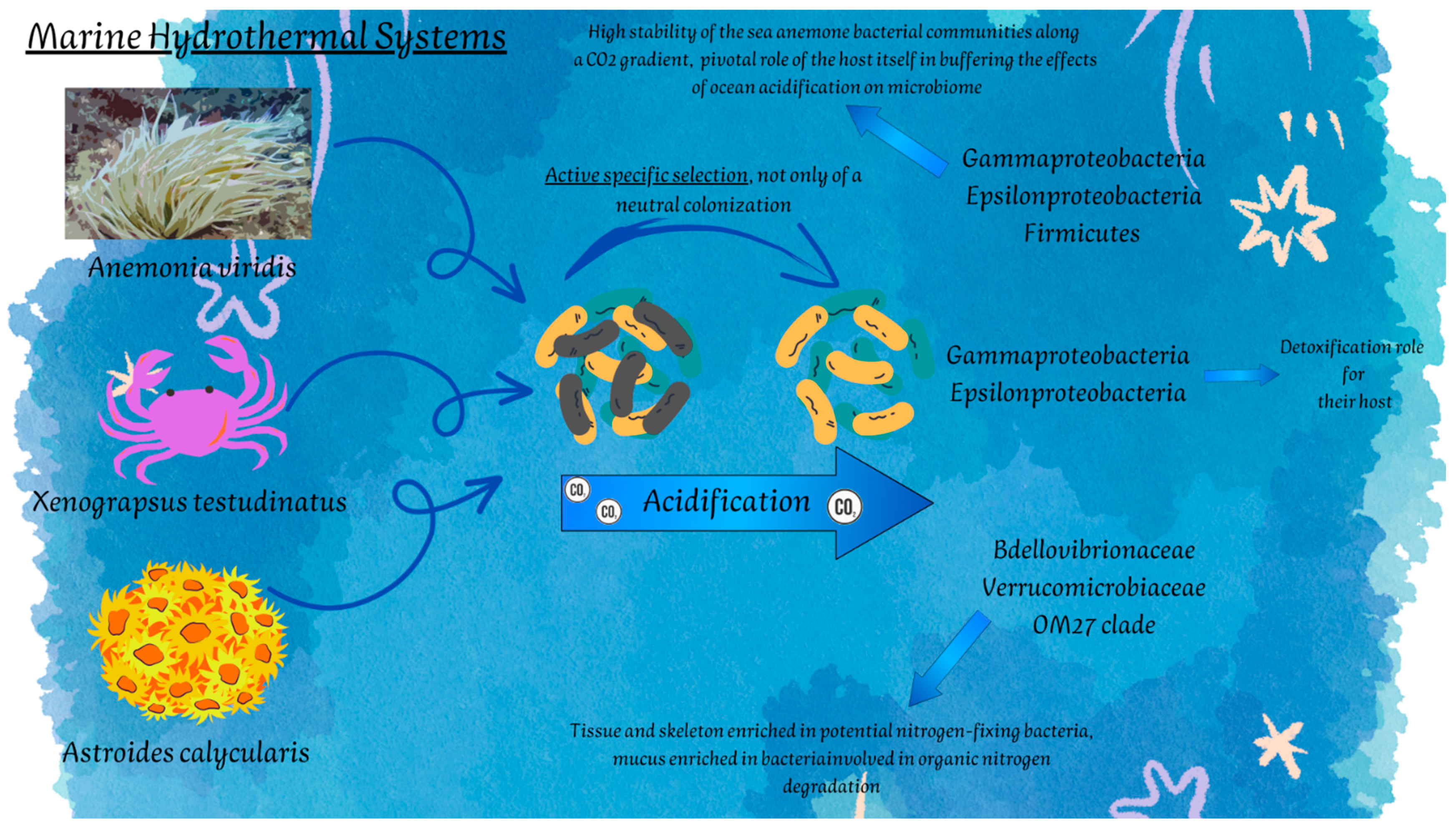
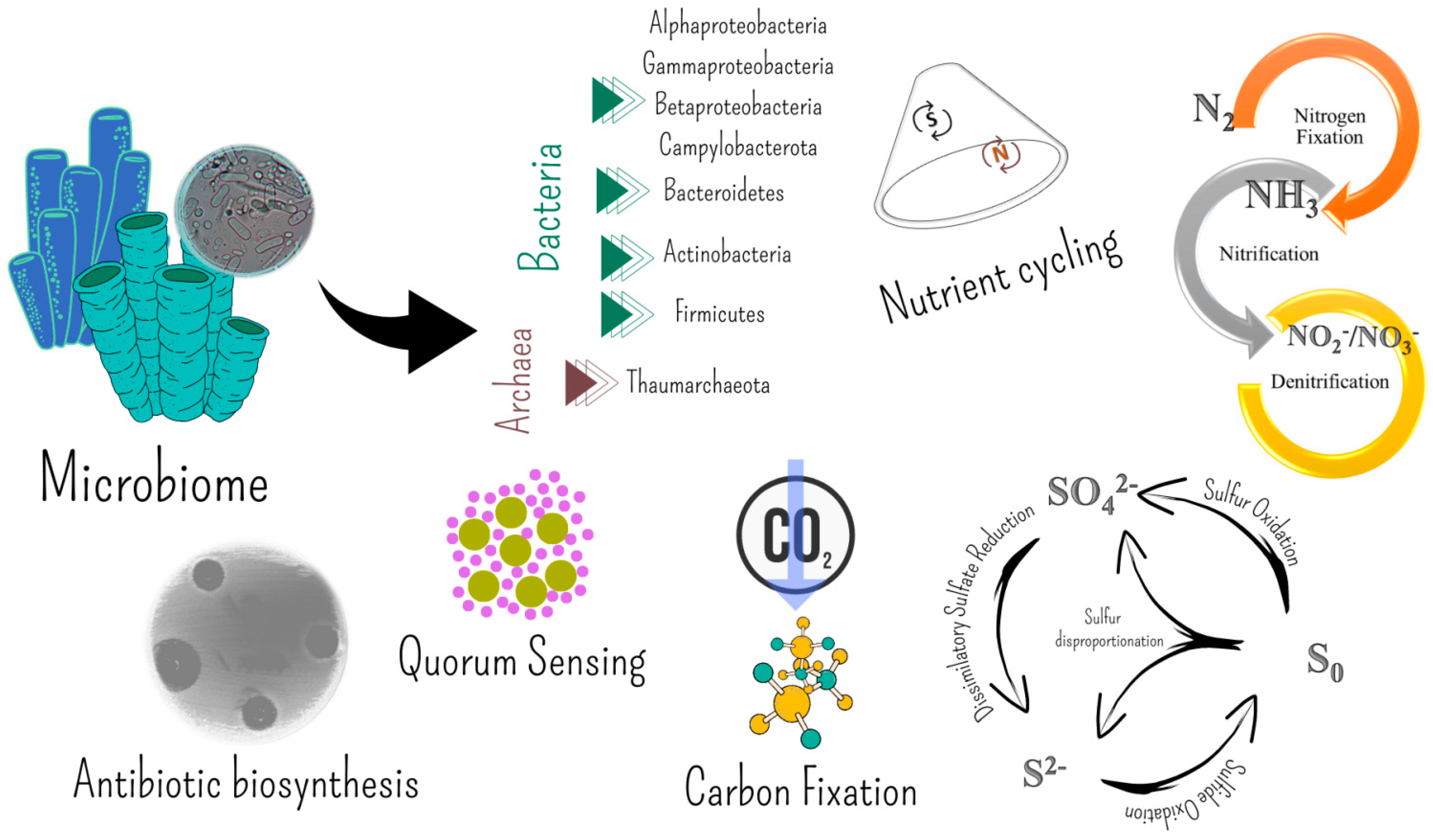
| Extreme Environment | Main Extreme Features (Alone or in Combination) | Stable, Unstable, or Temporary |
|---|---|---|
| Deep-sea | High pressure; low temperature; scarcity of food | Stable |
| Deep-sea hydrothermal vents | High temperatures; low O2 level; absence of light; presence of sulfide and heavy metals; high pressure | Temporary |
| Shallow hydrothermal vents | High temperatures; low O2 level; presence of sulfide and heavy metals | Temporary |
| Cold seeps | Low temperatures; high levels of sulfide, methane, and bicarbonate; high pressure; low O2 level | Temporary |
| Cold waters | Low and stable temperature; seasonal variations in light intensity and primary production; variations in salinity | Stable |
| Submarine canyons | Instability and physical disturbance; turbidity currents; low temperatures; high pressure; absence of light | Unstable |
| Hypersaline environments | High salinity; high UV irradiance; low nutrient availability; low O2 level | Unstable |
| Submarine caves | Darkness; low nutrient availability; limited accessibility; salinity gradient; presence of sulfides; deoxygenation | Stable |
| Extreme Environment | Invertebrate Host | Phylum or Sub-phylum | Main Associated Bacteria | Bacterial Function(s) in the Symbiosis | References |
|---|---|---|---|---|---|
| DSHVs | Alviniconcha sp. | Mollusca | Gammaproteobacteria Campylobacterota | Sulfur cycle | [12] |
| Alvinella pompejana Desbruyeres & Laubier, 1980 | Annelida | Gammaproteobacteria Campylobacterota | - Detoxification of sulfide and heavy metals - Sulfur oxidation | [98,99,100,101,103] | |
| Bathymodiolus azoricus Cosel & Comtet, 1999 | Mollusca | Gammaproteobacteria | Sulfur and methane oxidation | [110] | |
| Bathymodiolus puteoserpentis Cosel, Métivier & Hashimoto, 1994 | Mollusca | Gammaproteobacteria | Sulfur and methane oxidation | [110] | |
| Helicoradomenia sp. | Mollusca | Alphaproteobacteria Gammaproteobacteria | Not reported | [109] | |
| Kiwa spp. | Crustacea | Gammaproteobacteria Campylobacterota | -Sulfur oxidation - Detoxification | [96] | |
| Lamellibrachia anaximandri Southward, Andersen & Hourdez, 2011 | Annelida | Gammaproteobacteria | Sulfur oxidation | [112] | |
| Cold-seeps | Escarpia sp. | Annelida | Planctomycetes Proteobacteria | Ammonia oxidation | [116] |
| Hymedesmia (Stylopus) methanophila Cárdenas, 2019 | Porifera | Gammaproteobacteria | -Sulfur and methane oxidation -Hydrocarbon degradation | [30] | |
| Iophon methanophila Cárdenas, 2019 | Porifera | Gammaproteobacteria | -Sulfur and methane oxidation -Hydrocarbon degradation | [30] | |
| Lamellibrachia spp. | Annelida | Gammaproteobacteria | Sulfur oxidation | [110] | |
| SHVs | Actinia equina (Linnaeus, 1758) | Cnidaria | Gammaproteobacteria Campylobacterota | Sulfur oxidation | [130] |
| Anemonia viridis (Forsskål, 1775) | Cnidaria | Gammaproteobacteria | Sulfur oxidation | [132,133] | |
| Astroides calycularis (Pallas, 1766) | Cnidaria | Bdellovibrionaceae Verrucomicrobia | -Nitrogen fixation -Degradation of organic nitrogen | [134] | |
| Hymeniacidon sp. | Porifera | Alphaproteobacteria Gammaproteobacteria | Not reported | [130] | |
| Xenograpsus testudinatus Ng, Huang & Ho, 2000 | Crustacea | Gammaproteobacteria Campylobacterota | -Sulfur cycling -Detoxification | [126] | |
| Deep-sea | Characella sp. | Porifera | Gammaproteobacteria | Sulfur oxidation | [110] |
| Discodermia dissolute Schmidt, 1880 | Porifera | Entotheonellaeota | Not reported | [164,165] | |
| Geodia sp. | Porifera | Chloroflexi Acidobacteria Gammaproteobacteria Alphaproteobacteria | Not reported | [166,167] | |
| Inflatella pellicula Schmidt, 1875 | Porifera | Gammaproteobacteria Alphaproteobacteria Deltaproteobacteria Planctomycetes | Sulfur oxidation | [26,168] | |
| Lophelia pertusa (Linnaeus, 1758) | Cnidaria | Alphaproteobacteria Gammaproteobacteria Firmicutes Bacteroidetes Acidobacteria | Not reported | [170,171,172,173,174,175] | |
| Neamphius huxleyi (Sollas, 1888) | Porifera | Betaproteobacteria Campylobacterota | -Nitrogen cycling -CO2 fixation | [169] | |
| Poecillastra compressa (Bowerbank, 1866) | Porifera | Gammaproteobacteria Alphaproteobacteria Deltaproteobacteria Planctomycetes | Sulfur oxidation | [26,163] | |
| Stelletta normani Sollas, 1880 | Porifera | Gammaproteobacteria Alphaproteobacteria Deltaproteobacteria Planctomycetes | Sulfur oxidation | [26,163] | |
| Antarctic seawater | Abatus agassizii Mortensen, 1910 | Echinodermata | Gammaproteobacteria Bacteoidetes Actinobacteria | Sulfur cycling | [155] |
| Alcyonium antacticum Wright & Studer, 1889 | Cnidaria | Gammaproteobacteria Bacteroidetes Actinobacteria | Not reported | [152] | |
| Anoxycalyx (Scolymastra) joubini (Topsent, 1916) | Porifera | Gammaproteobacteria Bacteroidetes Actinobacteria | Not reported | [147,148,151] | |
| Grania sp. | Annelida | Gammaproteobacteria | Not reported | [156] | |
| Haliclonissa verrucosa Burton, 1932 | Porifera | Actinobacteria Gammaproteobacteria | Not reported | [147,151] | |
| Hemigellius pilosus (Kirkpatrick, 1907) | Porifera | Gammaproteobacteria Bacteroidetes Actinobacteria | Not reported | [141,142,146,148,151] | |
| Hymeniacidon torquata Topsent, 1916 | Porifera | Alphaproteobacteria Gammaproteobacteria | Not reported | [136,139,140,145] | |
| Isodictya bentarti Rios, Cristobo & Urgorri, 2004 | Porifera | Gammaproteobacteria | Not reported | [137] | |
| Lissodendoryx nobilis (Topsent, 1916) | Porifera | Gammaproteobacteria Bacteroidetes Actinobacteria | Not reported | [147,148,151] | |
| Mycale (Oxymycale) acerata Kirkpatrick, 1907 | Porifera | Gammaproteobacteria Bacteroidetes Actinobacteria | Not reported | [135,138,139,141,142,143,145] | |
| Sterechinus neumayeri (Meissner, 1900) | Echinodermata | Gammaproteobacteria Bacteroidetes | Not reported | [154] | |
| Tedania charcoti Topsent, 1907 | Porifera | Gammaproteobacteria Bacteroidetes Actinobacteria | Not reported | [151] | |
| Edswardsiella andrillae Daly, Rack & Zook, 2013 | Cnidaria | Proteobacteria Bacteroidetes | Not reported | [153] | |
| Synoicum adareanum (Herdman, 1902) | Chordata | Alphaproteobacteria Gammaproteobacteria Bacteroidetes Verrucomicrobia | Not reported | [157,158] | |
| Arctic seawater | Halichondria panicea (Pallas, 1766) | Porifera | Alphaproteobacteria Gammaproteobacteria Bacteroidetes Planctomycetes Cyanobacteria Verrucomicrobia | Not reported | [162] |
| Chondrocladia grandis (Verrill, 1879) | Porifera | Alphaproteobacteria Gammaproteobacteria Bacteroidetes | Not reported | [159] | |
| Cladorhiza oxeata Lundbeck, 1905 | Porifera | Alphaproteobacteria Gammaproteobacteria Bacteroidetes | Not reported | [159] |
| Invertebrate Host(s), Phylum or Subphylum | Bacterial Genus/Species/Strain * | Biomolecule(s) or (Potential) Bioactivity ** | Reference(s) |
|---|---|---|---|
| Deep-sea hydrothermal vents | |||
| Alvinella pompejana, Annelida | Alteromonas macleodii subsp. fijiensis biovar deepsane (strain HYD657) | High-molecular weight biopolymer DeepsaneTM with application in cosmetics | [220,221,222] |
| Alvinella pompejana, Annelida | Alteromonas macleodii strain HYD-1545 | Possible application of produced EPSs in the detoxification of metal-contaminated environments | [224] |
| Alvinella pompejana, Annelida | Vibrio diabolicus strain HE80 | Possible application of produced EPSs as a component of medicinal products due to its cicatrizing activity and bone regeneration | [225,226,227,228] |
| Riftia pachyptila, Annelida | Alteromonas infernus strain GY785 | Possible application of produced EPSs in wastewater treatment or in the recovery of metals; oversulfated low-molecular-weight EPS fractions showed: anticoagulant, antithrombotic and antitumor activities; possible application in cartilage regeneration | [225,232,233,234] |
| Shallow hydrothermal vents | |||
| Several invertebrates in the phyla Cnidaria, Tunicata, Porifera and Mollusca | Mainly Actinobacteria isolates | Antibiotic activity against pathogenic bacteria | [131] |
| Deep-sea and submarine canyons | |||
| Homaxinella balfourensis, Myxilla mollis, Radiella antarctica, Rossella nuda, Rossella racovitzae, Porifera | Several strains | Occurrence of genes coding for PKS | [150] |
| Unidentified Demosponge, Porifera | Micromonospora strain 28ISP2-46T | Production of antibiotic and antitumor compounds (e.g., kosinostatin and isoquinocycline B) | [189] |
| Inflatella pellicula, Poecillastra compressa, Stelletta normani, Porifera | Whole bacterial communities | Potential production of lipopeptides, glycopeptides, macrolides, and hepatotoxins | [163] |
| Forcepia, Discodermia, Gorgonacea, Leiodermatium, Porifera | Actinobacterial isolates; mainly Streptomyces strains | Antibiotic activity against MRSA and Candida albicans | [183] |
| Unidentified sponge (Fam. Oceanapiidae), Porifera | Salinispora strain M864 | Antibiotic activity against Clostridium difficile; antitumor activity | [183] |
| Lophelia pertusa, Cnidaria | Streptomyces sp. M-207 | Production of lobophorin K with antitumor and antibiotic activities | [196,197] |
| Unidentified coral (Fam. Caryophillidae), Cnidaria | Myceligenerans cantabricum strain M-201T | Antibiotic activity against pathogenic bacteria | [198] |
| Several invertebrates in the phyla Annelida, Echinodermata, Arthropoda, and Porifera | Streptomyces isolates | Production of compounds with antibiotic and/or cytotoxic activities (e.g., paulomycins A and B, maltophilins, antimycins, 6-epialteramides, fredericamycin) | [199] |
| Several invertebrates in the phyla Annelida, Echinodermata, Arthropoda, Porifera, Cnidaria | Actinobacteria isolates (genera Streptomyces, Myceligenerans, Micromonospora) | Antibiotic activity against a panel of resistant clinical pathogens | [197] |
| Colossendeis colossea, Arthropoda | Streptomyces strains M-231, M-157, M-192 | Production of compounds with antibacterial (e.g., paulomycins A and B, caboxamycin, aloesaponarin II, anthranilic acid), antifungal (bafilomycins B1 and C1, maltophilin), antitumor (caboxamycin, daunomycin, galtamycin), antiparasitic (paulomycins A and B, valinomycin), antiviral (valinomycin), and anti-inflammatory (lobophorin B) activities. | [197] |
| Antarctic waters | |||
| Anoxycalyx joubini, Haliclonissa verrucosa, Lissodendoryx nobilis, Porifera | Several strains; Pseudoalteromonas sp. TB41 | Potential production of antibiotic compounds against Bcc pathogens; occurrence of genes coding for PKS | [192] |
| Isodictya setifera, Porifera | Pseudomonas aeruginosa | Antibiotic compounds (phenazine alkaloid antibiotics) | [190] |
| Synoicum adareanum, Tunicata | Pseudovibrio and Microbulbifer strains | Antitumor compounds (e.g., palmerolide A); occurrence of genes coding for PKS | [157,187] |
| Hemigellius pilosus, Haliclona dancoi, Tedania charcoti, Haliclona virens, Anoxycalyx joubini, Calyx arcuarius, Haliclonissa verrucosa, Porifera | Several strains in the genera Colwellia, Pseudoalteromonas, Shewanella and Winogradskyella | Antibiofilm activity against Pseudomonas aeruginosa and Staphylococcus aureus | [211] |
| Tedania charcoti, Haliclonissa verrucosa, Hemigellius pilosus, Porifera | Strains in the genera Colwellia, Shewanella and Winogradskyella) | Possible application of produced EPSs in the detoxification of metal-contaminated environments and as cryoprotectant | [235] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo Giudice, A.; Rizzo, C. Bacteria Associated with Benthic Invertebrates from Extreme Marine Environments: Promising but Underexplored Sources of Biotechnologically Relevant Molecules. Mar. Drugs 2022, 20, 617. https://doi.org/10.3390/md20100617
Lo Giudice A, Rizzo C. Bacteria Associated with Benthic Invertebrates from Extreme Marine Environments: Promising but Underexplored Sources of Biotechnologically Relevant Molecules. Marine Drugs. 2022; 20(10):617. https://doi.org/10.3390/md20100617
Chicago/Turabian StyleLo Giudice, Angelina, and Carmen Rizzo. 2022. "Bacteria Associated with Benthic Invertebrates from Extreme Marine Environments: Promising but Underexplored Sources of Biotechnologically Relevant Molecules" Marine Drugs 20, no. 10: 617. https://doi.org/10.3390/md20100617
APA StyleLo Giudice, A., & Rizzo, C. (2022). Bacteria Associated with Benthic Invertebrates from Extreme Marine Environments: Promising but Underexplored Sources of Biotechnologically Relevant Molecules. Marine Drugs, 20(10), 617. https://doi.org/10.3390/md20100617







