Characterization of a Novel Alginate Lyase with Two Alginate Lyase Domains from the Marine Bacterium Vibrio sp. C42
Abstract
:1. Introduction
2. Results and Discussion
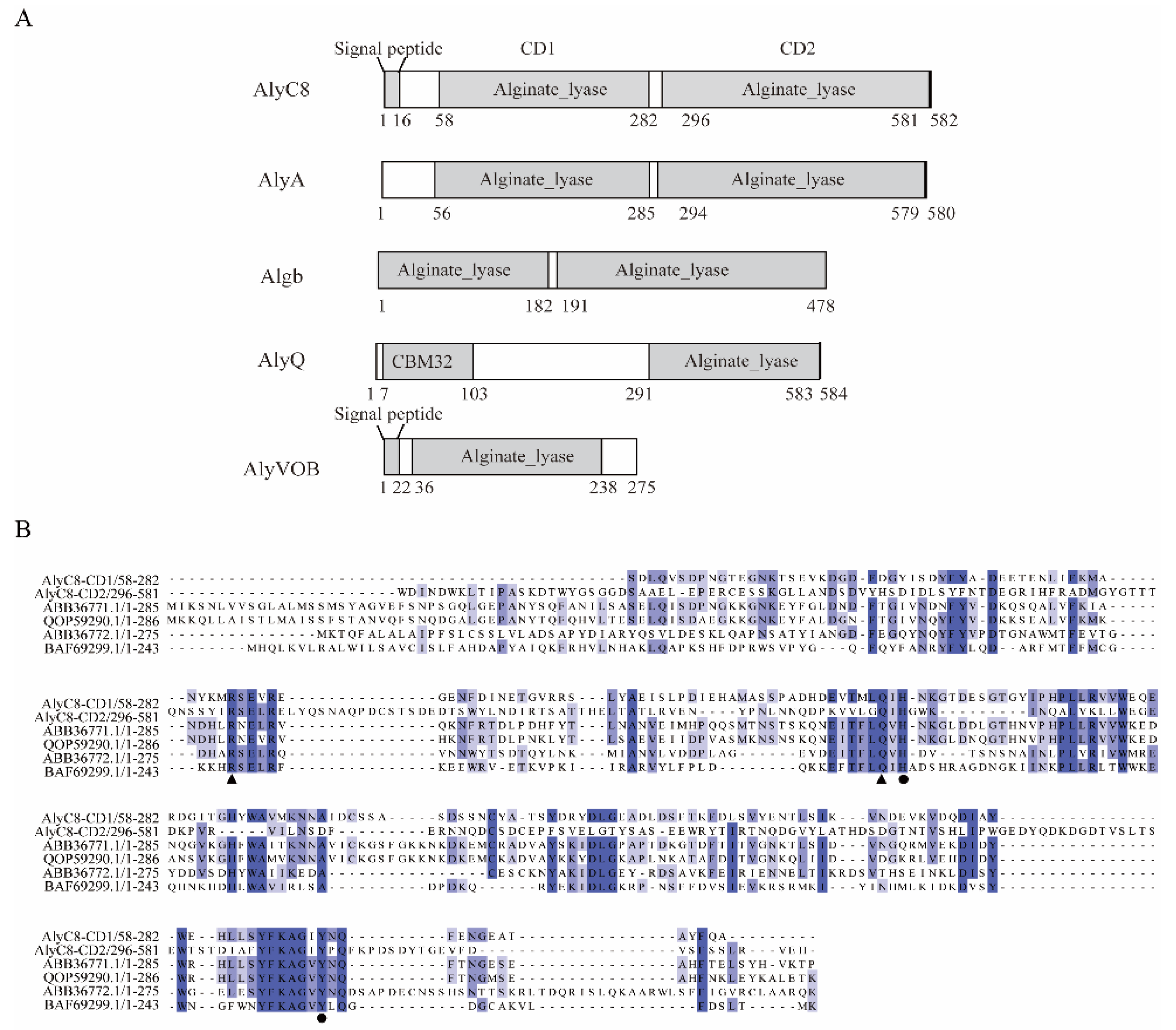
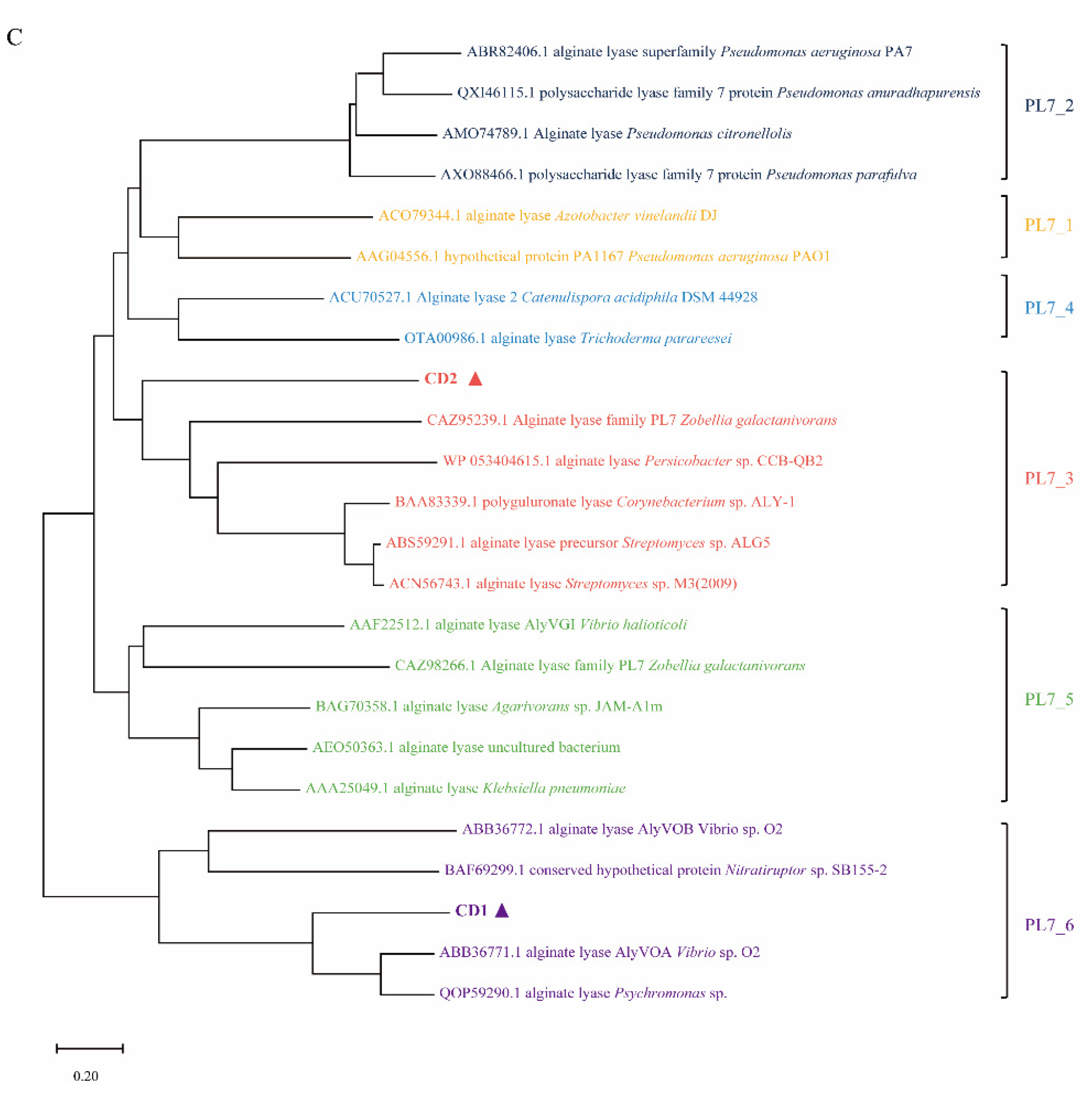
2.1. Sequence Analysis of AlyC8
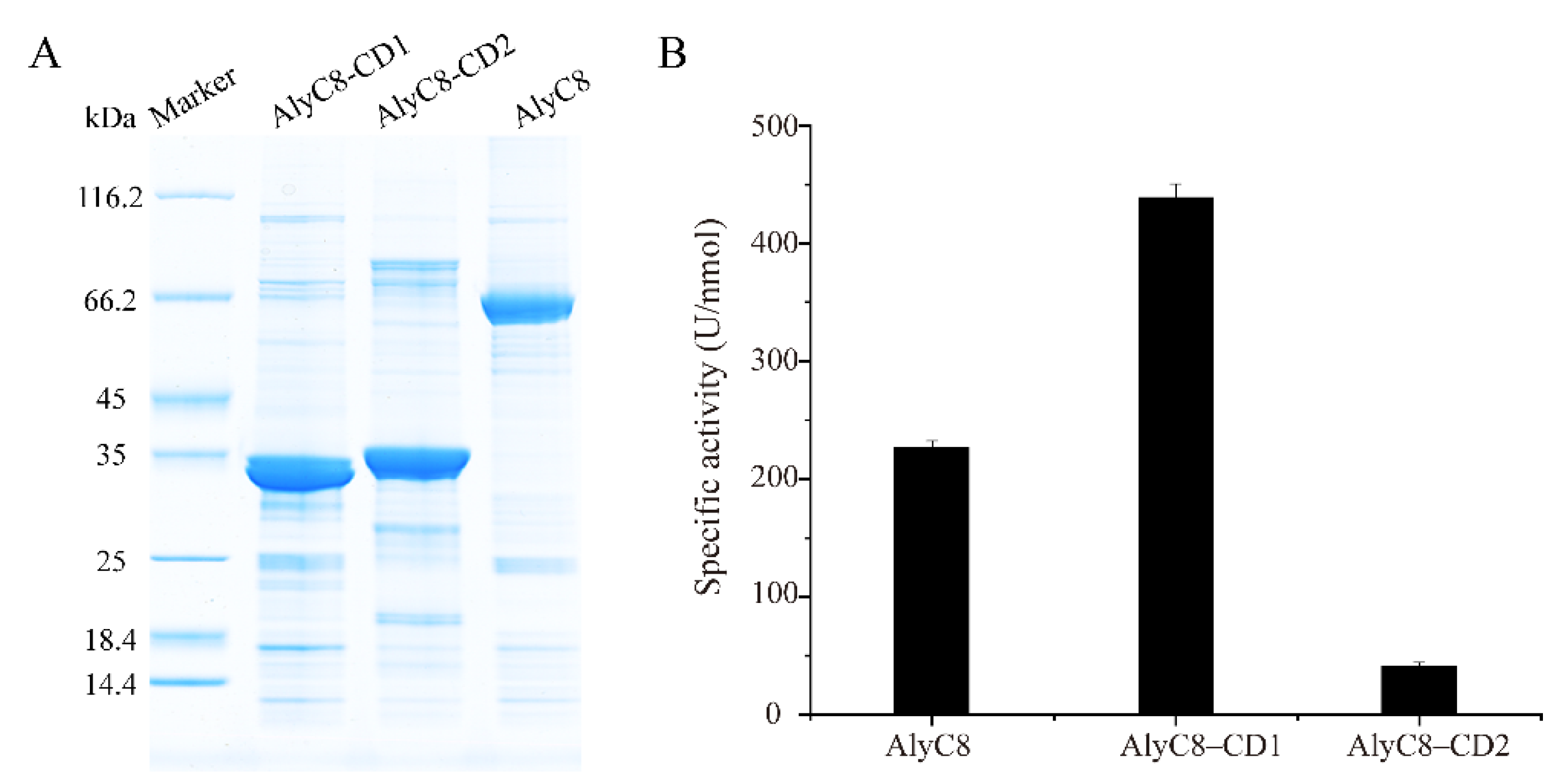
2.2. Activity Determination of AlyC8, AlyC8-CD1 and AlyC8-CD2
2.3. Effects of Temperature and pH on the Activities of AlyC8, AlyC8-CD1 and AlyC8-CD2
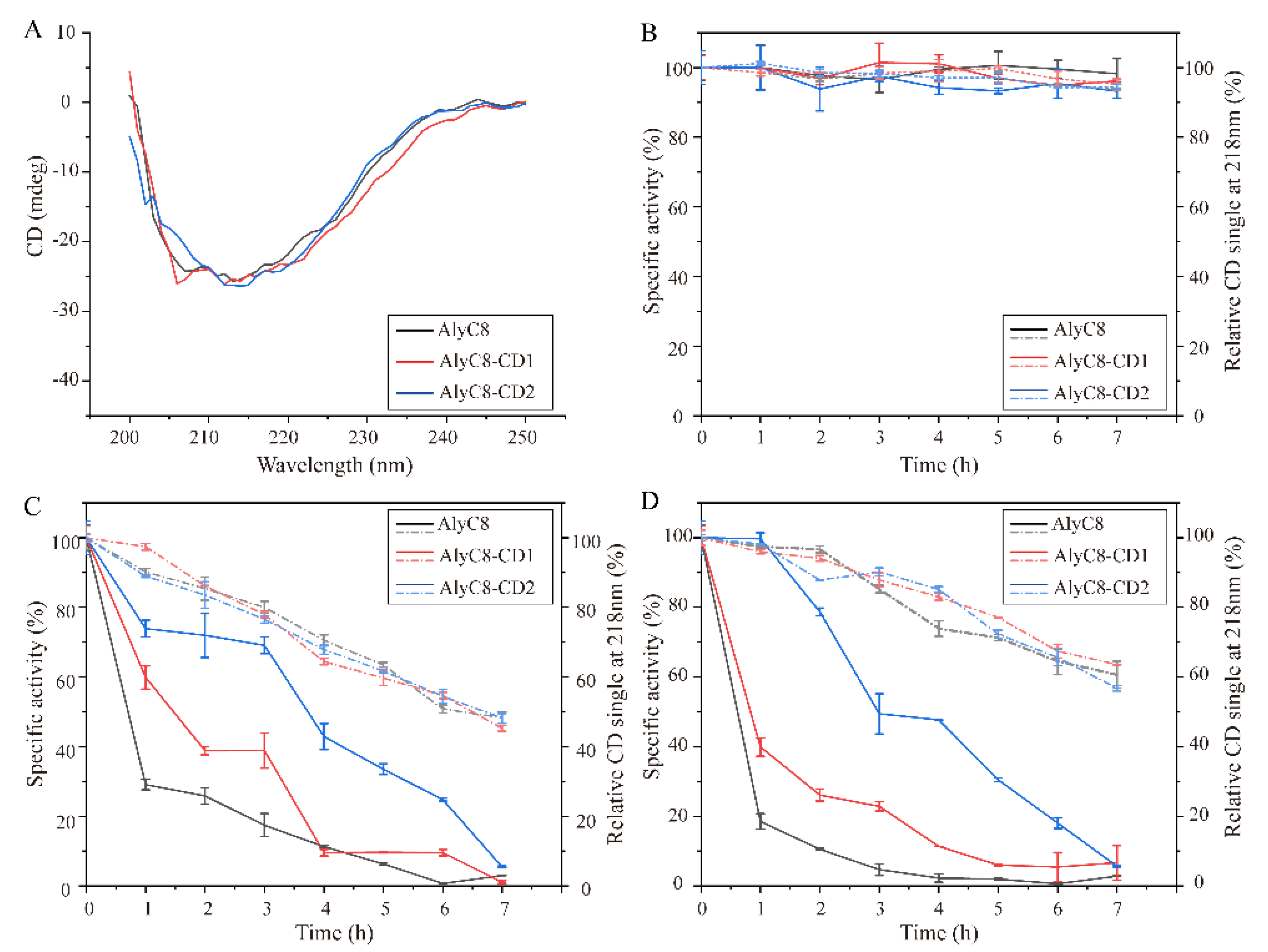
2.4. Thermal Stability of AlyC8, AlyC8-CD1 and AlyC8-CD2
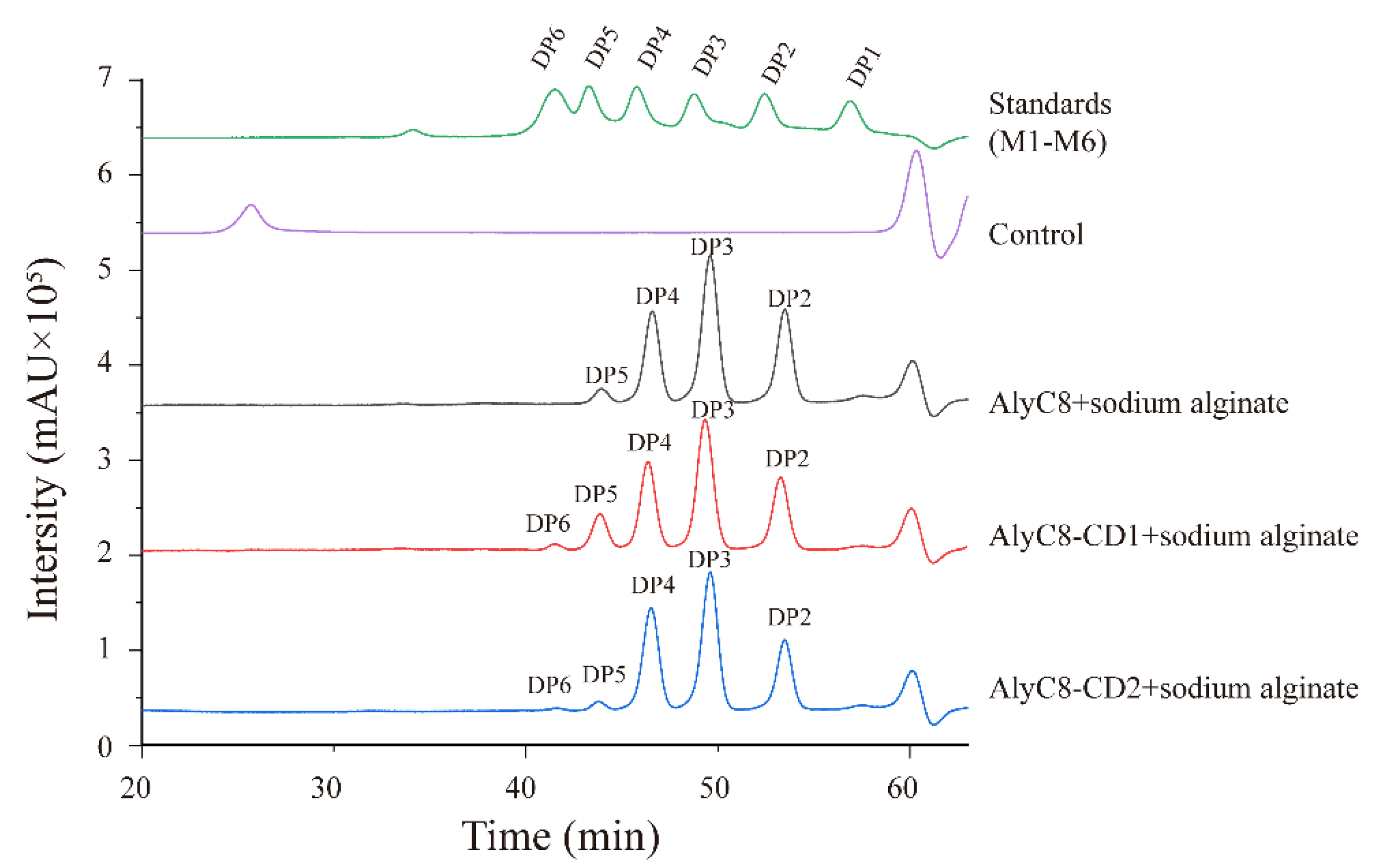
2.5. Substrate Specificities of AlyC8, AlyC8-CD1 and AlyC8-CD2
2.6. Kinetic Analysis of AlyC8-CD1 and AlyC8-CD2
2.7. The Synergy between AlyC8-CD1 and AlyC8-CD2
3. Conclusions
4. Materials and Methods
4.1. Materials and Strains
4.2. Bioinformatics
4.3. Gene Cloning and Mutagenesis
4.4. Protein Expression and Purification
4.5. Biochemical Characterization of AlyC8 and Its Mutants
4.6. Circular Dichroism Spectra
4.7. Degradation Products of the Alginate Lyases
4.8. Kinetic Analysis of the Alginate Lyases
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duarte, C.M.; Middelburg, J.J.; Caraco, N. Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2005, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Wang, P.; Zhang, Y.Z.; Chena, X.L. Diversity of three-dimensional structures and catalytic mechanisms of alginate Lyases. Appl. Environ. Microbiol. 2018, 84, e02040-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Danno, H.; Uchida, M.; Ishihara, K.; Suzuki, T.; Kaneniwa, M.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Analysis of extracellular alginate lyase and its gene from a marine bacterial strain, Pseudoalteromonas atlantica AR06. Appl. Microbiol. Biotechnol. 2010, 86, 567–576. [Google Scholar] [CrossRef]
- Li, J.W.; Dong, S.; Song, J.; Li, C.B.; Chen, X.L.; Xie, B.B.; Zhang, Y.Z. Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 2011, 9, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yang, S.Q.; Li, X.T.; Yan, Q.J.; Reaney, M.J.T.; Jiang, Z.Q. Alginate oligosaccharides: Production, biological activities, and potential applications. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1859–1881. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar]
- Jantarathina, S.; Borompichaichartkul, C.; Sanguandeekul, R. Microencapsulation of probiotic and prebiotic in alginate-chitosan capsules and its effect on viability under heat process in shrimp feeding. Mater. Today Proc. 2017, 4, 6166–6172. [Google Scholar] [CrossRef]
- Martins, E.; Poncelet, D.; Rodrigues, R.C.; Renard, D. Oil encapsulation techniques using alginate as encapsulating agent: Applications and drawbacks. J. Microencapsul. 2017, 34, 754–771. [Google Scholar] [CrossRef]
- Urtuvia, V.; Maturana, N.; Acevedo, F.; Pena, C.; Díaz-Barrera, A. Bacterial alginate production: An overview of its biosynthesis and potential industrial production. World J. Microb. Biot. 2017, 33, 198. [Google Scholar] [CrossRef]
- Inoue, A.; Mashino, C.; Uji, T.; Saga, N.; Mikami, K.; Ojima, T. Characterization of an eukaryotic PL-7 alginate lyase in the marine red alga Pyropia yezoensis. Curr. Biotechnol. 2015, 4, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T.; Katayose, M. A mode of action of alginate lyase from Turbo cornutus on sodium alginate. Agric. Biol. Chem. 1979, 43, 287–291. [Google Scholar]
- Zhu, B.W.; Tan, H.D.; Qin, Y.Q.; Xu, Q.S.; Du, Y.G.; Yin, H. Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int. J. Biol. Macromol. 2015, 75, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Ertesvåg, H.; Erlien, F.; Skjåk-Bræk, G.; Rehm, B.H.; Valla, S. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J. Bacteriol. 1998, 180, 3779–3784. [Google Scholar] [CrossRef] [Green Version]
- Pilgaard, B.; Vuillemin, M.; Holck, J.; Wilkens, C.; Meyer, A.S. Specificities and synergistic actions of novel PL8 and PL7 alginate lyases from the marine fungus Paradendryphiella salina. J. Fungi 2021, 7, 80. [Google Scholar] [CrossRef]
- Ogura, K.; Yamasaki, M.; Yamada, T.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action. J. Biol. Chem. 2009, 284, 35572–35579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helbert, W.; Poulet, L.; Drouillard, S.; Mathieu, S.; Loiodice, M.; Couturier, M.; Lombard, V.; Terrapon, N.; Turchetto, J.; Vincentelli, R.; et al. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc. Natl. Acad. Sci. USA 2019, 116, 6063–6068. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Hu, F.; Wang, M.Y.; Zhu, B.W.; Ni, F.; Yao, Z. Elucidation of degradation pattern and immobilization of a novel alginate lyase for preparation of alginate oligosaccharides. Int. J. Biol. Macromol. 2020, 146, 579–587. [Google Scholar] [CrossRef]
- Zhu, B.W.; Ni, F.; Sun, Y.; Ning, L.M.; Yao, Z. Elucidation of degrading pattern and substrate recognition of a novel bifunctional alginate lyase from Flammeovirga sp. NJ-04 and its use for preparation alginate oligosaccharides. Biotechnol. Biofuels 2019, 12, 13. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.T.; Chung, J.H.; Wang, D.; Lee, J.; Woo, H.C.; Choi, I.G.; Kim, K.H. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl. Microbiol. Biotechnol. 2012, 93, 2233–2239. [Google Scholar] [CrossRef]
- Suzuki, H.; Suzuki, K.; Inoue, A.; Ojima, T. A novel oligoalginate lyase from abalone, Haliotis discus hannai, that releases disaccharide from alginate polymer in an exolytic manner. Carbohydr. Res. 2006, 341, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- OStgaard, K.; Knutsen, S.H.; Dyrset, N.; Aasen, I.M. Production and characterization of guluronate lyase from Klebsiella pneumoniae for applications in seaweed biotechnology. Enzym. Microb. Technol. 1993, 15, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Kagaya, M.; Ojima, T. Preparation of protoplasts from Laminaria japonica using native and recombinant abalone alginate lyases. J. Appl. Phycol. 2008, 20, 633–640. [Google Scholar] [CrossRef]
- Barzkar, N.; Sheng, R.; Sohail, M.; Jahromi, S.T.; Babich, O.; Sukhikh, S.; Nahavandi, R. Alginate lyases from marine bacteria: An enzyme ocean for sustainable future. Molecules 2022, 27, 3375. [Google Scholar] [CrossRef]
- Islan, G.A.; Bosio, V.E.; Castro, G.R. Alginate lyase and ciprofloxacin co-immobilization on biopolymeric microspheres for cystic fibrosis treatment. Macromol. Biosci. 2013, 13, 1238–1248. [Google Scholar] [CrossRef]
- Chapman, V.J.; Chapman, D.J. Seaweeds and Their Uses, 3rd ed.; Chapman and Hall, Ltd.: London, UK, 1970. [Google Scholar]
- Zhang, Z.L.; Tang, L.Y.; Bao, M.M.; Liu, Z.G.; Yu, W.G.; Han, F. Functional characterization of carbohydrate-binding modules in a new alginate lyase, TsAly7B, from Thalassomonas sp. LD5. Mar. Drugs 2019, 18, 25. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Dong, F.; Wang, P.; Cao, H.Y.; Li, C.Y.; Li, P.Y.; Pang, X.H.; Zhang, Y.Z.; Chen, X.L. Novel molecular insights into the catalytic mechanism of marine bacterial alginate lyase AlyGC from polysaccharide lyase family 6. J. Biol. Chem. 2017, 292, 4457–4468. [Google Scholar] [CrossRef] [Green Version]
- Murata, K.; Inose, T.; Hisano, T.; Abe, S.; Yonemoto, Y.; Yamashita, T.; Takagi, M.; Sakaguchi, K.; Kimura, A.; Imanaka, T. Bacterial alginate lyase: Enzymology, genetics and application. J. Ferment. Bioeng. 1993, 76, 427–437. [Google Scholar] [CrossRef]
- Hisano, T.; Nishimura, M.; Yamashita, T.; Sakaguchi, K.; Murata, K. On the self-processing of bacterial alginate lyase. J. Ferment. Bioeng. 1994, 78, 109–110. [Google Scholar] [CrossRef]
- Miyake, O.; Ochiai, A.; Hashimoto, W.; Murata, K. Origin and Diversity of Alginate Lyases of Families PL-5 and -7 in Sphingomonas sp. Strain A1. J. Bacteriol. 2004, 186, 2891–2896. [Google Scholar] [CrossRef] [Green Version]
- Badur, A.H.; Jagtap, S.S.; Yalamanchili, G.; Lee, J.K.; Zhao, H.; Rao, C.V. Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl. Environ. Microbiol. 2015, 81, 1865–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belik, A.; Silchenko, A.; Malyarenko, O.; Rasin, A.; Kiseleva, M.; Kusaykin, M.; Ermakova, S. Two new alginate lyases of PL7 and PL6 families from polysaccharide-degrading bacterium Formosa algae KMM 3553T: Structure, properties, and products analysis. Mar. Drugs 2020, 18, 130. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.W.; Huang, L.S.; Tan, H.D.; Qin, Y.Q.; Du, Y.G.; Yin, H. Characterization of a new endo-type polyM-specific alginate lyase from Pseudomonas sp. Biotechnol. Lett. 2015, 37, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, M.; Moriwaki, S.; Miyake, O.; Hashimoto, W.; Murata, K.; Mikami, B. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: A novel alginate lyase with a beta-sandwich fold. J. Biol. Chem. 2004, 279, 31863–31872. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Chen, X.L.; Sun, X.H.; Dong, F.; Li, C.Y.; Li, P.Y.; Ding, H.; Chen, Y.; Zhang, Y.Z.; Wang, P. Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic. J. Biol. Chem. 2020, 295, 16380–16392. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Matsubara, Y.; Iwasaki, K.; Muramatsu, T. Action of poly (alpha-L-guluronate)lyase from Corynebacterium sp. ALY-1 strain on saturated oligoguluronates. Biosci. Biotechnol. Biochem. 1998, 62, 1055–1060. [Google Scholar] [CrossRef]
- Thomas, F.; Lundqvist, L.C.; Jam, M.; Jeudy, A.; Barbeyron, T.; Sandström, C.; Michel, G.; Czjzek, M. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 2013, 288, 23021–23037. [Google Scholar] [CrossRef] [Green Version]
- Ogura, K.; Yamasaki, M.; Mikami, B.; Hashimoto, W.; Murata, K. Substrate recognition by family 7 alginate lyase from Sphingomonas sp. A1. J. Mol. Biol. 2008, 380, 373–385. [Google Scholar] [CrossRef]
- Park, D.; Jagtap, S.; Nair, S.K. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J. Biol. Chem. 2014, 289, 8645–8655. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.R.; Zhang, Q.D.; Wang, S.M.; Guan, J.W.; Jiao, R.M.; Han, N.H.; Han, W.J.; Li, F.C. Biochemical characteristics and synergistic effect of two novel alginate lyases from Photobacterium sp. FC615. Biotechnol. Biofuels 2019, 12, 260. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, X.; Zhou, N.; Chen, Y.; Zhang, A.; Chen, K.; Ouyang, P. Property and function of a novel chitinase containing dual catalytic domains capable of converting chitin into N-Acetyl-D-Glucosamine. Front. Microbiol. 2022, 13, 790301. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Hao, Z.; Wang, K.; Tu, T.; Huang, H.; Wang, Y.; Bai, Y.G.; Wang, Y.; Luo, H.; Yao, B.; et al. The GH10 and GH48 dual-functional catalytic domains from a multimodular glycoside hydrolase synergize in hydrolyzing both cellulose and xylan. Biotechnol. Biofuels 2019, 12, 279. [Google Scholar] [CrossRef]
- Riedel, K.; Bronnenmeier, K. Intramolecular synergism in an engineered exo-endo-1,4-beta-glucanase fusion protein. Mol. Microbiol. 1998, 28, 767–775. [Google Scholar] [CrossRef]
- Haug, A.; Larsen, B.; Smidsrod, O. Studies on the sequence of uronic acid residues in alginic acid. Actachemscand 1967, 21, 691–704. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, H. Predicting secretory proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [PubMed] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Lu, S.N.; Wang, J.Y.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Orlando, M.; Pucciarelli, S.; Lotti, M. Endolysins from antarctic Pseudomonas display lysozyme activity at low temperature. Mar. Drugs 2020, 18, 579. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.M.; Hudgens, J.W.; Heselpoth, R.D.; Bales, P.M.; Nelson, D.C. Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A. PLoS ONE 2014, 9, e112939. [Google Scholar] [CrossRef] [PubMed]


| Enzyme | Km (mg/mL) | Vmax (U/mg) | kcat (s−1) | kcat/Km (s−1 mg−1 mL) |
|---|---|---|---|---|
| AlyC8-CD1 | 1.64 ± 0.24 | 24415.9 ± 1974.3 | 405.66 ± 32.81 | 247.35 |
| AlyC8-CD2 | 3.43 ± 0.52 | 2270.6 ± 160.2 | 7.81 ± 0.55 | 2.28 |
| Gene Product | Primer | Sequence (5′ to 3′) a |
|---|---|---|
| AlyC8 | AlyC8-F | AAGAAGGAGATATACATATGTGCGGTGGAAGCAGTTCAAA |
| AlyC8-R | TGGTGGTGGTGGTGCTCGAGGTTATGTTCAACCCTTAACG | |
| AlyC8-CD1 | AlyC8-CD1-F | AAGAAGGAGATATACATATGTGCGGTGGAAGCAGTTCAAA |
| AlyC8-CD1-R | TGGTGGTGGTGGTGCTCGAGGATACACCACAACCCAAATC | |
| AlyC8-CD2 | AlyC8-CD2-F | AAGAAGGAGATATACATATGATCAGTGGTTCAAACGATTG |
| AlyC8-CD2-R | TGGTGGTGGTGGTGCTCGAGGTTATGTTCAACCCTTAACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.-M.; Xue, Z.; Sun, M.-L.; Zhang, Y.; Zhang, Y.-Z.; Fu, H.-H.; Zhang, Y.-Q.; Wang, P. Characterization of a Novel Alginate Lyase with Two Alginate Lyase Domains from the Marine Bacterium Vibrio sp. C42. Mar. Drugs 2022, 20, 746. https://doi.org/10.3390/md20120746
Sun X-M, Xue Z, Sun M-L, Zhang Y, Zhang Y-Z, Fu H-H, Zhang Y-Q, Wang P. Characterization of a Novel Alginate Lyase with Two Alginate Lyase Domains from the Marine Bacterium Vibrio sp. C42. Marine Drugs. 2022; 20(12):746. https://doi.org/10.3390/md20120746
Chicago/Turabian StyleSun, Xiao-Meng, Zhao Xue, Mei-Ling Sun, Yi Zhang, Yu-Zhong Zhang, Hui-Hui Fu, Yu-Qiang Zhang, and Peng Wang. 2022. "Characterization of a Novel Alginate Lyase with Two Alginate Lyase Domains from the Marine Bacterium Vibrio sp. C42" Marine Drugs 20, no. 12: 746. https://doi.org/10.3390/md20120746
APA StyleSun, X.-M., Xue, Z., Sun, M.-L., Zhang, Y., Zhang, Y.-Z., Fu, H.-H., Zhang, Y.-Q., & Wang, P. (2022). Characterization of a Novel Alginate Lyase with Two Alginate Lyase Domains from the Marine Bacterium Vibrio sp. C42. Marine Drugs, 20(12), 746. https://doi.org/10.3390/md20120746






