Inflammation Regulation via an Agonist and Antagonists of α7 Nicotinic Acetylcholine Receptors in RAW264.7 Macrophages
Abstract
:1. Introduction
2. Results
2.1. Identification of nAChR Subunit Expression in RAW264.7 Cells
2.2. Differential Gene Expression Analysis of Various nAChR Subunits in Inflammatory RAW264.7 Cells
2.3. Synthesis of α7 nAChR Antagonist α-CTx [A10L]PnIA
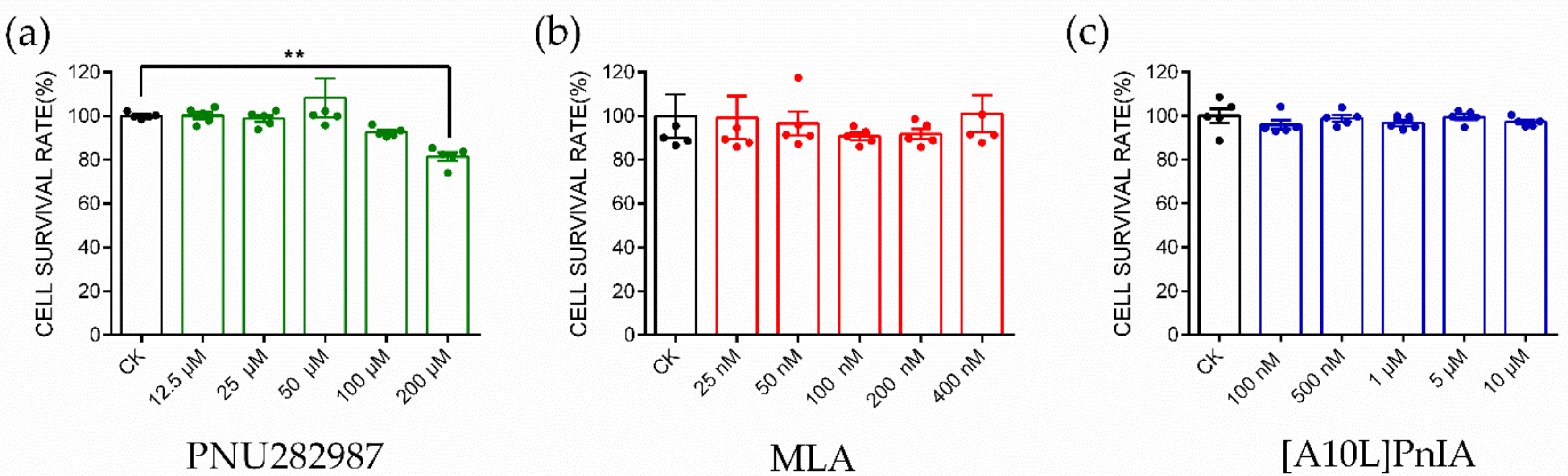
2.4. CCK-8 Assay to Identify Cytotoxicity
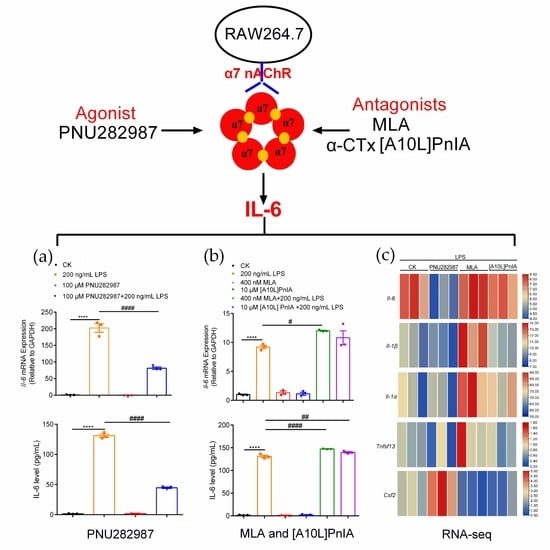
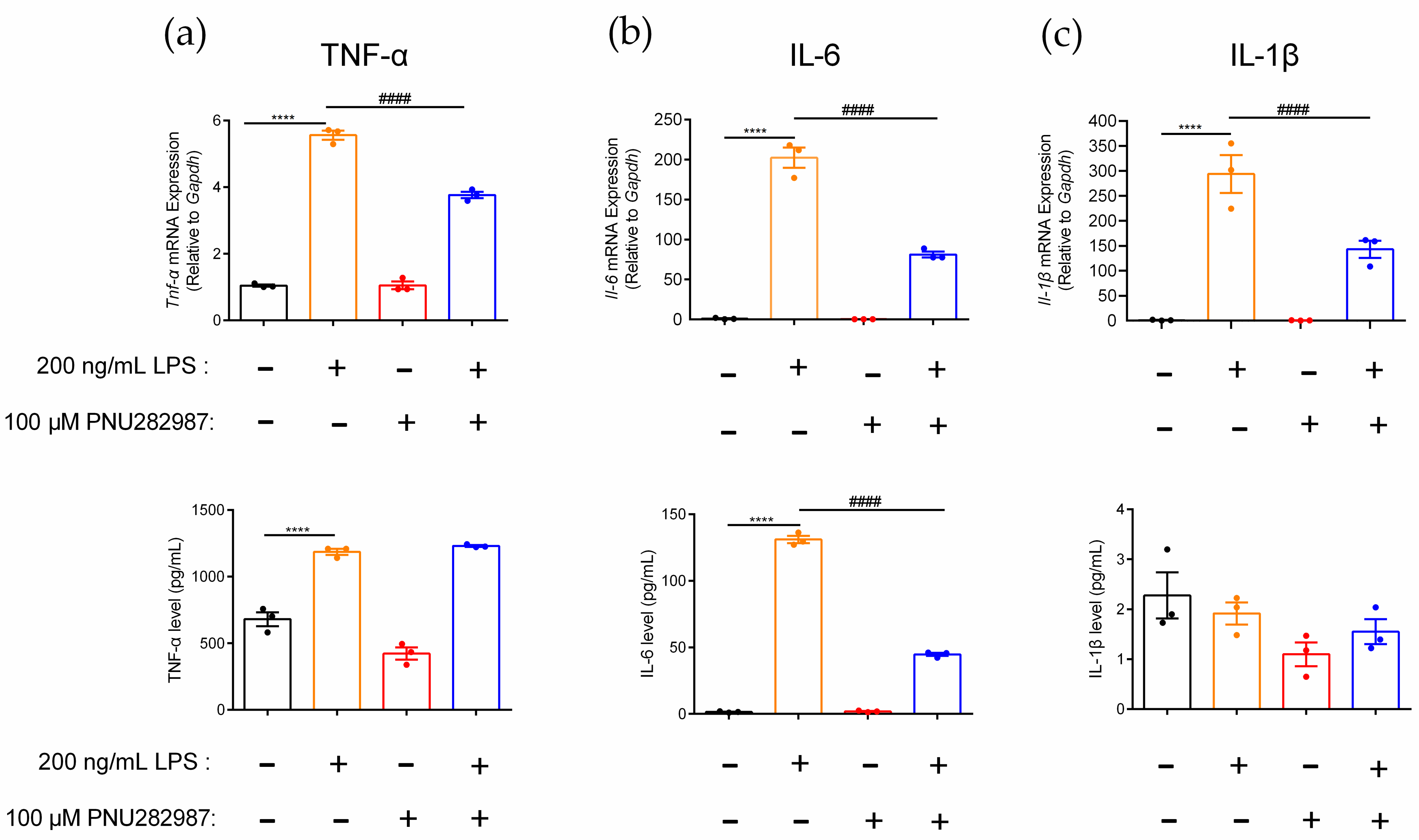
2.5. Regulation of Inflammatory Cytokines in RAW264.7 Cells Mediated through the α7 nAChR Agonist PNU282987
2.6. Regulation of Inflammatory Cytokines in RAW264.7 Cells via the α7 nAChR Antagonists MLA and α-CTx [A10L]PnIA
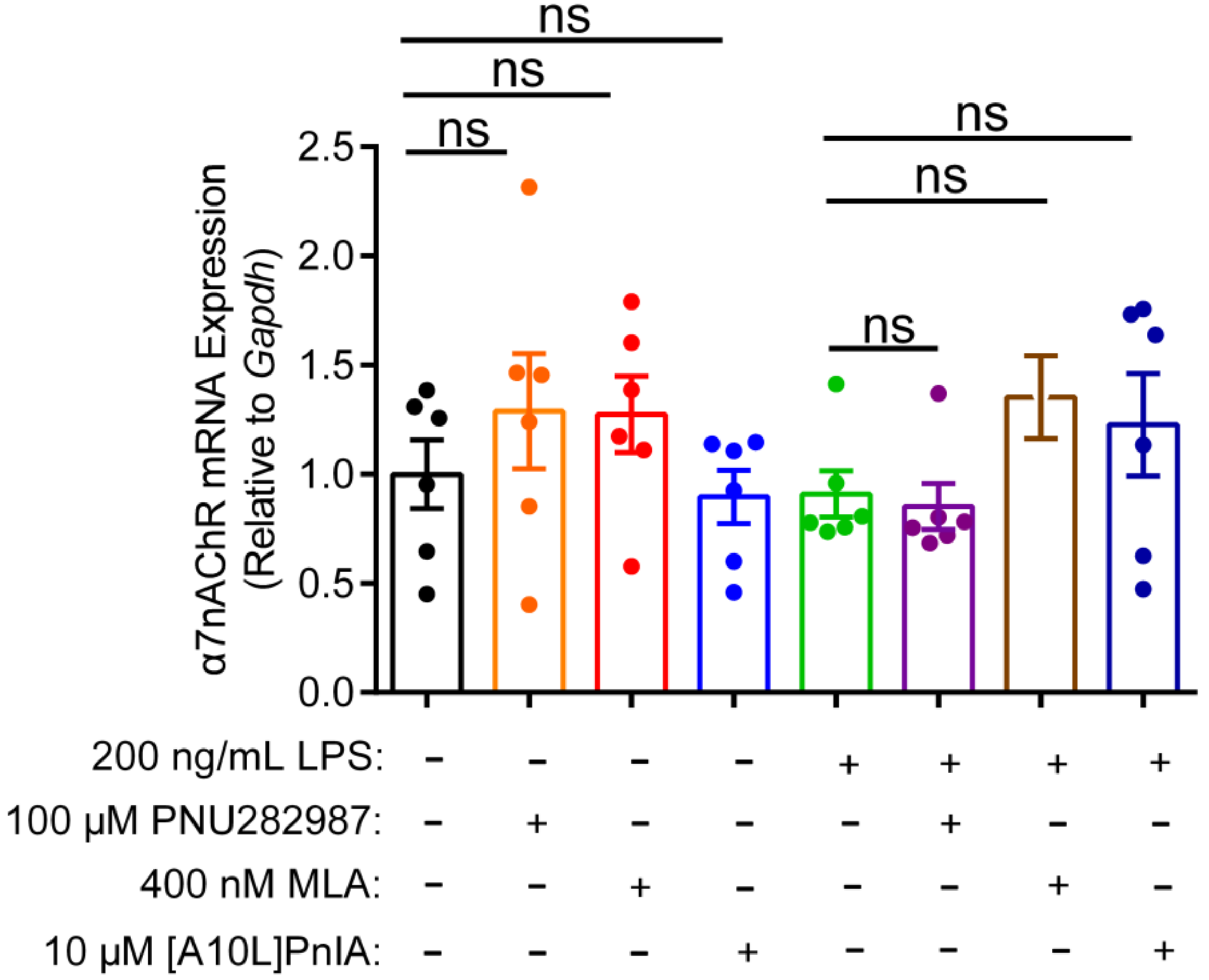
2.7. Effects of an Agonist and Antagonists on α7 nAChR Gene Expression in Inflammatory Macrophages RAW264.7
2.8. RNA-Seq Analysis of RAW264.7 Inflammatory Regulation Mediated via α7 nAChR Agonists and Antagonists
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Identification of nAChR Subunits on RAW264.7 Macrophages
4.4. Synthesis of the α7 nAChR Specific Antagonist [A10L]PnIA
4.5. CCK-8 Cytotoxicity Assay
4.6. qPCR and ELISAs
4.7. RNA-Seq Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| nAChR | Nicotinic acetylcholine receptor |
| ACh | Acetylcholine |
| CNS | Central nervous system |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| IκB | Inhibitor of nuclear factor kappa-B |
| NF-κB p65 | Nuclear factor kappa-B p65 |
| NF-κB | Nuclear factor kappa-B |
| JAK2 | Janus kinase 2 |
| JAK2/STAT3 | Janus Kinase 2/Signal Transducer and Activtor of Transcription 3 |
| IL-1β | Interleukin-1β |
| IL-6 | Interleukin-6 |
| IL-10 | Interleukin-10 |
| Il-2R | Interleukin-2 receptor |
| TNF-α | Tumor necrosis factor α |
| LPS | Lipopolysaccharide |
| MLA | Methyltaprotine |
| α-CTx [A10L]PnIA | α-conotoxin [A10L]PnIA |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| Tnfsf13 | Tumor Necrosis Factor Ligand Superfamily member 13 |
| Csf1 | Colony-stimulating factor 1 |
| Csf2 | Colony-stimulating factor 2 |
| GLAST | Glutamate transporter |
| IKK | Inhibitor of NF-kB (IkB) kinase |
| Arid5a | AT-rich interactive domain-containing protein 5a |
References
- Millar, N.S.; Gotti, C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology 2009, 56, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.B.; Bandyopadhyay, P.K.; Olivera, B.M.; McIntosh, J.M. αS-conotoxin GVIIIB potently and selectively blocks α9α10 nicotinic acetylcholine receptors. Biochem. Pharmacol. 2015, 96, 349–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabbani, N.; Nichols, R.A. Beyond the Channel: Metabotropic Signaling by Nicotinic Receptors. Trends Pharmacol. Sci. 2018, 39, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Kalkman, H.O.; Feuerbach, D. Modulatory effects of α7 nAChRs on the immune system and its relevance for CNS disorders. Cell. Mol. Life Sci. 2016, 73, 2511–2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zeng, X.; Hui, Y.; Zhu, C.; Wu, J.; Taylor, D.H.; Ji, J.; Fan, W.; Huang, Z.; Hu, J. Activation of α7 nicotinic acetylcholine receptors protects astrocytes against oxidative stress-induced apoptosis: Implications for Parkinson’s disease. Neuropharmacology 2015, 91, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.G.; Qian, Y.H. α7 nicotinic acetylcholine receptor and its effects on Alzheimer’s disease. Neuropeptides 2019, 73, 96–106. [Google Scholar] [CrossRef]
- Hajiasgharzadeh, K.; Somi, M.H.; Sadigh-Eteghad, S.; Mokhtarzadeh, A.; Shanehbandi, D.; Mansoori, B.; Mohammadi, A.; Doustvandi, M.A.; Baradaran, B. The dual role of α7 nicotinic acetylcholine receptor in inflammation-associated gastrointestinal cancers. Heliyon 2020, 6, e03611. [Google Scholar] [CrossRef]
- Ulleryd, M.A.; Mjörnstedt, F.; Panagaki, D.; Yang, L.J.; Engevall, K.; Gutiérrez, S.; Wang, Y.; Gan, L.M.; Nilsson, H.; Michaëlsson, E.; et al. Stimulation of α7 nicotinic acetylcholine receptor (α7 nAChR) inhibits atherosclerosis via immunomodulatory effects on myeloid cells. Atherosclerosis 2019, 287, 122–133. [Google Scholar] [CrossRef]
- Witayateeraporn, W.; Arunrungvichian, K.; Pothongsrisit, S.; Doungchawee, J.; Vajragupta, O.; Pongrakhananon, V. α7-Nicotinic acetylcholine receptor antagonist QND7 suppresses non-small cell lung cancer cell proliferation and migration via inhibition of Akt/mTOR signaling. Biochem. Biophys. Res. Commun. 2020, 521, 977–983. [Google Scholar] [CrossRef]
- Fujii, T.; Mashimo, M.; Moriwaki, Y.; Misawa, H.; Ono, S.; Horiguchi, K.; Kawashima, K. Expression and Function of the Cholinergic System in Immune Cells. Front. Immunol. 2017, 8, 1085. [Google Scholar] [CrossRef] [Green Version]
- Andersson, U. The cholinergic anti-inflammatory pathway alleviates acute lung injury. Mol. Med. 2020, 26, 64. [Google Scholar] [CrossRef]
- Mashimo, M.; Takeshima, S.; Okuyama, H.; Matsurida, A.; Murase, M.; Ono, S.; Kawashima, K.; Fujii, T. α7 nAChRs expressed on antigen presenting cells are insensitive to the conventional antagonists alpha-bungarotoxin and methyllycaconitine. Int. Immunopharmacol. 2020, 8, 106276. [Google Scholar] [CrossRef]
- De Jonge, W.J.; van der Zanden, E.P.; The, F.O.; Bijlsma, M.F.; van Westerloo, D.J.; Bennink, R.J.; Berthoud, H.R.; Uematsu, S.; Akira, S.; van den Wijngaard, R.M.; et al. Stimulation of the vagus nerve attenuates macrophage activation by activating the JAK2-STAT3 signaling pathway. Nat. Immunol. 2005, 6, 844–851. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Lv, Z.; Meng, L.; Xu, J.; Yuan, S.; Fu, Z. Activation of α7 Nicotinic Acetylcholine Receptors (α7 nAchR) Promotes the Protective Autophagy in LPS-Induced Acute Lung Injury (ALI) In Vitro and In Vivo. Inflammation 2019, 42, 2236–2245. [Google Scholar] [CrossRef]
- Egleton, R.D.; Brown, K.C.; Dasgupta, P. Nicotinic acetylcholine receptors in cancer: Multiple roles in proliferation and inhibition of apoptosis. Trends Pharmacol. Sci. 2008, 29, 151–158. [Google Scholar] [CrossRef]
- Paleari, L.; Catassi, A.; Ciarlo, M.; Cavalieri, Z.; Bruzzo, C.; Servent, D.; Cesario, A.; Chessa, L.; Cilli, M.; Piccardi, F.; et al. Role of α7 nicotinic acetylcholine receptor in human non-small cell lung cancer proliferation. Cell Prolif. 2008, 41, 936–959. [Google Scholar] [CrossRef]
- Bai, S.; Wen, W.; Hou, X.; Wu, J.; Yi, L.; Zhi, Y.; Lv, Y.; Tan, X.; Liu, L.; Wang, P.; et al. Inhibitory effect of sinomenine on lung cancer cells via negative regulation of α7 nicotinic acetylcholine receptor. J. Leukoc. Biol. 2021, 109, 843–852. [Google Scholar] [CrossRef]
- Dinarello, C.A.; Simon, A.; Van Der Meer, J.W.M. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat. Rev. Drug Discov. 2012, 11, 633–652. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.J.; Wang, L.; Ji, C.F.; Gu, S.F.; Yin, Q.; Zuo, J. The Role of α7 nAChR-Mediated Cholinergic Anti-inflammatory Pathway in Immune Cells. Inflammation 2021, 44, 821–834. [Google Scholar] [CrossRef]
- Bajpai, G.; Schneider, C.; Wong, N.; Bredemeyer, A.; Hulsmans, M.; Nahrendorf, M.; Epelman, S.; Kreisel, D.; Liu, Y.; Itoh, A.; et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat. Med. 2018, 24, 1234–1245. [Google Scholar] [CrossRef]
- França, C.N.; Izar, M.C.O.; Hortêncio, M.N.S.; do Amaral, J.B.; Ferreira, C.E.S.; Tuleta, I.D.; Fonseca, F.A.H. Monocyte subtypes and the CCR2 chemokine receptor in cardiovascular disease. Clin. Sci. 2017, 131, 1215–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epelman, S.; Lavine, K.J.; Beaudin, A.E.; Sojka, D.K.; Carrero, J.A.; Calderon, B.; Brija, T.; Gautier, E.L.; Ivanov, S.; Satpathy, A.T.; et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 2014, 40, 91–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funes, S.C.; Rios, M.; Escobar-Vera, J.; Kalergis, A.M. Implications of macrophage polarization in autoimmunity. Immunology 2018, 154, 186–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginhoux, F.; Schultze, J.L.; Murray, P.J.; Ochando, J.; Biswas, S.K. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 2016, 17, 34–40. [Google Scholar] [CrossRef]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef] [Green Version]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS−) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Barrett, T.J. Macrophages in Atherosclerosis Regression. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 20–33. [Google Scholar] [CrossRef]
- Shaikh, S.; Brittenden, J.; Lahiri, R.; Brown, P.A.; Thies, F.; Wilson, H.M. Macrophage subtypes in symptomatic carotid artery and femoral artery plaques. Eur. J. Vasc. Endovasc. Surg. 2012, 44, 491–497. [Google Scholar] [CrossRef]
- Pirola, L.; Ferraz, J.C. Role of pro- and anti-inflammatory phenomena in the physiopathology of type 2 diabetes and obesity. World J. Biol. Chem. 2017, 8, 120–128. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.; Zhu, L.; Yang, X.; He, F.; Wang, T.; Bao, T.; Lu, H.; Wang, H.; Yang, S. Inulin alleviates inflammation of alcoholic liver disease via SCFAs-inducing suppression of M1 and facilitation of M2 macrophages in mice. Int. Immunopharmacol. 2020, 78, 106062. [Google Scholar] [CrossRef]
- Wallace, T.L.; Porter, R.H.P. Targeting the nicotinic α7 acetylcholine receptor to enhance cognition in disease. Biochem. Pharmacol. 2011, 82, 891–903. [Google Scholar] [CrossRef]
- Morioka, N.; Hisaoka-Nakashima, K.; Nakata, Y. Regulation by Nicotinic Acetylcholine Receptors of Microglial Glutamate Transporters: Role of Microglia in Neuroprotection. In Nicotinic Acetylcholine Receptor Signaling in Neuroprotection; Springer: Singapore, 2018; pp. 73–88. [Google Scholar]
- Blank, T.; Prinz, M. Microglia as modulators of cognition and neuropsychiatric disorders. Glia 2013, 61, 62–70. [Google Scholar] [CrossRef]
- Feuerbach, D.; Lingenhoehl, K.; Olpe, H.R.; Vassout, A.; Gentsch, C.; Chaperon, F.; Nozulak, J.; Enz, A.; Bilbe, G.; McAllister, K.; et al. The selective nicotinic acetylcholine receptor α7 agonist JN403 is active in animal models of cognition, sensory gating, epilepsy and pain. Neuropharmacology 2009, 56, 254–263. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Z.; Xue, B.; Shi, H. Activation of the cholinergic antiinflammatory pathway ameliorates obesity-induced inflammation and insulin resistance. Endocrinology 2011, 152, 836–846. [Google Scholar] [CrossRef] [Green Version]
- Dash, P.K.; Zhao, J.; Kobori, N.; Redell, J.B.; Hylin, M.J.; Hood, K.N.; Moore, A.N. Activation of α7 Cholinergic Nicotinic Receptors Reduce Blood-Brain Barrier Permeability following Experimental Traumatic Brain Injury. J. Neurosci. 2016, 36, 2809–2818. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Sun, Q.; Gao, X.; Jiang, Y.; Li, R.; Ye, J. Anti-inflammation of spirocyclopiperazinium salt compound LXM-10 targeting α7 nAChR and M4 mAChR and inhibiting JAK2/STAT3 pathway in rats. PLoS ONE 2013, 8, e66895. [Google Scholar] [CrossRef]
- Han, Q.Q.; Deng, M.Y.; Liu, H.; Ali, U.; Li, X.Y.; Wang, Y.X. Cynandione A and PHA-543613 inhibit inflammation and stimulate macrophageal IL-10 expression following α7 nAChR activation. Biochem. Pharmacol. 2021, 190, 114600. [Google Scholar] [CrossRef]
- Kishimoto, T. IL-6: From its discovery to clinical applications. Int. Immunol. 2010, 22, 347–352. [Google Scholar] [CrossRef] [Green Version]
- Lokau, J.; Schoeder, V.; Haybaeck, J.; Garbers, C. Jak-Stat Signaling Induced by Interleukin-6 Family Cytokines in Hepatocellular Carcinoma. Cancers 2019, 11, 1704. [Google Scholar] [CrossRef] [Green Version]
- Yahagi, A.; Saika, T.; Hirano, H.; Takai-Imamura, M.; Tsuji, F.; Aono, H.; Iseki, M.; Morita, Y.; Igarashi, H.; Saeki, Y.; et al. IL-6-PAD4 axis in the earliest phase of arthritis in knock-in gp130F759 mice, a model for rheumatoid arthritis. RMD Open 2019, 5, e000853. [Google Scholar] [CrossRef]
- Liu, C.; Fang, C.; He, Q.; Xie, L. The value of interleukin-6 (IL-6) within 6 h after birth in the prompt diagnosis of early-onset neonatal sepsis. Transl. Pediatr. 2020, 9, 629–635. [Google Scholar] [CrossRef]
- Ma, J.; Sun, L.; Liu, Y.; Ren, H.; Shen, Y.; Bi, F.; Zhang, T.; Wang, X. Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. 2020, 20, 82. [Google Scholar] [CrossRef]
- Armishaw, C.; Jensen, A.A.; Balle, T.; Clark, R.J.; Harpsøe, K.; Skonberg, C.; Liljefors, T.; Strømgaard, K. Rational design of alpha-conotoxin analogues targeting α7 nicotinic acetylcholine receptors: Improved antagonistic activity by incorporation of proline derivatives. J. Biol. Chem. 2009, 284, 9498–9512. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Palanisamy, V.; Jakymiw, A.; Van Tubergen, E.A.; D’Silva, N.J.; Kirkwood, K.L. Control of cytokine mRNA expression by RNA-binding proteins and microRNAs. J. Dent. Res. 2012, 91, 651–658. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, H.; Takeuchi, O.; Teraguchi, S.; Matsushita, K.; Uehata, T.; Kuniyoshi, K.; Satoh, T.; Saitoh, T.; Matsushita, M.; Standley, D.M.; et al. The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat. Immunol. 2011, 12, 1167–1175. [Google Scholar] [CrossRef]
- Masuda, K.; Ripley, B.; Nishimura, R.; Mino, T.; Takeuchi, O.; Shioi, G.; Kiyonari, H.; Kishimoto, T. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 9409–9414. [Google Scholar] [CrossRef] [Green Version]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 123–147. [Google Scholar] [CrossRef] [Green Version]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.Y.; Xu, J.Y.; Shi, X.Y.; Huang, W.; Ruan, T.Y.; Xie, P.; Ding, J.L. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Investig. 2013, 93, 844–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitu, V.; Gokhan, S.; Nandi, S.; Mehler, M.F.; Stanley, E.R. Emerging Roles for CSF-1 Receptor and its Ligands in the Nervous System. Trends Neurosci. 2016, 39, 378–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waller, E.K. The role of sargramostim (rhGM-CSF) as immunotherapy. Oncologist 2007, 12 (Suppl. 2), 22–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.; Zhangsun, D.; Wu, Y.; Zhu, X.; Hu, Y.; McIntyre, M.; Christensen, S.; Akcan, M.; Craik, D.J.; McIntosh, J.M. Characterization of a novel α-conotoxin from conus textile that selectively targets α6/α3β2β3 nicotinic acetylcholine receptors. J. Biol. Chem. 2013, 288, 894–902. [Google Scholar] [CrossRef] [Green Version]

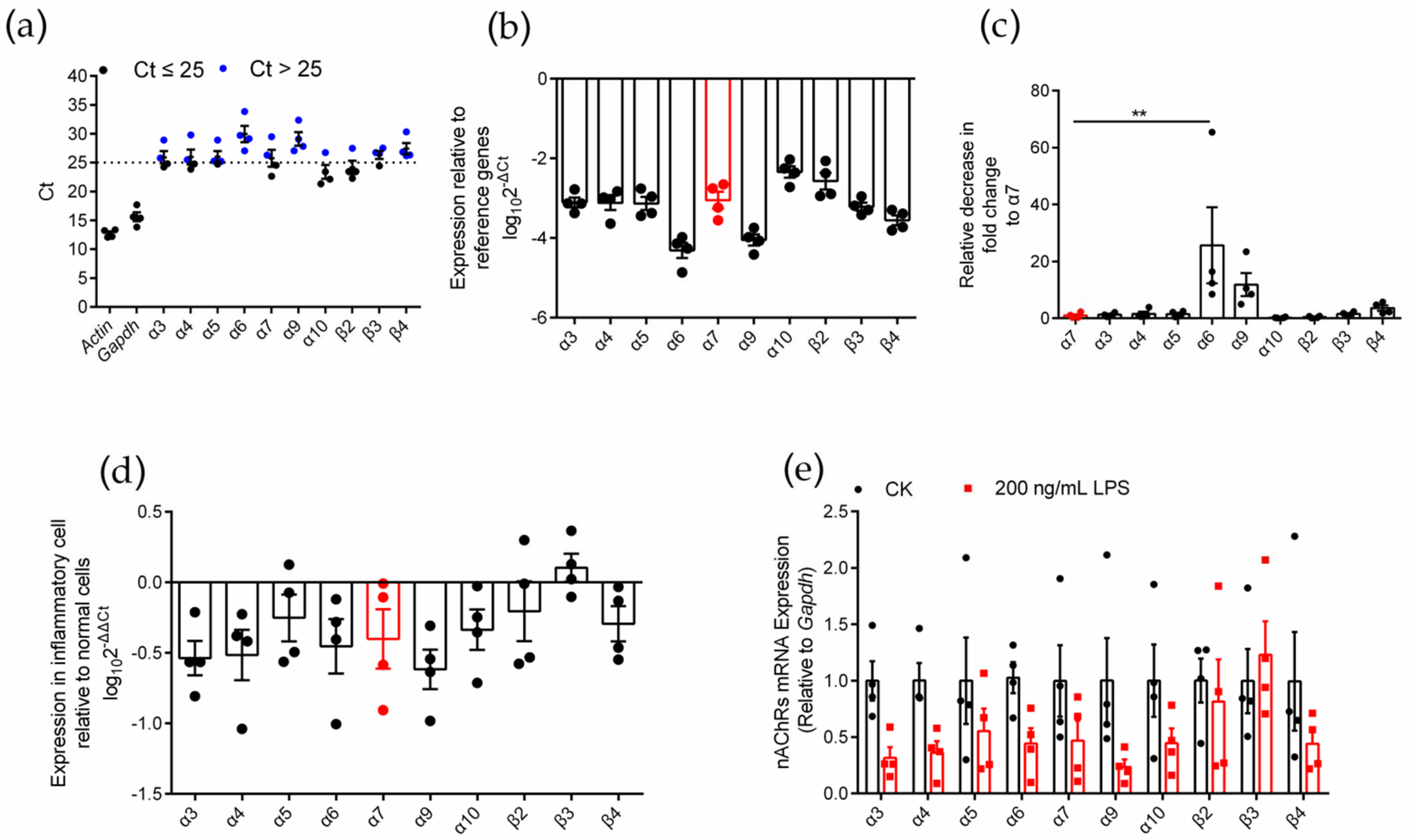



| Normal Cell | Inflammatory Cell | Inflammatory Cell Relative to Normal Cell | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Ct | n | log102−(ΔCt) | Relative decrease in fold change to α7 | Ct | n | log102-(ΔCt) | Relative decrease in fold change to α7 | log102−(ΔΔCt) | Fold-difference |
| Actin | 13.0 ± 0.7 | 4 | nd | nd | 12.7 ± 0.3 | 4 | nd | nd | nd | nd |
| Gapdh | 15.5 ± 0.9 | 4 | nd | nd | 15.6 ± 0.8 | 4 | nd | nd | nd | nd |
| Chrna-3 | 24.1 ± 0.3 | 4 | −2.6 ± 0.1 | 0.9 ± 0.1 ns | 25.9 ± 1.0 | 4 | −3.1 ± 0.1 | 1.3 ± 0.3 ns | −0.5 ± 0.1 | −3.9 ± 1.0 ns |
| Chrna-4 | 24.2 ± 0.5 | 4 | −2.6 ± 0.1 | 0.9 ± 0.1 ns | 26.0 ± 1.3 | 4 | −3.1 ± 0.2 | 1.6 ± 0.8 ns | −0.5 ± 0.2 | −4.4 ± 2.2 ns |
| Chrna-5 | 25.1 ± 0.5 | 4 | −2.9 ± 0.2 | 2.2 ± 0.8 ns | 26.0 ± 1.0 | 4 | −3.1 ± 0.2 | 1.5 ± 0.5 ns | −0.3 ± 0.2 | −1.7 ± 1.1 ns |
| Chrna-6 | 28.3 ± 0.6 | 4 | −3.9 ± 0.1 | 16.8 ± 2.6 **** | 29.9 ± 1.4 | 4 | −4.3 ± 0.2 | 25.7 ± 13.3 ** | −0.5 ± 0.2 | −3.0 ± 2.1 ns |
| Chrna-7 | 24.3 ± 0.7 | 4 | −2.6 ± 0.1 | 1.0 ± 0.2 | 25.8 ± 1.5 | 4 | −3.0 ± 0.2 | 1.0 ± 0.5 | −0.4 ± 0.2 | −3.5 ± 1.6 ns |
| Chrna-9 | 26.9 ± 0.4 | 4 | −3.4 ± 0.1 | 7.0 ± 1.7 * | 29.1 ± 1.2 | 4 | −4.0 ± 0.1 | 11.8 ± 4.0 ns | −0.6 ± 0.1 | −4.7 ± 1.7 ns |
| Chrna-10 | 22.2 ± 1.0 | 4 | −2.0 ± 0.2 | 0.3 ± 0.1 ns | 23.4 ± 1.2 | 4 | −2.3 ± 0.1 | 0.2 ± 0.1 ns | −0.3 ± 0.1 | −2.6 ± 0.9 ns |
| Chrnb-2 | 23.4 ± 0.8 | 4 | −2.4 ± 0.1 | 0.6 ± 0.2 ns | 24.2 ± 1.1 | 4 | −2.6 ± 0.2 | 0.5 ± 0.2 ns | −0.2 ± 0.2 | −1.6 ± 1.3 ns |
| Chrnb-3 | 26.5 ± 0.4 | 4 | −3.3 ± 0.1 | 5.2 ± 1.2 ns | 26.3 ± 0.7 | 4 | −3.2 ± 0.1 | 1.6 ± 0.3 ns | 0.1 ± 0.1 | 0.9 ± 0.8 ns |
| Chrnb-4 | 26.4 ± 0.4 | 4 | −3.3 ± 0.2 | 5.1 ± 1.7 ns | 27.4 ± 1.0 | 4 | −3.6 ± 0.1 | 3.6 ± 1.0 ns | −0.3 ± 0.1 | −2.2 ± 0.6 ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Y.; Chu, Z.; Shan, H.; Zhangsun, D.; Zhu, X.; Luo, S. Inflammation Regulation via an Agonist and Antagonists of α7 Nicotinic Acetylcholine Receptors in RAW264.7 Macrophages. Mar. Drugs 2022, 20, 200. https://doi.org/10.3390/md20030200
Tan Y, Chu Z, Shan H, Zhangsun D, Zhu X, Luo S. Inflammation Regulation via an Agonist and Antagonists of α7 Nicotinic Acetylcholine Receptors in RAW264.7 Macrophages. Marine Drugs. 2022; 20(3):200. https://doi.org/10.3390/md20030200
Chicago/Turabian StyleTan, Yao, Zhaoli Chu, Hongyu Shan, Dongting Zhangsun, Xiaopeng Zhu, and Sulan Luo. 2022. "Inflammation Regulation via an Agonist and Antagonists of α7 Nicotinic Acetylcholine Receptors in RAW264.7 Macrophages" Marine Drugs 20, no. 3: 200. https://doi.org/10.3390/md20030200
APA StyleTan, Y., Chu, Z., Shan, H., Zhangsun, D., Zhu, X., & Luo, S. (2022). Inflammation Regulation via an Agonist and Antagonists of α7 Nicotinic Acetylcholine Receptors in RAW264.7 Macrophages. Marine Drugs, 20(3), 200. https://doi.org/10.3390/md20030200