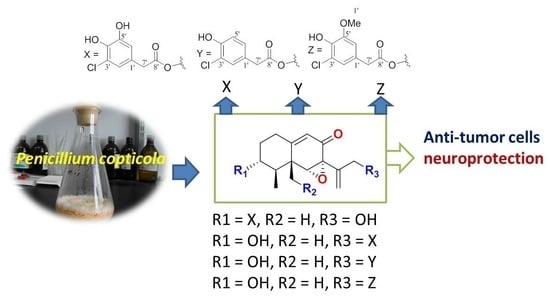
Eremophilane-Type Sesquiterpenes from a Marine-Derived Fungus Penicillium Copticola with Antitumor and Neuroprotective Activities †
Abstract
:1. Introduction
2. Results
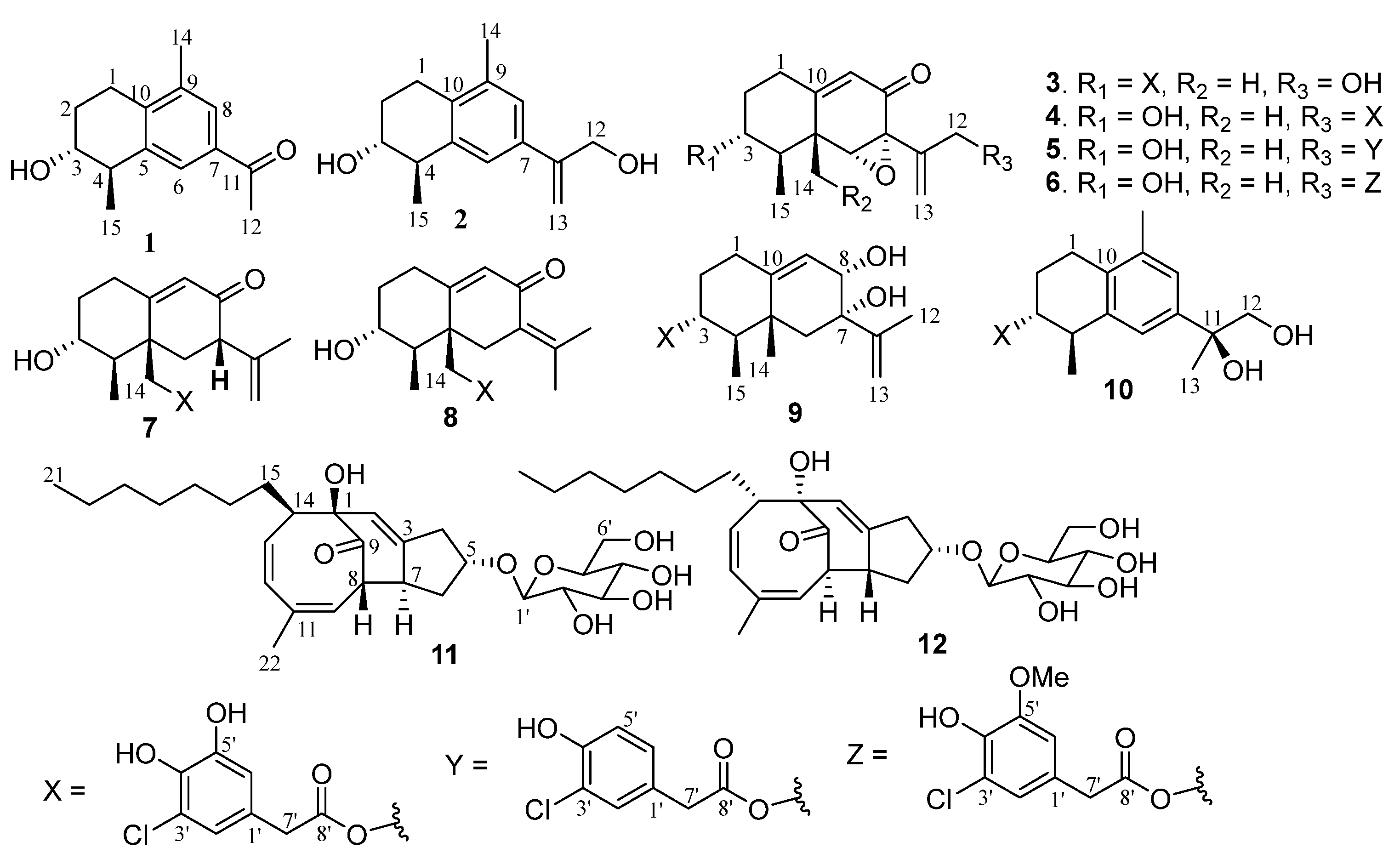
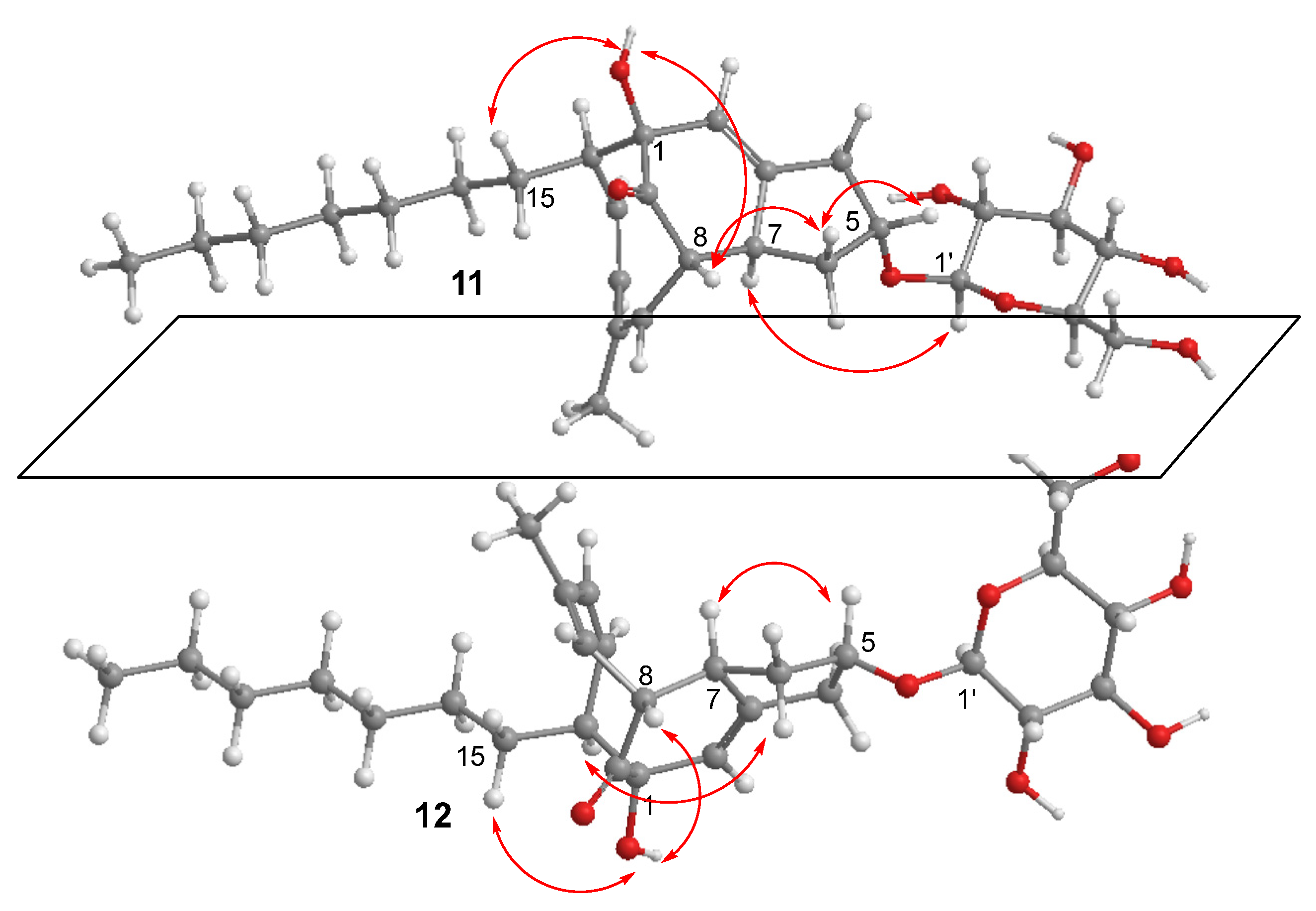
2.1. Structure Elucidation of New Compounds
2.2. Bioassays
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Fungal Strain and Identification
4.3. Fermentation and Extraction
4.4. Hydrolysis of 11 and 12
4.5. Mosher Reaction
4.6. Snatzke Method
4.7. Cytotoxic Detection
4.8. Cell Viability Assay
4.9. Measurement of Malondialdehyde (MDA)
4.10. Lactate Dehydrogenase (LDH) Release Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yuyama, K.T.; Fortkamp, D.; Abraham, W.R. Eremophilane-type sesquiterpenes from fungi and their medicinal potential. Biol. Chem. 2018, 399, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Fraga, B.M. Natural sesquiterpenoids. Nat. Prod. Rep. 2008, 25, 1180–1209. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liao, Z.; Liu, C.; Jia, H.; Sun, J. Eremophilane sesquiterpenes from the genus Ligularia. Chem. Biodivers. 2016, 13, 645–671. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Wang, J.; Wang, J.; Shi, L.; Li, K.; Lin, X.; Min, Y.; Yang, B.; Tang, L.; Liu, Y.; et al. Cytotoxic and antibacterial eremophilane sesquiterpenes from the marine-derived fungus Cochliobolus lunatus SCSIO41401. J. Nat. Prod. 2018, 81, 1405–1410. [Google Scholar] [CrossRef]
- Wang, L.; Li, M.; Tang, J.; Li, X. Eremophilane sesquiterpenes from a deep marine-derived fungus, Aspergillus sp. SCSIOW2, cultivated in the presence of epigenetic modifying agents. Molecules 2016, 21, 473. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Zhao, J.; Liu, D.; Proksch, P.; Zhao, Z.; Lin, W. Eremophilane-type sesquiterpenoids from an Acremonium sp. fungus isolated from deep-sea sediments. J. Nat. Prod. 2016, 79, 1035–1047. [Google Scholar] [CrossRef]
- Huang, Y.; Qiao, L.; Lv, A.; Pei, Y.; Tian, L. Eremophilane sesquiterenes from the marine fungus Penicillium sp. BL27-2. Chin. Chem. Lett. 2008, 19, 562–564. [Google Scholar] [CrossRef]
- Wu, G.; Lin, A.; Gu, Q.; Zhu, T.; Li, D. Four new chloro-eremophilane sesquiterpenes from an Antarctic deep-sea derived fungus, Penicillium sp. PR19N-1. Mar. Drugs 2013, 11, 1399–1408. [Google Scholar]
- Motohashi, K.; Hashimoto, J.; Inaba, S.; Khan, S.T.; Komaki, H.; Nagai, A.; Takagi, M.; Shin-ya, K. New sesquiterpenes, JBIR-27 and -28, isolated from a tunicate-derived fungus, Penicillium sp. SS080624SCf1. J. Antibiot. 2009, 62, 247–250. [Google Scholar] [CrossRef]
- Xu, J.; Liu, H.; Chen, Y.; Tan, H.; Guo, H.; Xu, L.; Li, S.; Huang, Z.; Li, H.; Gao, X.; et al. Highly substituted benzophenone aldehydes and eremophilane derivatives from the deep-sea derived fungus Phomopsis lithocarpus FS508. Mar. Drugs 2018, 16, 329. [Google Scholar] [CrossRef] [Green Version]
- Oh, H.; Jensen, P.R.; Murphy, B.T.; Fiorilla, C.; Sullivan, J.F.; Ramsey, T.; Fenical, W. Cryptosphaerolide, a cytotoxic Mcl-1 inhibitor from a marine-derived ascomycete related to the genus Cryptosphaeria. J. Nat. Prod. 2010, 73, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Riche, C.; Pascard-Billy, C.; Devys, M.; Gaudemer, A.; Barbier, M.; Bousquet, J.F. Crystal and molecular structure of phomenone, phytotoxin from the mushroom Phoma exigua. Tetrahedron Lett. 1974, 32, 2765–2766. [Google Scholar] [CrossRef]
- Iwamoto, C.; Minoura, K.; Hagishita, S.; Nomoto, K.; Numata, A. Penostatins F-I, novel cytotoxic metabolites from a Penicillium species separated from an Enteromorpha marine alga. J. Chem. Soc. Perkin Trans. 1998, 3, 449–456. [Google Scholar] [CrossRef]
- Isaka, M.; Srisanoh, U.; Veeranondha, S.; Choowong, W.; Lumyong, S. Cytotoxic eremophilane sesquiterpenoids from the saprobic fungus Berkleasmium nigroapicale BCC 8220. Tetrahedron 2009, 65, 8808–8815. [Google Scholar] [CrossRef]
- Isaka, M.; Jaturapat, A.; Kladwang, W.; Punya, J.; Lertwerawat, Y.; Tanticharoen, M.; Thebtaranonth, Y. Antiplasmodial compounds from the wood-decayed fungus Xylaria sp. BCC 1067. Planta Med. 2000, 66, 473–475. [Google Scholar] [CrossRef]
- McDonald, L.A.; Barbieri, L.R.; Bernan, V.S.; Janso, J.; Lassota, P.; Carter, G.T. 07H239-A, a new cytotoxic eremophilane sesquiterpene from the marine-derived xylariaceous fungus LL-07H239. J. Nat. Prod. 2004, 67, 1565–1567. [Google Scholar] [CrossRef]
- Yuan, W.; Goto, M.; Hsieh, K.; Yuan, B.; Zhao, Y.; Morris-Natschke, S.L.; Lee, K. Selective cytotoxic eremophilane-type sesquiterpenes from Penicillium citreonigrum. J. Asian Nat. Prod. Res. 2015, 17, 1239–1244. [Google Scholar]
- Chen, Z.; Zhong, C. Oxidative stress in Alzheimer’s disease. Neurosci. Bull. 2014, 30, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Bonda, D.J.; Wang, X.; Perry, G.; Nunomura, A.; Tabaton, M.; Zhu, X.; Smith, M.A. Oxidative stress in Alzheimer disease: A possibility for prevention. Neuropharmacology 2010, 59, 290–294. [Google Scholar] [CrossRef]
- Sisodia, S.S.; Price, D.L. Role of the β-amyloid protein in Alzheimer’s disease. FASEB J. 1995, 9, 366–370. [Google Scholar] [CrossRef]
- LaFerla, F.M.; Green, K.N.; Oddo, S. Intracellular amyloid-β in Alzheimer’s disease. Nature Rev. Neurosci. 2007, 8, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Westerink, R.H.S.; Ewing, A.G. The PC12 cell as model for neurosecretion. Acta Physiol. 2008, 192, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhu, L.; Li, H.; Xie, W.; Liu, J.; Zhang, Y.; Li, Y.; Wang, C. In vivo and in vitro neuroprotective effects of maca polysaccharide. Front. Biosci. 2022, 27, e8. [Google Scholar] [CrossRef]
- Zhang, L.; Kong, X.; Wang, Z.; Xu, F.; Zhu, Y. A study on neuroprotective effects of curcumin on the diabetic rat brain. J. Nutr. Health Aging 2016, 20, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Rani, R.; Kumar, V. When will small molecule lactate dehydrogenase inhibitors realize their potential in the cancer clinic? Future Med. Chem. 2017, 11, 1113–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Pinto, B.M. Human lactate dehydrogenase a inhibitors: A molecular dynamics investigation. PLoS ONE 2014, 9, e86365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusumi, T.; Hamada, T.; Ishitsuka, M.O.; Ohtani, I.; Kakisawa, H. Elucidation of the relative and absolute stereochemistry of lobatriene, a marine diterpene, by a modified Mosher method. J. Org. Chem. 1992, 57, 1033–1035. [Google Scholar] [CrossRef]
- Di Bari, L.; Pescitelli, G.; Pratelli, C.; Pini, D.; Salvadori, P. Determination of absolute configuration of acyclic 1,2-diols with Mo2(OAc)4. 1. Snatzke’s method revisited. J. Org. Chem. 2001, 66, 4819–4825. [Google Scholar] [CrossRef]
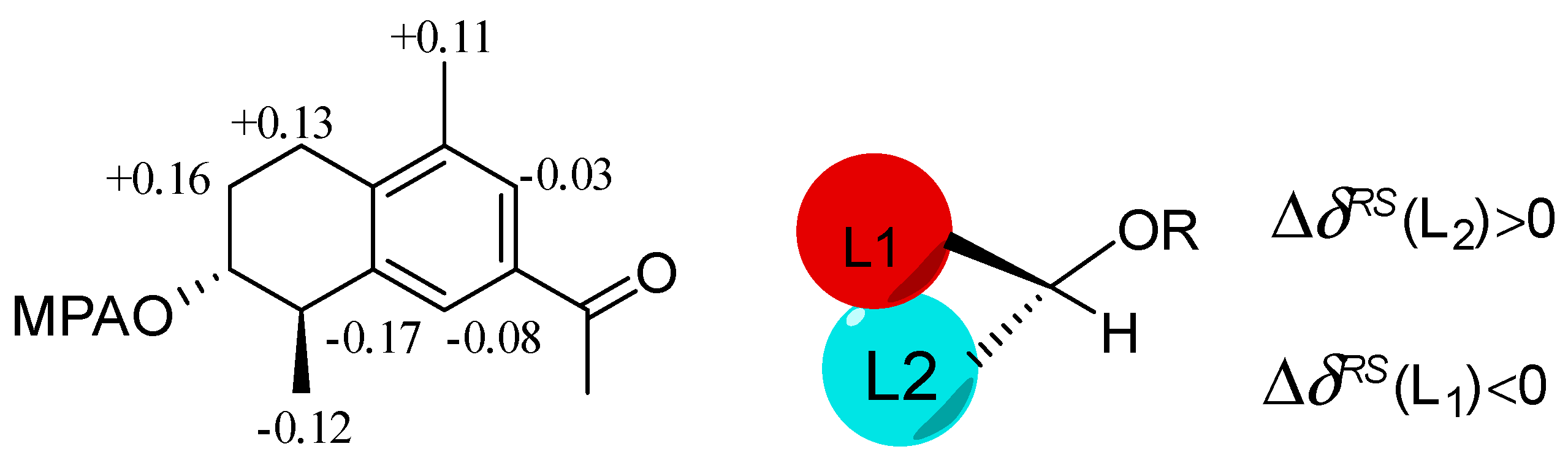






| 1 | 2 | 10 | ||||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | 2.62, dt (4.0, 12.0) 2.69, td (6.0, 12.0) | 23.7 | 2.50, dt (3.5, 12.0) 2.63, td (5.0, 12.0) | 23.3 | 2.55, dt (4.0, 12.0) 2.62, td (4.0, 12.0) | 22.4 |
| 2 | 1.80, m; 1.95, m | 27.1 | 1.71, m; 1.90, m | 27.7 | 1.90, m; 1.98, m | 23.9 |
| 3 | 3.66, dt (2.6, 4.9) | 69.8 | 3.60, dt (3.0, 5.0) | 70.3 | 4.87, dt (4.4, 6.3) | 74.6 |
| 4 | 2.78, dq (2.6, 7.2) | 41.2 | 2.67, dq (3.0, 7.2) | 41.2 | 2.91, dq (4.4, 7.2) | 38.1 |
| 5 | 141.1 | 140.5 | 138.0 | |||
| 6 | 7.61, brs | 127.1 | 7.06, brs | 124.7 | 7.12, s | 123.8 |
| 7 | 140.8 | 136.3 | 145.0 | |||
| 8 | 7.55, brs | 126.8 | 7.03, brs | 124.2 | 7.06, s | 125.1 |
| 9 | 136.4 | 135.6 | 134.9 | |||
| 10 | 134.8 | 134.1 | 131.4 | |||
| 11 | 198.3 | 148.3 | 73.9 | |||
| 12 | 2.53, s | 27.1 | 4.28, s | 63.0 | 3.35, dd (4.0, 12.0) 3.37, dd (4.0, 12.0) | 71.0 |
| 13 | 5.22, d (1.7) 5.34, d (1.7) | 110.1 | 1.36, s | 26.6 | ||
| 14 | 2.24, s | 19.8 | 2.16, s | 21.3 | 2.17, s | 20.1 |
| 15 | 1.23, d (7.2) | 21.4 | 1.12, d (7.2) | 19.9 | 1.21, d (7.2) | 21.7 |
| 1′ | 125.51 | |||||
| 2′ | 6.66, d (1.9) | 121.0 | ||||
| 3′ | 120.3 | |||||
| 4′ | 141.1 | |||||
| 5′ | 146.9 | |||||
| 6′ | 6.63, d (1.9) | 115.6 | ||||
| 7′ | 3.45, s | 40.1 | ||||
| 8′ | 171.4 | |||||
| OH-12 | 4.59, t (4.0) | |||||
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|
| 1 | 2.63, dt (2.0, 14.5) 2.36, td (2.0, 14.5) | 2.28, td (3.0, 12.0) 2.54, dt (3.0, 12.0) | 2.26, td (3.2, 14.1) 2.51, dt (3.2, 14.1) | 2.25, td (3.4, 11.2) 2.50, dt (3.4, 11.2) | 2.30, m 2.33, m | 2.30, td (3.4, 12.0) 2.39 dt (3.4, 11.2) | 2.07, m 2.29, m |
| 2 | 1.36, m 2.04, m | 1.25, m 1.99, m | 1.19, m 1.98, m | 1.20, m 1.98, m | 1.31, m 2.00, m | 1.29, m 2.01, m | 1.27, m 1.97, m |
| 3 | 4.82, dt (4.4, 11.2) | 3.44, dt (5.0, 12.0) | 3.42, dt (5.0, 10.5) | 3.43, dt (5.0, 10.5) | 3.49, m | 3.46, td (5.4, 10.6) | 4.73, m |
| 4 | 1.82, dq (6.8,11.2) | 1.53, dq (6.8, 12.0) | 1.54, dq (6.8, 10.5) | 1.54, dq (6.8, 10.5) | 1.29, dq (6.9, 10.5) | 1.37, dq (6.8, 10.6) | 1.44, dq (6.8, 11.0) |
| 6 | 3.40, s | 3.38, s | 3.38, s | 3.41, s | 1.80, t (11.0) 2.20, dd (4.0, 11.0) | 2.24, d (15.0) 3.08, d (15.0) | 1.68, d (12.6) 1.69, d (12.6) |
| 7 | 3.19, dd (4.0, 11.0) | ||||||
| 8 | 3.60, brd (4.5) | ||||||
| 9 | 5.75, d (1.7) | 5.71, d (1.7) | 5.70, d (1.7) | 5.68, d (1.7) | 5.78, d (1.3) | 5.82, d (1.3) | 5.40, d (4.5) |
| 12 | 4.05, dd (4.0, 12.0) 4.12, dd (4.0, 12.0) | 4.70, d (12.0) 4.71, d (12.0) | 4.67, d (12.0) 4.74, d (12.0) | 4.70, d (12.0) 4.76, d (12.0) | 1.60, s | 1.80, d (1.5) | 1.76, s |
| 13 | 5.08, d (1.5) 5.20, d (1.5) | 5.29, d (0.9) 5.36, d (0.9) | 5.28, d (1.1) 5.36, d (1.1) | 5.38, d (1.1) 5.29, d (1.1) | 4.68, brs 4.83, brs | 2.02, d (1.5) | 4.76, d (1.0) 4.85, d (1.0) |
| 14 | 1.22, s | 1.12, s | 1.12, s | 1.11, s | 4.19, d (11.5) 4.43, d (11.5) | 4.15, s | 1.08, s |
| 15 | 0.90, d (6.8) | 1.11, d (6.8) | 1.11, d (6.8) | 1.11, d (6.8) | 0.98, br d (6.9) | 1.06, d (6.8) | 0.79, d (6.8) |
| 2′ | 6.70, d (2.1) | 6.64, d (2.0) | 6.97, dd (1.5, 8,2) | 6.79, d (1.9) | 6.61, d (1.6) | 6.61, d (1.9) | 6.68, d (1.9) |
| 3′ | 6.88, d (8.2) | ||||||
| 6′ | 6.67, d (2.1) | 6.62, d (2.0) | 7.18, d (1.5) | 6.77, d (1.9) | 6.58, d (1.6) | 6.57, d (1.9) | 6.65, d (1.9) |
| 7′ | 3.50, s | 3.47, s | 3.54, s | 3.55, s | 3.45, s | 3.40, s | 3.47, s |
| OH-3 | 4.70, br | ||||||
| OH-12 | 4.83 t (4.0) | ||||||
| OH-4′ | 9.01, brs | 9.02, s | 10.0, brs | 9.00, s | 8.99, s | ||
| OH-5′ | 9.76, brs | 9.74, s | 9.80, s | 9.69, s | |||
| MeO | 3.78, s |
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|
| 1 | 29.9 | 30.9 | 30.9 | 30.3 | 31.4 | 31.2 | 30.1 |
| 2 | 31.7 | 35.8 | 35.8 | 35.4 | 35.7 | 35.8 | 32.9 |
| 3 | 73.2 | 69.0 | 69.2 | 68.4 | 69.5 | 69.6 | 74.7 |
| 4 | 41.9 | 44.8 | 44.8 | 44.3 | 50.7 | 49.3 | 47.9 |
| 5 | 41.2 | 41.2 | 41.2 | 40.7 | 43.1 | 44.8 | 38.6 |
| 6 | 68.3 | 69.2 | 68.9 | 68.6 | 39.1 | 38.3 | 40.6 |
| 7 | 61.7 | 61.2 | 61.2 | 60.7 | 50.7 | 127.3 | 73.7 |
| 8 | 192.2 | 192.1 | 192.2 | 191.6 | 198.0 | 190.2 | 68.2 |
| 9 | 120.7 | 120.0 | 120.0 | 119.4 | 126.4 | 128.6 | 121.2 |
| 10 | 163.7 | 166.2 | 166.6 | 165.6 | 164.3 | 163.1 | 143.0 |
| 11 | 145.3 | 139.3 | 139.3 | 138.8 | 144.0 | 142.9 | 150.9 |
| 12 | 62.1 | 64.9 | 64.8 | 64.5 | 20.3 | 22.6 | 19.4 |
| 13 | 111.9 | 117.1 | 117.2 | 116.8 | 114.3 | 23.0 | 110.4 |
| 14 | 18.1 | 18.4 | 18.4 | 17.9 | 66.6 | 66.6 | 20.2 |
| 15 | 11.3 | 11.7 | 11.8 | 11.2 | 11.4 | 11.9 | 11.0 |
| 1′ | 126.0 | 125.6 | 126.1 | 125.1 | 125.5 | 125.5 | 126.2 |
| 2′ | 121.0 | 120.3 | 131.0 | 122.1 | 120.9 | 121.0 | 121.0 |
| 3′ | 120.3 | 121.0 | 119.8 | 119.5 | 120.4 | 120.4 | 120.3 |
| 4′ | 141.2 | 141.3 | 152.5 | 141.6 | 141.3 | 141.2 | 141.1 |
| 5′ | 146.9 | 146.9 | 116.9 | 148.4 | 146.9 | 146.9 | 146.9 |
| 6′ | 115.5 | 115.6 | 129.4 | 111.8 | 115.5 | 115.6 | 115.4 |
| 7′ | 39.9 | 39.6 | 39.2 | 39.7 | 39.0 | 40.0 | 40.0 |
| 8′ | 171.2 | 171.1 | 171.2 | 170.6 | 171.2 | 171.2 | 171.3 |
| OMe | 56.6 |
| No | 11 | 12 | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 82.1 | 82.3 | ||
| 2 | 124.9 | 5.50, br d (2.6) | 125.3 | 5.50, br d (2.6) |
| 3 | 146.1 | 144.9 | ||
| 4 | 37.9 | 2.46, m 2.60, m | 37.7 | 2.27, m 2.68, m |
| 5 | 78.7 | 4.38, t (5.0) | 78.1 | 4.34, m |
| 6 | 39.6 | 1.44, m 2.43, m | 39.5 | 1.45, m 2.57, m |
| 7 | 48.6 | 3.11, m | 48.5 | 2.79, m |
| 8 | 50.4 | 2.86, t (5.6) | 50.4 | 2.91, m |
| 9 | 211.2 | 210.6 | ||
| 10 | 128.3 | 5.58, dd (0.9, 6.4) | 128.2 | 5.61, dd (0.9, 6.4) |
| 11 | 129.4 | 129.5 | ||
| 12 | 130.2 | 5.67, d (11.5) | 130.3 | 5.68, d (11.5) |
| 13 | 133.9 | 5.60, dd (9.1, 11.5) | 133.9 | 5.34, dd (9.2, 11.5) |
| 14 | 43.3 | 2.57, m | 42.7 | 2.53, m |
| 15 | 28.4 | 1.55, m | 28.2 | 1.53, m |
| 16 | 28.0 | 1.09, m 1.22, m | 27.8 | 1.08, m 1.21, m |
| 17 | 29.6 | 1.22, m | 29.2 | 1.20, m |
| 18 | 29.1 | 1.22, m | 29.0 | 1.20, m |
| 19 | 31.7 | 1.22, m | 31.7 | 1.21, m |
| 20 | 22.5 | 1.25, m | 22.5 | 1.25, m |
| 21 | 14.4 | 0.87, t (7.1) | 14.4 | 0.85, t (7.1) |
| 22 | 25.6 | 1.75, s | 25.6 | 1.74, s |
| 1’ | 102.1 | 4.22, d (7.9) | 102.5 | 4.20, d (7.7) |
| 2’ | 73.8 | 2.93, m | 73.8 | 2.89, m |
| 3’ | 77.2 | 3.16, m | 77.3 | 3.13, m |
| 4’ | 70.6 | 3.05, m | 70.5 | 3.03, m |
| 5’ | 77.4 | 3.08, m | 77.4 | 3.03, m 3.08, m |
| 6’ | 61.6 | 3.44, dt (5.7, 11.8) 3.67, dd (2.0, 11.8) | 61.6 | 3.43, m 3.67, dd (2.0, 11.8) |
| IC50 (μM) | |||
|---|---|---|---|
| Comps | A549 | HCT-8 | MCF-7 |
| 1 | >10 | >10 | >10 |
| 2 | >10 | >10 | >10 |
| 3 | >10 | >10 | >10 |
| 4 | >10 | 5.4 ± 0.1 | >10 |
| 5 | >10 | 7.3 ± 0.1 | >10 |
| 6 | >10 | >10 | >10 |
| 7 | >10 | >10 | >10 |
| 8 | 3.2 ± 0.1 | >10 | >10 |
| 9 | >10 | >10 | >10 |
| 10 | >10 | >10 | >10 |
| taxol | 0.2 ± 0.1 | 0.7 ± 0.3 | 0.2 ± 0.1 |
| % PC12 Cell Viability to Control | ||||||
|---|---|---|---|---|---|---|
| μM | 0 | 5.0 | 10.0 | 20.0 | 30.0 | 40.0 |
| Aβ25-35 | 100 | 92.5 | 81.0 | 74.6 | 64.8 | 49.5 |
| Aβ25-35 + 3 | 50.0 | 53.7 | 55.8 | 62.1 | 63.7 | 65.5 |
| Aβ25-35 + 4 | 50.0 | 55.2 | 52.4 | 67.8 | 72.2 | 75.5 |
| Aβ25-35 + 5 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 |
| Aβ25-35 + 6 | 50.0 | 51.0 | 53.0 | 55.0 | 57.0 | 59.2 |
| Aβ25-35 + 7 | 50.0 | 56.2 | 60.5 | 74.6 | 78.6 | 84.3 |
| Aβ25-35 + 8 | 50.0 | 52.6 | 55.5 | 58.4 | 62.7 | 65.5 |
| Aβ25-35 + 9 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 | 50.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, D.; Fan, A.; Huang, J.; Lin, W. Eremophilane-Type Sesquiterpenes from a Marine-Derived Fungus Penicillium Copticola with Antitumor and Neuroprotective Activities. Mar. Drugs 2022, 20, 712. https://doi.org/10.3390/md20110712
Zhang J, Liu D, Fan A, Huang J, Lin W. Eremophilane-Type Sesquiterpenes from a Marine-Derived Fungus Penicillium Copticola with Antitumor and Neuroprotective Activities. Marine Drugs. 2022; 20(11):712. https://doi.org/10.3390/md20110712
Chicago/Turabian StyleZhang, Jianping, Dong Liu, Aili Fan, Jian Huang, and Wenhan Lin. 2022. "Eremophilane-Type Sesquiterpenes from a Marine-Derived Fungus Penicillium Copticola with Antitumor and Neuroprotective Activities" Marine Drugs 20, no. 11: 712. https://doi.org/10.3390/md20110712
APA StyleZhang, J., Liu, D., Fan, A., Huang, J., & Lin, W. (2022). Eremophilane-Type Sesquiterpenes from a Marine-Derived Fungus Penicillium Copticola with Antitumor and Neuroprotective Activities. Marine Drugs, 20(11), 712. https://doi.org/10.3390/md20110712