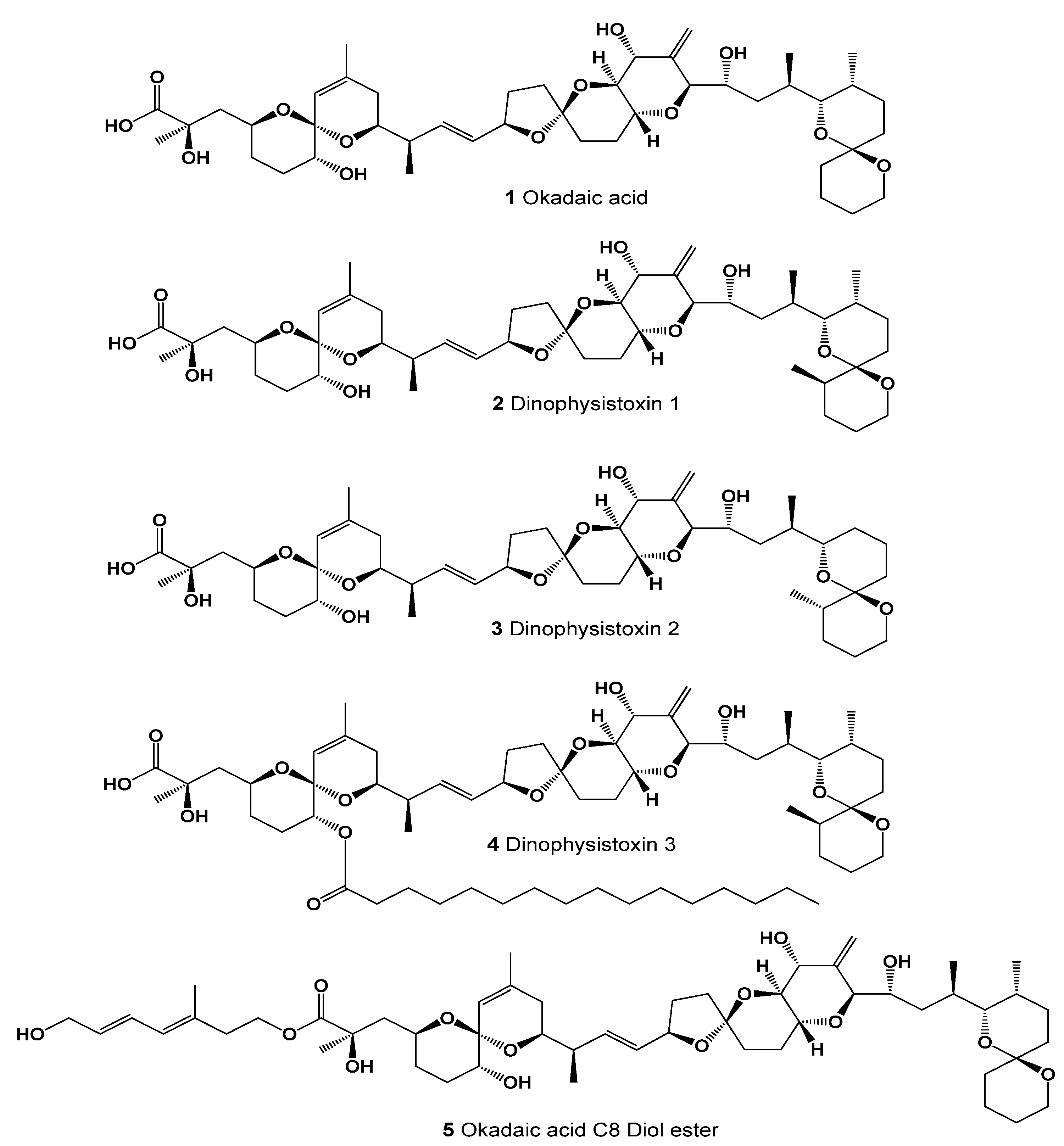
Natural Polyether Ionophores and Their Pharmacological Profile
Abstract
1. Introduction
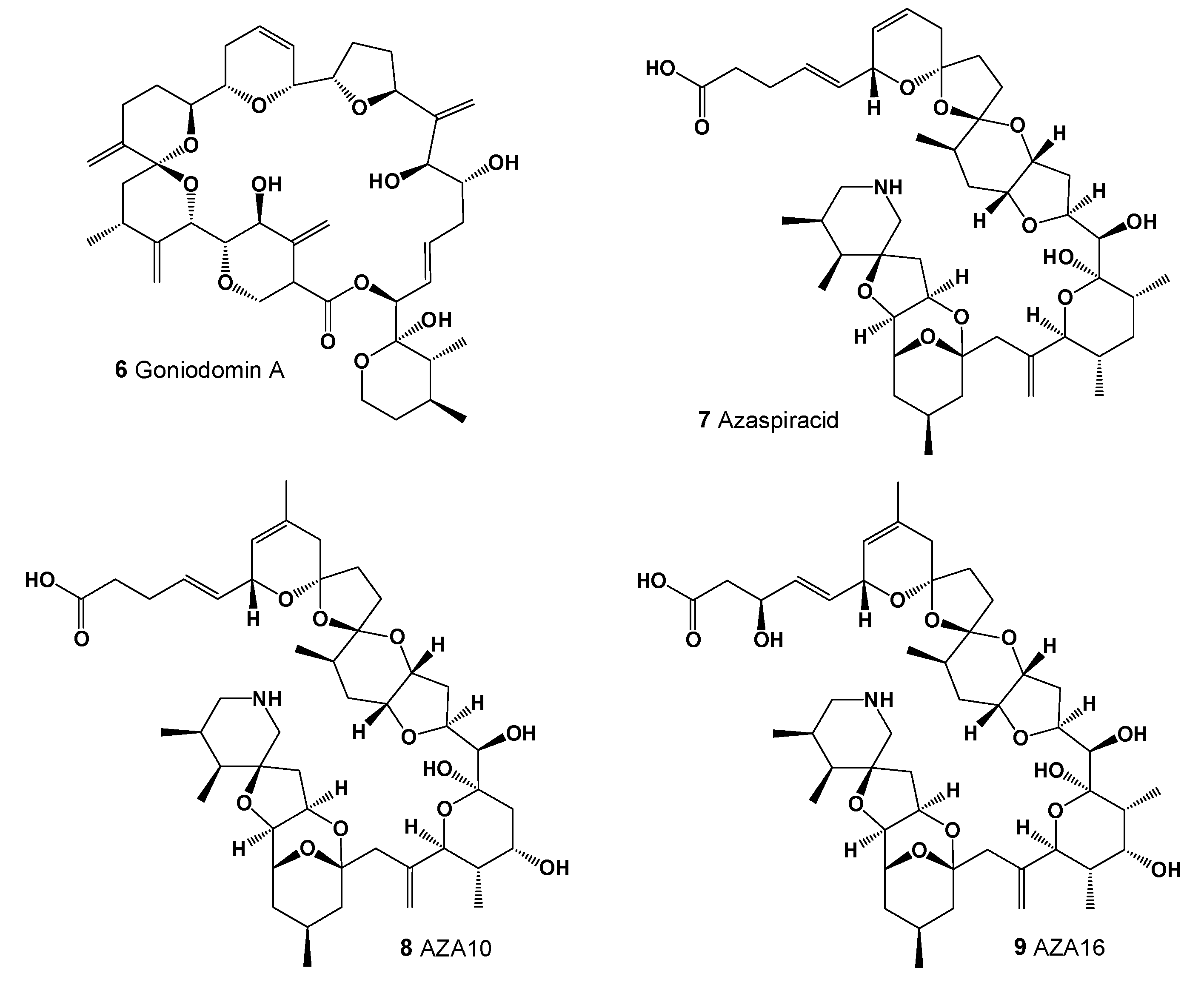
2. Polyether Metabolites Produced by Dinoflagellates
3. Polyether Ionophores Derived from Marine Algae and Invertebrates
4. Polyether Ionophores Produced by Actinomycetes
5. Structure–Activity Relationships and Biological Activities of Natural Polyether Ionophores
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Dembitsky, V.M. Astonishing diversity of natural surfactants: 2. Polyether glycosidic ionophores and macrocyclic glycosides. Lipids 2005, 40, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M. Chemistry and biodiversity of the biologically active natural glycosides. Chem. Biodivers. 2004, 1, 673–781. [Google Scholar] [CrossRef]
- Westley, J.W. Polyether Antibiotics: Naturally Occurring Acid Ionophores; Marcel Dekker Inc.: New York, NY, USA, 1982. [Google Scholar]
- Fuwa, H. Synthesis-driven stereochemical assignment of marine polycyclic ether natural products. Mar. Drugs 2021, 19, 257. [Google Scholar] [CrossRef] [PubMed]
- Chekan, J.R.; Fallon, T.R.; Moore, B.S. Biosynthesis of marine toxins. Curr. Opin. Chem. Biol. 2020, 59, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, J.J.; Souto, M.L.; Norte, M. Marine polyether triterpenes. Nat. Prod. Rep. 2000, 17, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Vilotijevic, I.; Jamison, T.F. Epoxide-opening cascades in the synthesis of polycyclic polyether natural products. Angew. Chem. Int. Ed. 2009, 48, 5250–5281. [Google Scholar] [CrossRef]
- Nakanishi, K. The chemistry of brevetoxins: A review. Toxicon 1985, 23, 473–479. [Google Scholar] [CrossRef]
- Vilotijevic, I.; Jamison, T.E. Synthesis of marine polycyclic polyethers via endo-selective epoxide-opening cascades. Mar. Drugs 2010, 8, 763–809. [Google Scholar] [CrossRef]
- Satake, M. Marine polyether compounds. In Marine Natural Products. Topics in Heterocyclic Chemistry; Kiyota, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 5. [Google Scholar] [CrossRef]
- Wan, X.; Yao, G.; Liu, Y.; Chen, J.; Jiang, H. Research progress in the biosynthetic mechanisms of marine polyether toxins. Mar. Drugs 2019, 17, 594. [Google Scholar] [CrossRef]
- Whittle, K.; Gallacher, S. Marine toxins. British Med. Bull. 2000, 56, 236–253. [Google Scholar] [CrossRef]
- Van Dolah, F.M. Marine algal toxins: Origins, health effects, and their increased occurrence. Environ. Health Perspect. 2000, 108, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Kellmann, R.; Stüken, A.; Orr, R.J.; Svendsen, H.M. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs 2010, 8, 1011–1048. [Google Scholar] [CrossRef] [PubMed]
- Rein, K.S.; Borrone, J. Polyketides from dinoflagellates: Origins, pharmacology, and biosynthesis. Comp. Biochem. Physiol. 1999, 124B, 117–131. [Google Scholar] [CrossRef]
- Nagai, H.; Satake, M.; Yasumoto, T. Antimicrobial activities of polyether compounds of dinoflagellate origins. J. Appl. Phycol. 1990, 2, 305–308. [Google Scholar] [CrossRef]
- Backer, L.C.; McGillicuddy, D.J., Jr. Harmful algal blooms: At the interface between coastal oceanography and human health. Oceanography 2006, 19, 94–106. [Google Scholar] [CrossRef]
- Dolah, F.M.V.; Roelke, D.; Greene, R.M. Health, and ecological impacts of harmful algal blooms: Risk assessment needs. Hum. Ecol. Risk Assess. Int. J. 2001, 7, 329–1345. [Google Scholar] [CrossRef]
- Ganesan, P.; Devadharshini, S. Marine toxins—A potential threat to human life. Biot. Res. Today 2021, 3, 797–799. [Google Scholar]
- Poroikov, V.V. Computer-aided drug design: From discovery of novel pharmaceutical agents to systems pharmacology. Biochemistry 2020, 14, 216–227. [Google Scholar]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.V. QSAR without borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Gloriozova, T.A.; Poroikov, V.V. Antitumor profile of carbon-bridged steroids (CBS) and triterpenoids. Mar. Drugs 2021, 19, 324. [Google Scholar] [CrossRef]
- Fusetani, N. Toxins produced by marine organisms. J. Synth. Org. Chem. Japan 1986, 87, 674–685. [Google Scholar] [CrossRef]
- Tachibana, K. Structural Studies on Marine Toxins. Ph.D. Thesis, Hawaii University, Honolulu, HI, USA, 1980. [Google Scholar]
- Scheuer, P.J. Some marine ecological phenomena: Chemical basis and biomedical potential. Science 1990, 248, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Hallegraeff, G.M. Harmful algal blooms: A global overview. In Manual on Harmful Marine Microalgae; Hallegraeff, G.M., Anderson, D.M., Cembella, A.D., Eds.; UNESCO: Paris, France, 1995; pp. 1–22. [Google Scholar]
- Silva, M.; Barreiro, A.; Rodriguez, P.; Otero, P.; Azevedo, J.; Alfonso, A.; Botana, L.M.; Vasconcelos, V. New invertebrate vectors for PST, spirolides and okadaic acid in the North Atlantic. Mar. Drugs 2013, 11, 1936–1960. [Google Scholar] [CrossRef] [PubMed]
- Sassolas, A.; Hayat, A.; Catanante, G.; Marty, J.L. Detection of the marine toxin okadaic acid: Assessing seafood safety. Talanta 2013, 105, 306–316. [Google Scholar] [CrossRef]
- Valdiglesias, V.; Prego-Faraldo, M.V.; Pásaro, E.; Méndez, J.; Laffon, B. Okadaic acid: More than a diarrheic toxin. Mar. Drugs 2013, 11, 4328–4349. [Google Scholar] [CrossRef]
- Corriere, M.; Soliño, L.; Costa, P.R. Effects of the marine biotoxins okadaic acid and dinophysistoxins on Fish. J. Mar. Sci. Eng. 2021, 9, 293. [Google Scholar] [CrossRef]
- Fernandez, J.; Suarez-Gomez, B.; Souto, M.L.; Norte, M. Identification of new okadaic acid derivatives from laboratory cultures of Prorocentrum lima. J. Nat. Prod. 2003, 66, 1294–1296. [Google Scholar] [CrossRef]
- López-Rosales, L.; Gallardo-Rodríguez, J.; Sánchez-Mirón, A.; Cerón-García, M.; Belarbi, E.; García-Camacho, F.; Molina-Grima, E. Simultaneous effect of temperature and irradiance on growth and okadaic acid production from the marine dinoflagellate Prorocentrum belizeanum. Toxins 2014, 6, 229–253. [Google Scholar] [CrossRef]
- Morton, S.L.; Moeller, P.D.R.; Young, K.A.; Lanoue, B. Okadaic acid production from the marine dinoflagellate Prorocentrum belizeanum Faust isolated from the Belizean coral reef ecosystem. Toxicon 1998, 36, 201–206. [Google Scholar] [CrossRef]
- Hu, W.; Xu, J.; Sinkkonen, J.; Wu, J. Polyketides from marine dinoflagellates of the genus Prorocentrum, biosynthetic origin and bioactivity of their okadaic acid analogues. Mini-Rev. Med. Chem. 2010, 10, 51–56. [Google Scholar]
- Reizopoulou, S.; Strogyloudi, E.; Giannakourou, A. Okadaic acid accumulation in macrofilter feeders subjected to natural blooms of Dinophysis acuminate. Harmful Algae 2008, 7, 228–234. [Google Scholar] [CrossRef]
- Yasumoto, T.; Oshima, Y.; Sugawara, W.; Fukuyo, Y.; Oguri, H.; Igarashi, T.; Fujita, N. Identification of Dinophysis fortii as the causative organism of diarrhetic shellfish poisoning. Bull. Jpn. Soc. Sci. Fish. 1980, 46, 1405–1411. [Google Scholar] [CrossRef]
- Reguera, B.; Riobó, P.; Rodríguez, F.; Díaz, P.A.; Pizarro, G.; Paz, B.; Franco, J.M.; Blanco, J. Dinophysis toxins: Causative organisms, distribution, and fate in shellfish. Mar. Drugs 2014, 12, 394–461. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.M.; Marr, J.; De Freitas, A.S.W.; Quilliam, M.A.; Walter, J.A.; Wright, J.L.C.; Pleasance, S. New diol esters isolated from cultures of the dinoflagellates Prorocentrum lima and Prorocentrum concavum. J. Nat. Prod. 1992, 55, 1631–1637. [Google Scholar] [CrossRef]
- Pizarro, G.; Paz, B.; Franco, J.M.; Suzuki, T.; Reguera, B. First detection of pectenotoxin-11 and confirmation of OA-D8 diol-ester in Dinophysis acuta from European waters by LC-MS/MS. Toxicon 2008, 52, 889–896. [Google Scholar] [CrossRef]
- Miles, C.O.; Wilkins, A.L.; Munday, R.; Dines, M.H.; Hawkes, A.D.; Briggs, L.R.; Sandvik, M.; Jensen, D.J.; Cooney, J.M.; Holland, P.T. Isolation of pectenotoxin-2 from Dinophysis acuta and its conversion to pectenotoxin-2 seco acid, and preliminary assessment of their acute toxicities. Toxicon 2004, 43, 1–9. [Google Scholar] [CrossRef]
- Miles, C.O.; Wilkins, A.L.; Hawkes, A.D.; Jensen, D.J.; Cooney, J.M.; Larsen, K.; Petersen, D.; Rise, F.; Beuzenberg, V.; Lincoln MacKenzie, A. Isolation, and identification of a cis-C8-diol-ester of okadaic acid from Dinophysis acuta in New Zealand. Toxicon 2006, 48, 195–203. [Google Scholar] [CrossRef]
- Uchida, H.; Watanabe, R.; Matsushima, R.; Oikawa, H.; Nagai, S. Toxin profiles of okadaic acid analogues and other lipophilic toxins in Dinophysis from Japanese coastal waters. Toxins 2018, 10, 457. [Google Scholar] [CrossRef]
- Murakami, M.; Makabe, K.; Yamaguchi, K.; Konosu, S.; Walchli, M.R. Goniodomin a, a novel polyether macrolide from the dinoflagellate Goniodoma pseudogoniaulax. Tetrahedron Lett. 1988, 29, 1149–1152. [Google Scholar] [CrossRef]
- Kita, T.; Fukuyo, Y. Description of the gonyaulcoid dinoflagellate Alexandrium hiranoi sp. nov. inhabiting tidepools on Japanese Pacific coast. Bull. Plankton Soc. Jpn. 1988, 1, 1–7. [Google Scholar]
- Murakami, M.; Okita, Y.; Matsuda, H.; Okino, T.; Yamaguchi, K. From the dinoflagellate Alexandrium hiranoi. Phytochemistry 1998, 48, 85–88. [Google Scholar] [CrossRef]
- MacKenzie, A.L.; De Salas, M.; Adamson, J.; Beuzenberg, V. The dinoflagellate genus Alexandrium (Halim) in New Zealand coastal waters: Comparative morphology, toxicity, and molecular genetics. Harmful Algae 2004, 3, 71–92. [Google Scholar] [CrossRef]
- Bravo, I.; Garcés, E.; Diogène, J.; Fraga, S.; Sampedro, N.; Figueroa, R.I. Resting cysts of the toxigenic dinoflagellate genus Alexandrium in recent sediments from the Western Mediterranean coast, including the first description of cysts of A. kutnerae and A. peruvianum. Eur. J. Phycol. 2006, 41, 293–302. [Google Scholar] [CrossRef]
- Hsia, M.H.; Morton, S.L.; Smith, L.L.; Beauchesne, K.R.; Huncik, K.M.; Moeller, P.D.R. Production of goniodomin A by the planktonic, chain-forming dinoflagellate Alexandrium monilatum (Howell) Balech isolated from the gulf coast of the United States. Harmful Algae 2006, 5, 290–299. [Google Scholar] [CrossRef]
- Zmerli Triki, H.; Laabir, M.; Moeller, P.; Chomérat, N.; Daly-Yahia, O.K. First report of goniodomin A production by the dinoflagellate Alexandrium pseudogonyaulax developing in southern Mediterranean (Bizerte Lagoon, Tunisia). Toxicon 2016, 111, 91–99. [Google Scholar] [CrossRef]
- Abe, M.; Inoue, D.; Matsunaga, K.; Ohizumi, Y.; Ueda, H.; Asano, T.; Murakami, M.; Sato, M. Goniodomin A, an antifungal polyether macrolide, exhibits antiangiogenic activities via inhibition of actin reorganization in endothelial cells. J. Cell. Physiol. 2002, 190, 109–116. [Google Scholar] [CrossRef]
- Twiner, M.J.; Rehmann, N.; Hess, P.; Doucette, G.J. Azaspiracid shellfish poisoning: A review on the chemistry, ecology, and toxicology with an emphasis on human health impacts. Mar. Drugs 2008, 6, 39–72. [Google Scholar] [CrossRef]
- Twiner, M.J.; Hess, P.; Doucette, G.J. Azaspiracids: Toxicology, pharmacology, and risk assessment. In Seafood and Freshwater Toxins, 3rd ed.; Botana, L., Ed.; CRC Press: Boca Raton, FL, USA, 2013; Chapter 28. [Google Scholar]
- Krock, B.; Tillmann, U.; John, U.; Cembella, A.D. Characterization of azaspiracids in plankton size-fractions and isolation of an azaspiracid-producing dinoflagellate from the North Sea. Harmful Algae 2009, 8, 254–263. [Google Scholar] [CrossRef]
- James, K.J.; Moroney, C.; Roden, C. Ubiquitous ‘benign’ alga emerges as the cause of shellfish contamination responsible for the human toxic syndrome, azaspiracid poisoning. Toxicon 2002, 41, 145–151. [Google Scholar] [CrossRef]
- Boente-Juncal, A.; Raposo-García, S.; Carmen Louzao, M.; Vale, C.; Botana, L.M. Targeting chloride ion channels: New insights into the mechanism of action of the marine toxin azaspiracid. Chem. Res. Toxicol. 2021, 34, 865–879. [Google Scholar] [CrossRef]
- Yasumoto, T.; Murata, M. Polyether toxins involved in seafood poisoning. ACS Symp. Ser. 1990, 418, 120–132. [Google Scholar]
- Yasumoto, T.; Murata, M. Marine toxins. Chem. Rev. 1993, 93, 1897–1909. [Google Scholar] [CrossRef]
- Yasumoto, T.; Murata, M.; Oshima, Y.; Sano, M. Diarrhetic shellfish poisons. Tetrahedron 1985, 41, 1019–1025. [Google Scholar] [CrossRef]
- Lee, J.-S.; Igarashi, T.; Fraga, S.; Dahl, E.; Hovgaard, P.Y.; Yasumoto, T. Determination of diarrhetic shellfish toxins in various dinoflagellate species. J. Appl. Phycol. 1989, 1, 147–152. [Google Scholar] [CrossRef]
- Pillet, S.; Houvenaghel, G. Influence of experimental toxification by DSP producing microalgae, Prorocentrum lima, on clearance rate in blue mussels Mytilus edulis. In Harmful Marine Algal Blooms; Intercept: Nantes, France, 1995; pp. 481–486. [Google Scholar]
- Pillet, S.; Pereira, A.; Braekman, J.C.; Houvenaghel, G. Patterns in long term accumulation of okadaic acid and DTX1 in blue mussels, Mytilus edulis, experimentally fed with the DSP-containing alga Prorocentrum lima. In Harmful Marine Algal Blooms; Intercept: Nantes, France, 1995; pp. 487–492. [Google Scholar]
- Burgess, V.; Shaw, G. Pectenotoxins—An issue for public health: A review of their comparative toxicology and metabolism. Environ. Int. 2001, 27, 275–283. [Google Scholar] [CrossRef]
- Gaillard, S.; Le Goïc, N.; Malo, F.; Boulais, M. Cultures of Dinophysis sacculus, D. acuminata and pectenotoxin 2 affect gametes and fertilization success of the Pacific oyster, Crassostrea gigas. Environ. Pollut. 2020, 265, 114840. [Google Scholar]
- Lu, C.K.; Chou, H.N.; Lee, C.-K.; Lee, T.-H. Prorocentin, a new polyketide from the marine dinoflagellate Prorocentrum lima. Org. Lett. 2005, 7, 3893–3896. [Google Scholar] [CrossRef]
- Cuypers, E.; Abdel-Mottaleb, Y.; Kopljar, I.; Rainier, J.D.; Raes, A.L.; Snyders, D.J.; Tytgata, J. Gambierol, a toxin produced by the dinoflagellate Gambierdiscus toxicus, is a potent blocker of voltage-gated potassium channels. Toxicon 2008, 51, 974–983. [Google Scholar] [CrossRef]
- Yasumoto, T. The chemistry and biological function of natural marine toxins. Chem. Rec. 2001, 1, 228–242. [Google Scholar] [CrossRef]
- Ito, E.; Suzuki-Toyota, F.; Toshimori, K.; Fuwa, H.; Tachibana, K.; Satake, M.; Sasaki, M. Pathological effects on mice by gambierol, possibly one of the ciguatera toxins. Toxicon 2003, 42, 733–740. [Google Scholar] [CrossRef]
- Rodríguez, I.; Genta-Jouve, G.; Alfonso, C.; Calabro, K.; Alonso, E.; Sanchez, J.A.; Alfonso, A.; Thomas, O.P.; Botana, L.M. Gambierone, a ladder-shaped polyether from the dinoflagellate Gambierdiscus belizeanus. Org. Lett. 2015, 17, 2392–2395. [Google Scholar] [CrossRef] [PubMed]
- Murray, I.S.; Selwood, A.I.; Harwood, T.D.; Ginkel, R.; Puddick, J.; Rhodes, L.L.; Rise, F.; Wilkins, A.L. 44-Methylgambierone, a new gambierone analogue isolated from Gambierdiscus australes. Tetrahedron Lett. 2019, 60, 621–625. [Google Scholar] [CrossRef]
- Morohashi, A.; Satake, M.; Nagai, H.; Oshima, Y.; Yasumoto, T. The absolute configuration of gambieric acids A–D, potent antifungal polyethers, isolated from the marine dinoflagellate Gambierdiscus toxicus. Tetrahedron 2000, 56, 8995–9001. [Google Scholar] [CrossRef]
- Adachi, R.; Fukuyo, Y. The thecal structure of a marine toxic dinoflagellate Gambierdiscus toxicus gen. et sp. nov. collected in a ciguatera-endemic area. Nippon Suisan Gakkaishi 1978, 45, 67–71. [Google Scholar] [CrossRef]
- Scheuer, P.J.; Takahashi, W.; Tsutsumi, J.; Yoshida, T. Ciguatoxin: Isolation and chemical nature. Science 1967, 155, 1267–1268. [Google Scholar] [CrossRef]
- Yasumoto, T.; Nakajima, I.; Bagnis, R.; Adachi, R. Finding of a dinoflagellate as a likely culprit of ciguatera. Nippon Suisan Gakkaishi 1977, 43, 1021–1026. [Google Scholar] [CrossRef]
- Satake, M.; Ishibashi, Y.; Legrand, A.-M.; Yasumoto, T. Isolation and structure of ciguatoxin-4A, a new ciguatoxin precursor, from cultures of dinoflagellate Gambierdiscus toxicus and parrotfish Scarus gibbus. Biosci. Biotech. Biochem. 1996, 60, 2103–2105. [Google Scholar] [CrossRef]
- Legrand, A.M.; Litaudon, M.; Genthon, J.N. Isolation and some properties of ciguatoxin. J. Appl. Phycol. 1989, 1, 183–188. [Google Scholar] [CrossRef]
- Lewis, R.J.; Sellin, M. Multiple ciguatoxins in the flesh of fish. Toxicon 1992, 30, 915–919. [Google Scholar] [CrossRef]
- Pasinszki, T.; Lako, J.; Dennis, T.E. Advances in detecting ciguatoxins in fish. Toxins 2020, 12, 494. [Google Scholar] [CrossRef]
- Lewis, R.J. Ciguatoxins are potent ichthyotoxins. Toxicon 1992, 30, 207–211. [Google Scholar] [CrossRef]
- Fusetani, N.; Kem, W. Marine toxins: An overview. Prog. Mol. Subcell. Biol. 2009, 46, 1–44. [Google Scholar] [PubMed]
- Murata, M.; Yasumoto, T. The structure elucidation and biological activities of high molecular weight algal toxins: Maitotoxin, prymnesins and zooxanthellatoxins. Nat. Prod. Rep. 2000, 17, 293–314. [Google Scholar] [CrossRef] [PubMed]
- Dechraoui, M.Y.; Naar, J.; Pauillac, S.; Legrand, A.M. Ciguatoxins and brevetoxins, neurotoxic polyether compounds active on sodium channels. Toxicon 1999, 37, 125–143. [Google Scholar] [CrossRef]
- Khan, U.; Benabderrazik, N.; Bourdelais, A.J.; Baden, D.G.; Rein, K.; Gardinali, P.R.; Arroyo, L.; O’Shea, K.E. UV and solar TiO2 photocatalysis of brevetoxins (PbTxs). Toxicon 2010, 55, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, C.R.; Bodi, D.; Tartaglione, L.; Dell ‘Aversano, C. Improving in vitro ciguatoxin and brevetoxin detection: Selecting neuroblastoma (Neuro-2a) cells with lower sensitivity to ouabain and veratridine (OV-LS). Harmful Algae 2021, 103, 101994. [Google Scholar] [CrossRef]
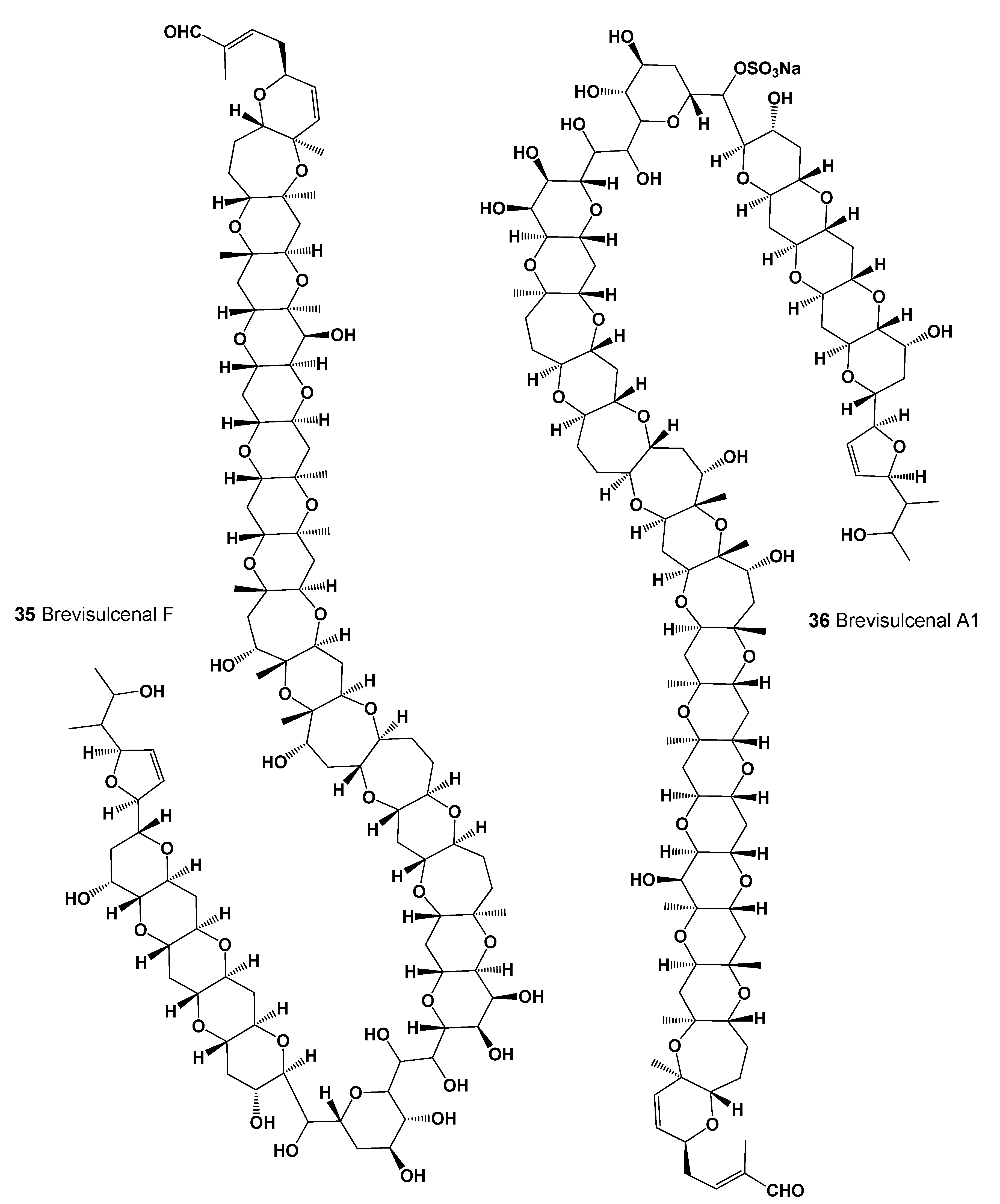
- Hamamoto, Y.; Tachibana, K.; Holland, P.T.; Shi, F.; Beuzenberg, V.; Itoh, Y.; Satake, M. Brevisulcenal-F: A polycyclic ether toxin associated with massive fish-kills in New Zealand. J. Am. Chem. Soc. 2012, 134, 4963–4968. [Google Scholar] [CrossRef]
- Satake, M.; Irie, R.; Holland, P.T.; Harwood, D.T.; Shi, F.; Itoh, Y.; Hayashi, F.; Zhang, H. Brevisulcenals-A1 and A2, sulfate esters of brevisulcenals, isolated from the red tide dinoflagellate Karenia brevisulcata. Toxins 2021, 13, 82. [Google Scholar] [CrossRef]
- Satake, M.; Shoji, M.; Oshima, Y.; Naoki, H.; Fujita, T.; Yasumoto, T. Gymnocin-A, a cytotoxic polyether from the notorious red tide dinoflagellate, Gymnodinium mikimotoi. Tetrahedron Lett. 2002, 43, 5829–5832. [Google Scholar] [CrossRef]
- Satake, M.; Shoji, M.; Oshima, Y.; Naoki, H.; Fujita, T.; Yasumoto, T. Gymnocin-B with the largest contiguous polyether rings from the red tide dinoflagellate, Karenia (formerly Gymnodinium) mikimotoi. Tetrahedron Lett. 2005, 46, 3537–3540. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; John, A.; Andersen, C.; Andersen, N.G.; Nielsen, K.F.; Hansen, P.J.; Larsen, T.O. Chemical diversity, origin, and analysis of phycotoxins. J. Nat. Prod. 2016, 79, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, G.M.; Lewis, R.J. Ciguatoxins: Cyclic polyether modulators of voltage-gated ion. Channel Function. Mar. Drugs 2006, 4, 82–118. [Google Scholar] [CrossRef]
- Ramos-Sosa, M.J.; García-Álvarez, N.; Sanchez-Henao, A.; Silva Sergent, F.; Padilla, D.; Estévez, P.; Caballero, M.J.; Martín-Barrasa, J.L.; Gago-Martínez, A.; Diogène, J. Ciguatoxin detection in flesh and liver of relevant fish species from the Canary Islands. Toxins 2022, 14, 46. [Google Scholar] [CrossRef]
- Suzuki, T.; Ha, D.V.; Uesugi, A.; Uchida, H. Analytical challenges to ciguatoxins. Curr. Opin. Food Sci. 2017, 18, 37–42. [Google Scholar] [CrossRef]
- Uemura, D.; Chou, T.; Haito, T.; Nagatsu, A.; Fukuzawa, S.; Zheng, A.-Z.; Chen, H.-S. Pinnatoxin A: A toxic amphoteric macrocycle from the Okinawan bivalve Pinna muricata. J. Am. Chem. Soc. 1995, 117, 1155–1156. [Google Scholar] [CrossRef]
- Chou, T.; Haino, T.; Kuramoto, M.; Uemura, D. Isolation and structure of pinnatoxin D, a new shellfish poison from the Okinawan bivalve Pinna muricata. Tetrahedron Lett. 1996, 37, 4027–4030. [Google Scholar] [CrossRef]
- Noboru, T.; Umemura, N.; Suenaga, K.; Chou, T.; Nagatsu, A.; Haino, T.; Yamada, K.; Uemura, D. Pinnatoxins B and C, the most toxic components in the pinnatoxin series from the Okinawan bivalve Pinna muricata. Tetrahedron Lett. 2001, 42, 3491–3494. [Google Scholar]
- Selwood, A.I.; Moles, C.O.; Wilkins, A.L.; Van Ginkel, R.; Munday, R.; Rise, F.; McNabb, P. Isolation and structural determination, and acute toxicity of novel pinnatoxins E, F and G. J. Agric. Food Chem. 2010, 58, 6532–6542. [Google Scholar] [CrossRef]
- Satta, C.T.; Angles, S.; Garce, S.E.; Luglie, A.; Sechi, N.; Camp, J. Resting stages in the genus Bysmatrum. In Proceedings of the 14th International Conference on Harmful Algae, Crete, Greece, 1–5 November 2010. [Google Scholar]
- Rhodes, L.; Smith, K.; Selwood, A.; McNabb, P.; Munday, R.; Susa, S.; Molenaar, S.; Hallgraeff, G. Dinoflagellate Vulcanodinium rugosum identified as the causative organism of pinnatoxins in Australia, New Zealand and Japan. Phycologia 2011, 50, 624–628. [Google Scholar] [CrossRef]
- Clarke, M.R.; Jones, B.; Squires, C.L.M.; Imhoff, F.M.; Harwood, D.T.; Rhodes, L.; Selwood, A.I.; McNabb, P.S.; Baird, S.K. Cyclic imine pinnatoxin G is cytotoxic to cancer cell lines via nicotinic acetylcholine receptor-driven classical apoptosis. J. Nat. Prod. 2021, 84, 2035–2042. [Google Scholar] [CrossRef]
- Tubaro, A.; Dell’Ovo, V.; Sosa, S.; Florio, C. Yessotoxins: A toxicological overview. Toxicon 2010, 56, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Korsnes, M.S. Yessotoxin as a tool to study induction of multiple cell death pathways. Toxins 2012, 4, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, A.; Vieytes, M.R.; Botana, L.M. Yessotoxin, a promising therapeutic tool. Mar. Drugs 2016, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Paz, B.; Daranas, A.H.; Norte, M.; Riobó, P.; Franco, J.M.; Fernández, J.J. Yessotoxins, a group of marine polyether toxins: An overview. Mar. Drugs 2008, 6, 73–102. [Google Scholar] [CrossRef] [PubMed]
- Rubini, S.; Albonetti, S.; Menotta, S.; Cervo, A.; Callegari, E.; Cangini, M.; Dall’Ara, S.; Baldini, E.; Vertuani, S.; Manfredini, S. New trends in the occurrence of yessotoxins in the Northwestern Adriatic Sea. Toxins 2021, 13, 634. [Google Scholar] [CrossRef]
- Cho, K.; Heo, J.; Han, J.; Hong, H.D.; Jeon, H.; Hwang, H.J. Industrial applications of dinoflagellate phycotoxins based on their modes of action: A review. Toxins 2020, 12, 805. [Google Scholar] [CrossRef]
- Satake, M.; Murata, M.; Yasumoto, T.; Fujita, T.; Naoki, H. Amphidinol, a polyhydroxy-polyene antifungal agent with an unprecedented structure, from a marine dinoflagellate, Amphidinium klebsii. J. Am. Chem. Soc. 1991, 113, 9859–9861. [Google Scholar] [CrossRef]
- Paul, G.K.; Matsumori, N.; Murata, M.; Tachibana, K. Isolation and chemical structure of amphidinol 2, a potent hemolytic compound from marine dinoflagellate Amphidinium klebsii. Tetrahedron Lett. 1995, 36, 6279–6282. [Google Scholar] [CrossRef]
- Kobayashi, J.; Tsuda, M. Amphidinolides, bioactive macrolides from symbiotic marine dinoflagellates. Nat. Prod. Rep. 2004, 21, 77–93. [Google Scholar] [CrossRef]
- Martínez, K.A.; Lauritano, C.; Druka, D.; Romano, G.; Grohmann, T. Amphidinol 22, a new cytotoxic and antifungal amphidinol from the dinoflagellate Amphidinium carterae. Mar. Drugs 2019, 17, 385. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Shi, Q.W.; Dong, M. Progress in marine toxins. Nat. Prod. Res. Dev. 2011, 23, 582–589. [Google Scholar]
- Van Wagoner, R.M.; Deeds, J.R.; Tatters, A.O.; Place, A.R.; Tomas, C.R.; Wright, J.L.C. Structure and relative potency of several karlotoxins from Karlodinium veneficum. J. Nat. Prod. 2010, 73, 1360–1365. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Place, A.R.; Yoshida, W.; Anklin, C.; Hamann, M.T. Structure and absolute configuration of karlotoxin-2, an ichthyotoxin from the marine dinoflagellate Karlodinium veneficum. J. Am. Chem. Soc. 2010, 132, 3277–3279. [Google Scholar] [CrossRef] [PubMed]
- Cousseau, A.; Siano, R.; Probert, I.; Bach, S. Marine dinoflagellates as a source of new bioactive structures. Stud. Nat. Prod. Chem. 2020, 65, 125–171. [Google Scholar]
- Dembitsky, V.M.; Pechenkina-Shubina, E.E.; Rozentsvet, O.A. Glycolipids and fatty acids of some seaweeds and marine grasses from the Black Sea. Phytochemistry 1991, 30, 2279–2283. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Rozentsvet, O.A.; Pechenkina, E.E. Glycolipids, phospholipids and fatty acids of brown algae species. Phytochemistry 1990, 29, 3417–3421. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Rezanka, T. Natural occurrence of arseno compounds in plants, lichens, fungi, algal species, and microorganisms. Plant Sci. 2003, 165, 1177–1192. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Oxidation, epoxidation and sulfoxidation reactions catalysed by haloperoxidases. Tetrahedron 2003, 26, 4701–4720. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Srebnik, M. Natural halogenated fatty acids: Their analogues and derivatives. Prog. Lipid Res. 2002, 41, 315–367. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Maoka, T. Allenic and cumulenic lipids. Prog. Lipid Res. 2007, 46, 328–375. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Betaine ether-linked glycerolipids: Chemistry and biology. Prog. Lipid Res. 1996, 35, 1–51. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Anticancer activity of natural and synthetic acetylenic lipids. Lipids 2006, 41, 883–924. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M.; Levitsky, D.O. Arsenolipids. Prog. Lipid Res. 2004, 43, 403–448. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Bioactive cyclobutane-containing alkaloids. J. Nat. Med. 2008, 62, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Manning, S.R.; La Claire, J.W., II. Prymnesins: Toxic metabolites of the golden alga, Prymnesium parvum Carter (Haptophyta). Mar. Drugs 2010, 8, 678–704. [Google Scholar] [CrossRef] [PubMed]
- Medić, N.; Varga, E.; Van de Waal, D.B.; Larsen, T.O.; Hansen, P.J. The coupling between irradiance, growth, photosynthesis and prymnesin cell quota and production in two strains of the bloom-forming haptophyte, Prymnesium parvum. Harmful Algae 2022, 112, 102173. [Google Scholar] [CrossRef]
- Anestis, K.; Kohli, G.S.; Wohlrab, S.; Varga, E. Polyketide synthase genes and molecular trade-offs in the ichthyotoxic species Prymnesium parvum. Sci. Total Environ. 2021, 795, 148878. [Google Scholar] [CrossRef]
- Taylor, R.B.; Hill, B.N.; Bobbitt, J.M.; Hering, A.S. Suspect and non-target screening of acutely toxic Prymnesium parvum. Sci. Total Environ. 2020, 715, 136835. [Google Scholar] [CrossRef]
- Kan, Y.; Uemura, D.; Hirata, Y.; Ishiguro, M.; Iwashita, T. Complete NMR signal assignment of palytoxin and N-acetylpalytoxin. Tetrahedron Lett. 2001, 42, 3197–3202. [Google Scholar] [CrossRef]
- Katikou, P. Chemistry of palytoxins and ostreocins. In Phycotoxins, Chemistry and Biochemistry; Botana, L.M., Ed.; Blackwell Publishing: Ames, IA, USA, 2007; pp. 75–93. [Google Scholar]
- Moore, R.E.; Scheuer, P.J. Palytoxin–New marine toxin from a coelenterate. Science 1971, 172, 495–498. [Google Scholar] [CrossRef]
- Katikou, P. Palytoxin and analogs: Ecobiology and origin, chemistry, metabolism, and chemical analysis. In Seafood and Freshwater Toxins: Pharmacology, Physiology, and Detection, 2nd ed.; Botana, L.M., Ed.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2008; pp. 631–663. [Google Scholar]
- Gleibs, S.; Mebs, D. Distribution and sequestration of palytoxin in coral reef animals. Toxicon 1999, 37, 1521–1527. [Google Scholar] [CrossRef]
- Ramos, V.; Vasconcelos, V. Palytoxin and analogs: Biological and ecological effects. Mar. Drugs 2010, 8, 2021–2037. [Google Scholar] [CrossRef] [PubMed]
- Maeda, M.; Kodama, R.; Tanaka, T.; Yohizumi, H.; Nomyoto, K.; Takemoto, T.; Fujita, M. Structures of insecticidal substances isolated from a red alga, Chondria armata. In Proceedings of the 27th Symposium on the Chemistry of Natural Products, Symposium Organizing Committee, Hiroshima, Japan, 2 August 1985; p. 616. [Google Scholar]
- Mori, S.; Sugahara, K.; Maeda, M.; Nomoto, K.; Iwashita, T. Insecticidal activity guided isolation of palytoxin from a red alga, Chondria armata. Tetrahedron Lett. 2016, 57, 3612–3617. [Google Scholar] [CrossRef]
- Usami, M.; Satake, M.; Ishida, S.; Inoue, A.; Kan, Y.; Yasumoto, T. Palytoxin analogs from the dinoflagellate Ostreopsis siamensis. J. Am. Chem. Soc. 1995, 117, 5389–5390. [Google Scholar] [CrossRef]
- Ukena, T.; Satake, M.; Usami, M.; Oshima, Y.; Naoki, H.; Fujita, T.; Kan, Y.; Yasumoto, T. Structure elucidation of ostreocin D, a palytoxin analog isolated from the dinoflagellate Ostreopsis siamensis. Biosci. Biotechnol. Biochem. 2001, 65, 2585–2588. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, S.; Ten-Hage, L.; Turquet, J.; Quod, J.P.; Bernard, C.; Hennion, M.C. First evidence of palytoxin analogs from an Ostreopsis mascarenensis (Dinophyceae) benthic bloom in Southwestern Indian Ocean. J. Phycol. 2004, 40, 1042–1051. [Google Scholar] [CrossRef]
- Lenoir, S.; Ten-Hage, L.; Turquet, J.; Quod, J.P.; Hennion, M.C. Characterization of new analogs of palytoxin isolated from an Ostreopsis mascarenensis bloom in the south-western Indian Ocean. Afr. J. Mar. Sci. 2006, 28, 389–391. [Google Scholar] [CrossRef]
- Carballeira, N.M.; Emiliano, A.; Sostre, A.; Restituyo, J.A.; González, I.M.; Colón, G.M.; Tosteson, C.G.; Tosteson, T.R. Fatty acid composition of bacteria associated with the toxic dinoflagellate Ostreopsis lenticularis and with Caribbean Palythoa species. Lipids 1998, 33, 627–632. [Google Scholar] [CrossRef]
- Seemann, P.; Gernert, C.; Schmitt, S.; Mebs, D.; Hentschel, U. Detection of hemolytic bacteria from Palythoa caribaeorum (Cnidaria, Zoantharia) using a novel palytoxin-screening assay. Anton. Leeuw. Int. J. G 2009, 96, 247–258. [Google Scholar] [CrossRef]
- Frolova, G.M.; Kuznetsova, T.A.; Mikhailov, V.V.; Elyakov, G.B. An enzyme linked immunosorbent assay for detecting palytoxin-producing bacteria. Russ. J. Bioorg. Chem. 2000, 26, 285–289. [Google Scholar] [CrossRef]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, X.; Liu, J.; Bao, K.; Zhang, G.; Tu, G.; Kieser, T.; Deng, Z. ‘Streptomyces nanchangensis’, a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 2002, 148, 361–371. [Google Scholar] [CrossRef] [PubMed]
- De Lima Procópio, R.E.; Da Silva, I.R.; Martins, M.K.; De Azevedo, J.L.; De Araújo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Chater, K.F.; Tian, Y.; Zhang, J.; Tan, H. Specialized metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 2016, 40, 554–573. [Google Scholar] [CrossRef]
- Mast, Y.; Stegmann, E. Actinomycetes: The antibiotics producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef]
- Rezanka, T.; Spížek, J.; Přikrylová, V.; Prell, A.; Dembitsky, V.M. Five new derivatives of nonactic and homo-nonactic acids from Streptomyces globisporus. Tetrahedron 2004, 60, 4781–4787. [Google Scholar] [CrossRef]
- Rezanka, T.; Spizek, J.; Prikrylova, V.; Dembitsky, V.M. Four new derivatives of trihomononactic acids from Streptomyces globisporus. Eur. J. Org. Chem. 2004, 2004, 4239–4244. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Kilimnik, A. Anti-melanoma agents derived from fungal species. Mathews J. Pharm. Sci. 2016, 1, 002. [Google Scholar]
- Dembitsky, V.M. Naturally occurring bioactive Cyclobutane-containing (CBC) alkaloids in fungi, fungal endophytes, and plants. Phytomedicine 2014, 21, 1559–1581. [Google Scholar] [CrossRef]
- Sproule, A.; Correa, H.; Decken, A.; Haltli, B.; Berrué, F.; Overy, D.P.; Kerr, R.G. Terrosamycins A and B, Bioactive Polyether Ionophores from Streptomyces sp. RKND004 from Prince Edward Island Sediment. Mar. Drugs 2019, 17, 347. [Google Scholar] [CrossRef]
- Khan, N.; Yilmaz, S.; Aksoy, S.; Uzel, A.; Tosun, C.; Kirmizibayrak, P.B.; Bedir, E. Polyethers isolated from the marine actinobacterium Streptomyces cacaoi inhibit autophagy and induce apoptosis in cancer cells. Chem. Biol. Interact. 2019, 307, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hermann, T.E. Characterization of ionomycin as a calcium ionophore. J. Biol. Chem. 1978, 253, 5892–5894. [Google Scholar] [CrossRef]
- Koskinen, A.M.P.; Karisalmi, K. Polyketide stereotetrads in natural products. Chem. Soc. Rev. 2005, 34, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Antoszczak, M.; Huczyński, A. Anticancer activity of polyether ionophore-salinomycin. Anti-Cancer Agents Med. Chem. 2015, 15, 575–591. [Google Scholar] [CrossRef]
- Kamiya, K. Development of artificial cell models using microfluidic technology and synthetic biology. Micromachines 2020, 11, 559. [Google Scholar] [CrossRef]
- Liu, W.; Slusarchyk, D.S.; Astle, G.; Trejo, W.H.; Brown, W.E.; Meyers, E. Ionomycin, a new polyether antibiotic. J. Antibiot. 1978, 31, 815–819. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Shibuya, M.; Sugawara, H.; Kawaguchi, O.; Hirose, C.; Nagatsu, J. Salinomycin, a new polyether antibiotic. J. Antibiot. 1974, 27, 814–821. [Google Scholar] [CrossRef]
- Paulus, E.F.; Vertesy, L. Crystal structure of 2-(6-[2-(5-ethyl-5-hydroxy-6-methyl-tetrahydro-pyran-2-yl)-15-oxo-2, 10, 12-trimethyl-1, 6, 8-trioxa-dispiro [4.1. 5.3] pentadec-13-en-9-yl]-2-hydroxy-1, 3-dimethyl-4-oxo-heptyl-5-methyl-tetrahydro-pyran-2-yl)-butyrate sodium, Na (C42H67O11), SY-9–antibiotic 20-oxo-salinomycin. Zeitsch. Kristallograph. 2004, 219, 184–186. [Google Scholar]
- Rutkowski, J.; Brzezinski, B. Structures and properties of naturally occurring polyether antibiotics. BioMed Res. Int. 2013, 17, 162513. [Google Scholar] [CrossRef]
- Berg, D.H.; Hamill, R.L. The isolation and characterization of narasin, a new polyether antibiotic. J. Antibiot. 1978, 31, 1–6. [Google Scholar] [CrossRef]
- Markiewicz, W.; Barski, D.; Burmanczuk, A.; Tomaszewska, E. Toxicity of salinomycin and narasin in turkeys. J. Elementol. 2014, 19, 903–914. [Google Scholar] [CrossRef]
- Golbek, T.W.; Schmüser, L.; Rasmussen, M.H.; Poulsen, T.B.; Weidner, T. Lasalocid acid antibiotic at a membrane surface probed by sum frequency generation spectroscopy. Langmuir 2020, 36, 3184–3192. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Rachlin, A.I.; Scott, W.E.; Sternbach, L.H.; Goldberg, M.W. The isolation of three new crystalline antibiotics from Streptomyces. J. Am. Chem. Soc. 1951, 73, 5295–5298. [Google Scholar] [CrossRef]
- Mitrovic, M.; Schildknecht, G. Anticoccidial activity of lasalocid (X-537A) in chicks. Poult. Sci. 1974, 53, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Keller-Juslen, C.; King, H.D.; Kuhn, M.; Loosli, H.R.; Pache, W.; Petcher, T.J.; Weber, H.P. Tetronomycin, a novel polyether of unusual structure. J. Antibiot. 1982, 35, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Keller-Juslen, C.; King, H.D.; Kuhn, M.; Loosli, H.R. Noboritomycins A and B, new polyether antibiotics. J. Antibiot. 1978, 31, 820–828. [Google Scholar] [CrossRef]
- Ebata, E.; Kasahara, H.; Sekine, K.; Inoue, Y. Lysocellin, a new polyether antibiotic. I. Isolation, purification, physicochemical and biological properties. J. Antibiot. 1975, 28, 118–121. [Google Scholar] [CrossRef][Green Version]
- Steinrauf, L.K.; Pinkerton, M.; Chamberlin, J.W. The structure of nigericin. Biochem. Biophys. Res. Commun. 1968, 33, 29–31. [Google Scholar] [CrossRef]
- Gachon, P.; Kergomard, A.; Veschambre, H.; Esteve, C.; Staron, T. Grisorixin, a new antibiotic related to nigericin. J. Chem. Soc. D 1970, 13, 1421–1422. [Google Scholar] [CrossRef]
- Gachon, P.; Kergomard, A.; Staron, T.; Esteve, C. Grisorixin, an ionophorous antibiotic of the nigericin group I. Fermentation, isolation, biological properties and structure. J. Antibiot. 1975, 28, 345–350. [Google Scholar] [CrossRef]
- Forbes, M.W.; Volmer, D.A.; Francis, G.J.; Böhme, D.K. A comparison of data analysis methods for determining gas phase stabilities by cid: Alkali metal complexes of polyether ionophore antibiotics. J. Am. Soc. Mass Spectrom. 2005, 16, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Blount, J.F.; Westley, J.W. Crystal and molecular structure of the free acid form of antibiotic X-206 hydrate. J. Chem. Soc. Chem. Commun. 1975, 4, 533. [Google Scholar] [CrossRef]
- Otoguro, K.; Kohana, A.; Manabe, C.; Ishiyama, A.; Ui, H.; Shiomi, K.; Yamada, H.; Omura, S. Potent antimalarial activities of polyether antibiotic, X-206. J. Antibiot. 2001, 54, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Svenningsen, E.B.; Thyrsted, J.; Blay-Cadanet, J.; Liu, H.; Lin, S.; Moyano-Villameriel, J.; Olagnier, D.; Idorn, M.; Paludan, S.R.; Holm, C.K.; et al. Ionophore antibiotic X-206 is a potent inhibitor of SARS-CoV-2 infection in vitro. Antivir. Res. 2021, 185, 104988. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.D.; Chaney, M.O.; Chamberlin, J.W.; Hamill, R.L.; Chen, S. Structure ofA204A, a new polyether antibiotic. J. Am. Chem. Soc. 1973, 95, 3399–4000. [Google Scholar] [CrossRef] [PubMed]
- Keller-Juslen, C.; King, H.D.; Kis, Z.L. Septamycin, a polyether antibiotic. J. Antibiot. 1975, 28, 854–859. [Google Scholar] [CrossRef][Green Version]
- Ricketts, A.P.; Dirlam, J.P.; Shively, J.E. Anticoccidial efficacy and chicken toleration of potent new polyether ionophores: 1. The septamycin relative CP-82,009. Poult. Sci. 1992, 71, 1626–1630. [Google Scholar] [CrossRef]
- Dirlam, J.P.; Belton, A.M.; Bordner, J.; Cullen, W.P.; Huang, L.H.; Kojima, Y.H. CP-82,009, a potent polyether anticoccidial related to septamycin and produced by Actinomadura sp. J. Antibiot. 1992, 45, 331–340. [Google Scholar] [CrossRef][Green Version]
- Liu, X.; Chu, J.; Gui, C.; Liao, W.; Qin, F.; Hu, X.; Liang, B.; Jiang, J.; Huang, J.; Ning, C. Polyether antibiotics K-41A and K-41Am inhibit HIV-1 replication via suppressing the activities of HIV-1 reverse transcriptase and integrase. Arch. Med. Sci. 2020, 16. [Google Scholar] [CrossRef]
- Irschik, H.; Jansen, R.; Gerth, K. The sorangicins, novel and powerful inhibitors of eubacterial RNA polymerase isolated from myxobacteria. J. Antibiot. 1987, 40, 7–13. [Google Scholar] [CrossRef]
- Agtarap, A.; Chamberlin, J.W.; Pinkerton, M.; Steinrauf, L. The structure of monensic acid, a new biologically active compound. J. Am. Chem. Soc. 1967, 89, 5737–5739. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.E., Jr.; Hoehn, M.M. Monensin, a new biologically active compound. I. Discovery and isolation. Antimicrob. Agent Chemother. 1968, 11, 349–352. [Google Scholar]
- Łowicki, D.; Huczyński, A. Structure and antimicrobial properties of monensin A and its derivatives: Summary of the achievements. BioMed Res. Int. 2013, 21, 742149. [Google Scholar] [CrossRef] [PubMed]
- Amano, S.I.; Morota, T.; Kano, Y. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J. Antibiot. 2010, 63, 486–491. [Google Scholar] [CrossRef]
- Fehr, T.; King, H.D.; Kuhn, M. Mutalomycin, a new polyether antibiotic taxonomy, fermentation, isolation and characterization. J. Antibiot. 1977, 30, 903–907. [Google Scholar] [CrossRef][Green Version]
- Hamill, R.L.; Nakatsukasa, W.M.; Yao, R.C. Novel Polyether Anbitiotic A80438 from Streptomyces. U.S. patent 004824863, 25 April 1989. [Google Scholar]
- Omura, S.; Shibata, M.; Machida, S.; Sawada, J. Isolation of a new polyether antibiotic, lonomycin. J. Antibiot. 1976, 29, 15–20. [Google Scholar] [CrossRef][Green Version]
- Funayama, S.; Nozoe, S.; Tronquet, C. Isolation and structure of a new polyether antibiotic, octacyclomycin. J. Antibiot. 1992, 45, 1686–1691. [Google Scholar] [CrossRef]
- Garcia-Princival, I.M.R.; Princival, J.L.; Da Silva, E.D. Streptomyces hygroscopicus UFPEDA 3370: A valuable source of the potent cytotoxic agent nigericin and its evaluation against human colorectal cancer cells. Chem. Biol. Interact. 2021, 333, 109316. [Google Scholar] [CrossRef]
- Vazquez-Hernandez, M.; Ceapa, C.D.; Rodríguez-Luna, S.D.; Rodríguez-Sanoja, R.; Sánchez, S. Draft genome sequence of Streptomyces scabrisporus NF3, an endophyte isolated from Amphipterygium adstringens. Genome Announ. 2017, 5, e00267-17. [Google Scholar] [CrossRef]
- Kusakabe, Y.; Mitsuoka, S.; Omuro, Y. Antibiotic No. 6016, a polyether antibiotic. J. Antibiot. 1980, 33, 1437–1442. [Google Scholar] [CrossRef]
- Delort, A.M.; Jeminet, G.; Sareth, S.; Riddle, F.G. Ionophore properties of cationomycin in large unilamellar vesicles studied by 23Na- and 39K-NMR. Chem. Pharm. Bull. 1998, 46, 1618–1620. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nakamura, G.; Kobayashi, K.; Sakurai, T.; Sono, K. Cationomycin, a new polyether ionophore antibiotic produced by Actinomadura nov. sp. J. Antibiot. 1981, 34, 1513–1514. [Google Scholar] [CrossRef] [PubMed]
- Oscarson, J.R.; Bordner, J.; Celmer, W.D. Endusamycin, a novel polycyclic ether antibiotic produced by a strain of Streptomyces endus subsp. Aureus. J. Antibiot. 1989, 42, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Mahtal, N.; Wu, Y.; Cintrat, J.C.; Barbier, J.; Lemichez, E. Revisiting old ionophore lasalocid as a novel inhibitor of multiple toxins. Toxins 2020, 12, 26. [Google Scholar] [CrossRef] [PubMed]
- Erwin, G.S.; Heikkinen, J.; Halima, P.; Haber, C.L. Streptomyces lasalocidi sp. nov. (formerly ‘Streptomyces lasaliensis’), an actinomycete isolated from soil which produces the polyether antibiotic lasalocid. Int. J. Syst. Evol. Microbiol. 2020, 70, 3076–3083. [Google Scholar] [CrossRef]
- Westley, J.W.; Benz, W.; Donahue, J. Biosynthesis of lasalocid. III Isolation and structure determination of four homologs of lasalocid A. J. Antibiot. 1974, 27, 744–753. [Google Scholar] [CrossRef][Green Version]
- Hoshi, M.; Shimotohno, K.W.; Endo, T. Microbial and chemical conversion of antibiotic K-41. I. Isolation and identification of conversion product. J. Antibiot. 1997, 50, 631–634. [Google Scholar] [CrossRef]
- Chen, J.; Gui, C.; Wei, Q.; Liu, J.; Ye, L.; Tian, X.; Gu, Y.C.; Li, Q.; Ju, J. Characterization of tailoring methyltransferases involved in K-41A biosynthesis: Modulating methylation to improve K-41A anti-infective activity. Org. Lett. 2020, 22, 4627–4632. [Google Scholar] [CrossRef]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef]
- Iorio, M.; Tocchetti, A.; Cruz, J.C.S.; Gatto, G.D.; Brunati, C. Novel polyethers from screening Actinoallomurus spp. Antibiotics 2018, 7, 47. [Google Scholar] [CrossRef]
- Wehbie, R.S.; Runsheng, C.A.I.; Lardy, H.A. The antibiotic W341C, its ion transport properties and inhibitory effects on mitochondrial substrate oxidation. J. Antibiot. 1987, 50, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.C.; Kim, J.H.; Ha, J.W.; Park, N.S.; Sohng, J.K. Production and biological activity of laidlomycin, anti-MRSA/VRE antibiotic from Streptomyces sp. CS684. J. Microbiol. 2007, 45, 6–10. [Google Scholar] [PubMed]
- Dirlam, J.P.; Belton, A.M.; Bordner, J.; Cullen, W.P. CP-84,657, A potent polyether Anticoccidal related to portmicin and produved by Actinomadura sp. J. Antibiot. 1990, 43, 668–679. [Google Scholar] [CrossRef]
- Cullen, W.P.; Celmer, W.D.; Chappel, L.R. CP-54,883 a novel chlorine-containing polyether antibiotic produced by a new species of Actinomadura: Taxonomy of the producing culture, fermentation, physico-chemical and biological properties of the antibiotic. J. Antibiot. 1987, 40, 1490–1495. [Google Scholar] [CrossRef] [PubMed]
- Dirlam, J.P.; Presseau-Linabury, L.; Koss, D.A. The structure of CP-80, 219, a new polyether antibiotic related to dianemycin. J. Antibiot. 1990, 43, 727–730. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alleaume, M.; Hickel, D. Crystal structure of the thallium salt of the antibiotic grisorixin. J. Chem. Soc. Chem. Commun. 1972, 2, 175–176. [Google Scholar] [CrossRef]
- Hatsu, M.; Sasaki, T.; Miyadoh, S. SF2487, a new polyether antibiotic produced by Actinomadura. J. Antibiot. 1990, 43, 259–266. [Google Scholar] [CrossRef]
- Westley, J.W.; Liu, C.M.; Sello, L.H. Isolation and characterization of antibiotic X-14931A, the naturally occurring 19-deoxyaglycone of dianemycin. J. Antibiot. 1984, 37, 813–815. [Google Scholar] [CrossRef][Green Version]
- Liu, C.M.; Westley, J.W.; Hermann, T.E. Novel polyether antibiotics X-14873A, G and H produced by a Streptomyces: Taxonomy of the producing culture, fermentation, biological and ionophorous properties of the antibiotics. J. Antibiot. 1986, 39, 1712–1718. [Google Scholar] [CrossRef]
- Westley, J.W. Antibiotic structure and biosynthesis. J. Nat. Prod. 1986, 49, 35–47. [Google Scholar] [CrossRef]
- Liu, C.; Hermann, T.E.; La Prosser, T.B. X-14766A, a halogen containing polyether antibiotic produced by Streptomyces malachitofuscus subsp. downeyi ATCC 31547. Discovery, fermentation, biological properties and taxonomy of the producing culture. J. Antibiot. 1981, 34, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Kevin, D.A.; Meujo, D.A.F.; Hamann, M.T. Polyether ionophores: Broad-spectrum and promising biologically active molecules for the control of drug-resistant bacteria and parasites. Expert Opin. Drug Discov. 2009, 4, 109–146. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Ayala, H.L.; Tabet, J.C. Abierixin, a new polyether antibiotic. Production, structural determination and biological activities. J. Antibiot. 1985, 38, 1655–1663. [Google Scholar] [CrossRef]
- Xia, H.Y.; Lin, K.C.; Yang, C.J.; Qin, Z.J. Cloning and functional studies of the chromosomal replication origin of Actinomadura yumaensis. Acta Microbiol. Sin. 2007, 47, 529–532. [Google Scholar]
- Fleck, W.F.; Strauss, D.G.; Meyer, J.; Porstendorfer, G. Fermentation, isolation, and biological activity of maduramycin: A new antibiotic from Actinomadura rubra. Zeitsch. Allgem. Mikrobiol. 1978, 18, 389–398. [Google Scholar] [CrossRef]
- Hossain, S.M.; Hossain, A.M.; Rahman, M.M.; Mondol, M.A.; Bhuiyan, M.S.A.; Gray, A.I.; Flores, M.E.; Rashid, M.A. Amides from the fungus Streptomyces hygroscopicus and their antimicrobial activity. Phytochemistry 2004, 65, 2147–2151. [Google Scholar] [CrossRef] [PubMed]
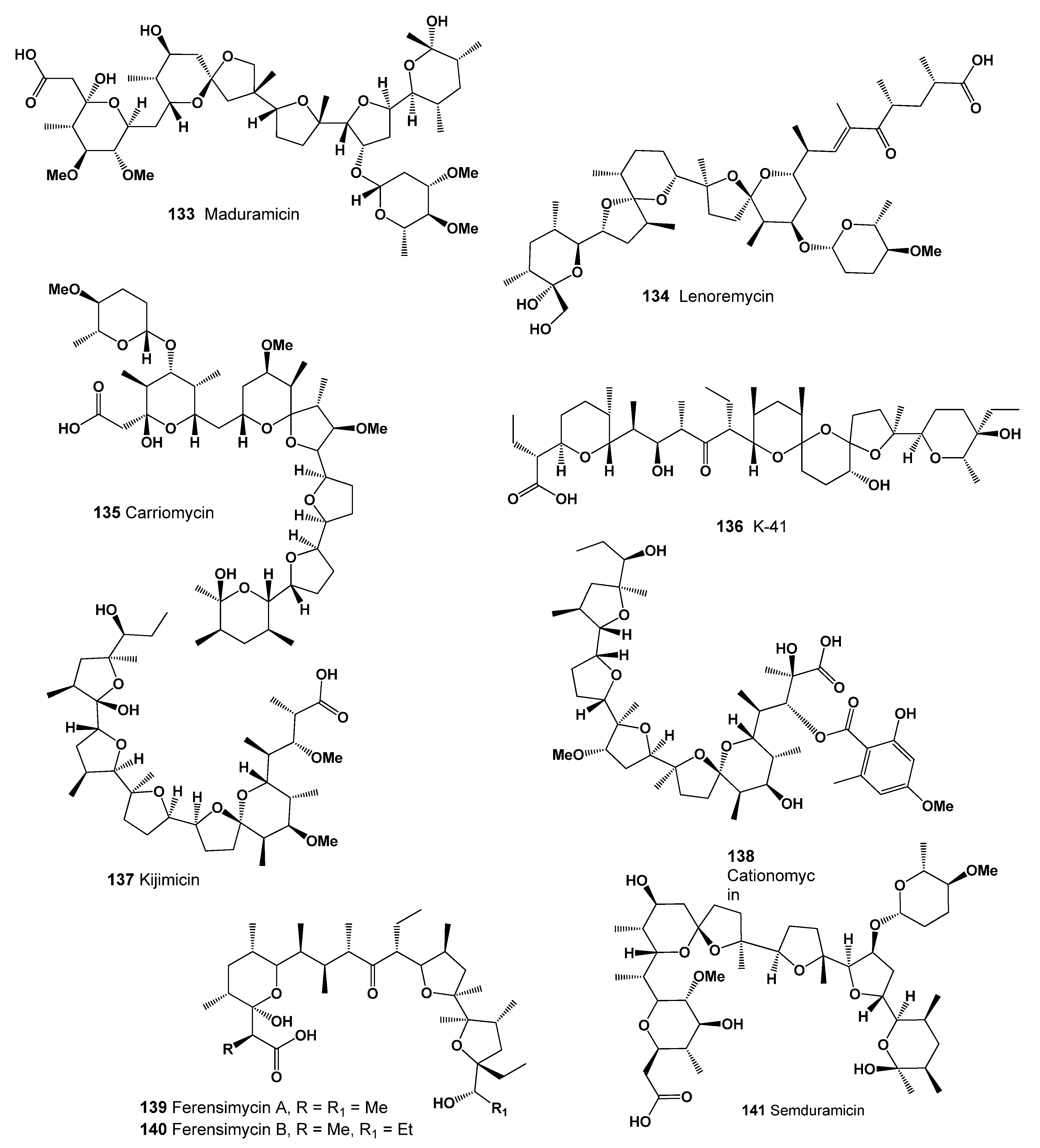
- Imada, A.; Nozaki, Y.; Hasegawa, T.; Mizuta, E.; Igarasi, S.; Yoneda, M. Carriomycin, a new polyether antibiotic produced by Streptomyces hygroscopicus. J. Antibiot. 1978, 31, 7–14. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nakamura, H.; Ogata, R. Kijimicin, a polyether antibiotic. J. Antibiot. 1991, 43, 441–443. [Google Scholar] [CrossRef][Green Version]
- Dutton, C.J.; Banks, B.J.; Cooper, C.B. Polyether ionophores. Nat. Prod. Rep. 1995, 12, 165–181. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Shibata, A.; Yahagi, T. Production of Salinomycin. Japanese Patent 61,247,398, 1 January 1986. [Google Scholar]
- Nakamura, G.; Isono, K. A new species of Actinomadura producing a polyether antibiotic, cationomycin. J. Antibiot. 1983, 36, 1468–1472. [Google Scholar] [CrossRef]
- Ubukata, M.; Uzawa, J.; Isono, K. Biosynthesis of cationomycin: Direct and indirect incorporation of 13C-acetate and application of homoscalar correlated 2-D carbon-13 NMR and double quantum coherence. J. Am. Chem. Soc. 1984, 106, 2213–2214. [Google Scholar] [CrossRef]
- Kusakabe, Y.; Mizuno, T.; Kawabata, S. Ferensimycins A and B, two polyether antibiotics. J. Antibiot. 1982, 35, 1119–1129. [Google Scholar] [CrossRef]
- Tynan, E.J.; Nelson, T.H.; Davies, R.A.; Wernau, W.C. The production of semduramicin by direct fermentation. J. Antibiot. 1992, 45, 813–815. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, A.P.; Glazer, E.A.; Migaki, T.T.; Olson, J.A. Anticoccidial efficacy of semduramicin in battery studies with laboratory isolates of coccidia. Poult. Sci. 1992, 71, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Fraser, T.R. The connection of chemical constitution and physiological action. Trans. R. Soc. Edinb. 1868, 25, 224–242. [Google Scholar]
- Cros, A.F.A. Action de l’Alcohol Amylique Sur l’Organisme. Ph.D. Thesis, University of Strasbourg, Strasbourg, France, 1863. [Google Scholar]
- Richet, M.C. Note sur le rapport entre la toxicité et les propriétes physiques des corps. Compt. Rend. Soc. Biol. 1893, 45, 775–776. [Google Scholar]
- Meyer, H. Zur Theorie der AIkoholnarkose. Arch. Exp. Path. Pharm. 1899, 42, 109–118. [Google Scholar] [CrossRef]
- Overton, C.E. Studien über die Narkose; Fischer: Jena, Germany, 1901. [Google Scholar]
- Hammett, L.P. Some relations between reaction rates and equilibrium constants. Chem. Rev. 1935, 17, 125–136. [Google Scholar] [CrossRef]
- Hammett, L.P. The effect of structure upon the reactions of organic compounds. Benzene derivatives. J. Am. Chem. Soc. 1937, 59, 96–103. [Google Scholar] [CrossRef]
- Taft, R.W. Separation of polar, steric and resonance effects in reactivity. In Steric Effects in Organic Chemistry; Newman, M.S., Ed.; Wiley: Hoboken, NJ, USA, 1956; pp. 556–675. [Google Scholar]
- Hansch, C.; Fujita, T. p-τ-p Analysis. A method for the correlation of biological activity and chemical structure. J. Am. Chem. Soc. 1964, 86, 1616–1626. [Google Scholar] [CrossRef]
- Hansch, C.; Leo, A. Exploring QSAR; American Chemical Society: Washington, DC, USA, 1995. [Google Scholar]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W., Jr. Computational methods in drug discovery. Pharm. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016, 12, 2694–2718. [Google Scholar] [CrossRef] [PubMed]
- Kokh, D.B.; Amaral, M.; Bomke, J.; Grädler, U.; Musil, D. Estimation of drug-target residence times by γ-random acceleration molecular dynamics simulations. J. Chem. Theor. Comput. 2018, 14, 3859–3869. [Google Scholar] [CrossRef] [PubMed]
- Cherkasov, A.M.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J. QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef] [PubMed]
- PASS Online URL. Available online: http://www.way2drug.com/passonline/ (accessed on 7 April 2022).
- Dembitsky, V.M.; Gloriozova, T.A.; Savidov, N. Steroid phosphate esters and phosphonosteroids and their biological activities. Appl. Microbiol. Biotechnol. 2018, 102, 7679–7692. [Google Scholar] [CrossRef] [PubMed]
- Vil, V.A.; Gloriozova, T.A.; Poroikov, V.V.; Terent’ev, A.O.; Savidov, N.; Dembitsky, V.M. Peroxy steroids derived from plant and fungi and their biological activities. Appl. Microbiol. Biotechnol. 2018, 102, 7657–7667. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M.; Savidov, N.; Gloriozova, T.A. Sulphur containing steroids: Structures and biological activities. Vietnam J. Chem. 2018, 56, 540–582. [Google Scholar] [CrossRef]
- Savidov, N.; Gloriozova, T.A.; Poroikov, V.V.; Dembitsky, V.M. Highly oxygenated isoprenoid lipids derived from fungi and fungal endophytes: Origin and biological activities. Steroids 2018, 140, 114–124. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Ermolenko, E.; Savidov, N.; Gloriozova, T.A.; Proroikov, V.V. Antiprotozoal and antitumor activity of natural polycyclic endoperoxides: Origin, structures, and biological activity. Molecules 2021, 26, 686. [Google Scholar] [CrossRef]
- Dembitsky, V.M. In silico prediction of steroids and triterpenoids as potential regulators of lipid metabolism. Mar. Drugs 2021, 19, 650. [Google Scholar] [CrossRef]
- Pounina, T.A.; Gloriozova, T.A.; Savidov, N.; Dembitsky, V.M. Sulfated and sulfur-containing steroids and their pharmacological profile. Mar. Drugs 2021, 19, 240. [Google Scholar] [CrossRef]
- Sikorsky, T.V.; Ermolenko, E.V.; Gloriozova, T.A.; Dembitsky, V.M. Mini Review: Anticancer activity of diterpenoid peroxides. Vietnam J. Chem. 2020, 58, 273–280. [Google Scholar] [CrossRef]
- Ermolenko, E.V.; Imbs, A.B.; Gloriozova, T.A.; Poroikov, V.V.; Sikorskaya, T.V.; Dembitsky, V.M. Chemical diversity of soft coral steroids and their pharmacological activities. Mar. Drugs 2020, 18, 613. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M.; Gloriozova, T.A.; Poroikov, V.V. Pharmacological profile of natural and synthetic compounds with rigid adamantane-based scaffolds as potential agents for the treatment of neurodegenerative diseases. Biochem. Biophys. Res. Commun. 2020, 529, 1225–1241. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M. Antitumor and hepatoprotective activity of natural and synthetic neo steroids. Prog. Lipid Res. 2020, 79, 101048. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M.; Dzhemileva, L.; Gloriozova, T.; D’yakonov, D. Natural and synthetic drugs used for the treatment of the dementia. Biochem. Biophys. Res. Commun. 2020, 524, 772–783. [Google Scholar] [CrossRef] [PubMed]

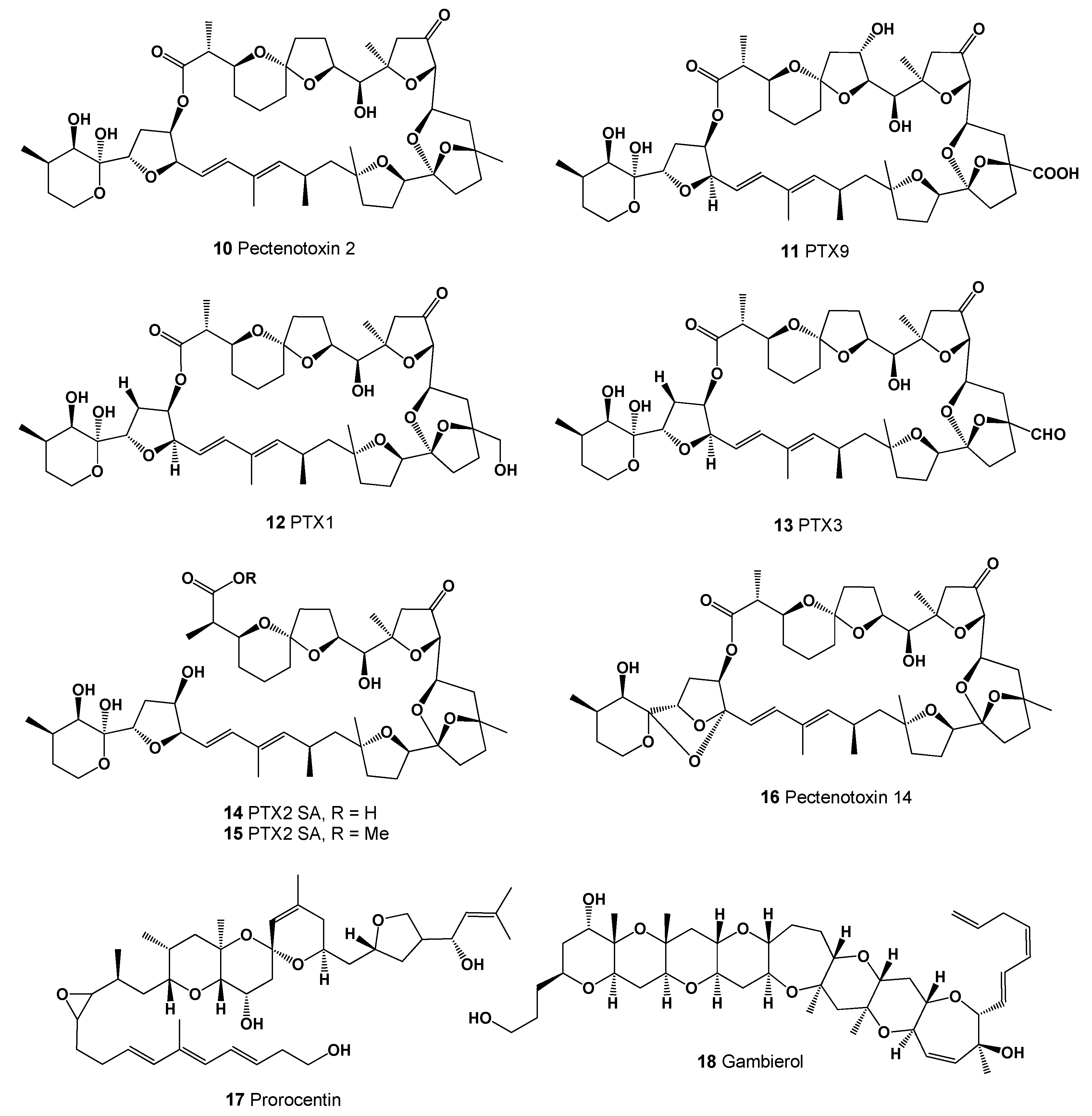

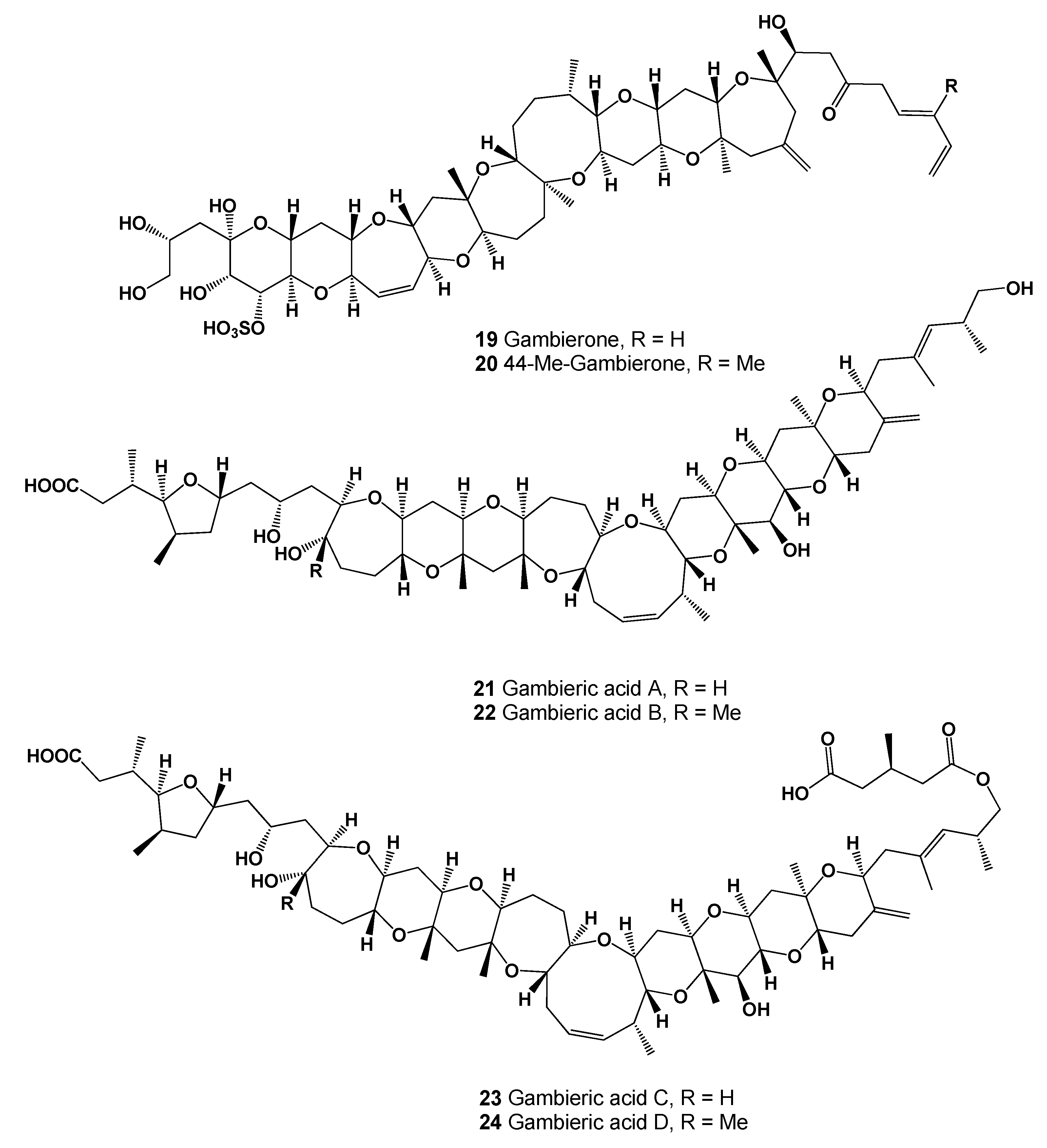




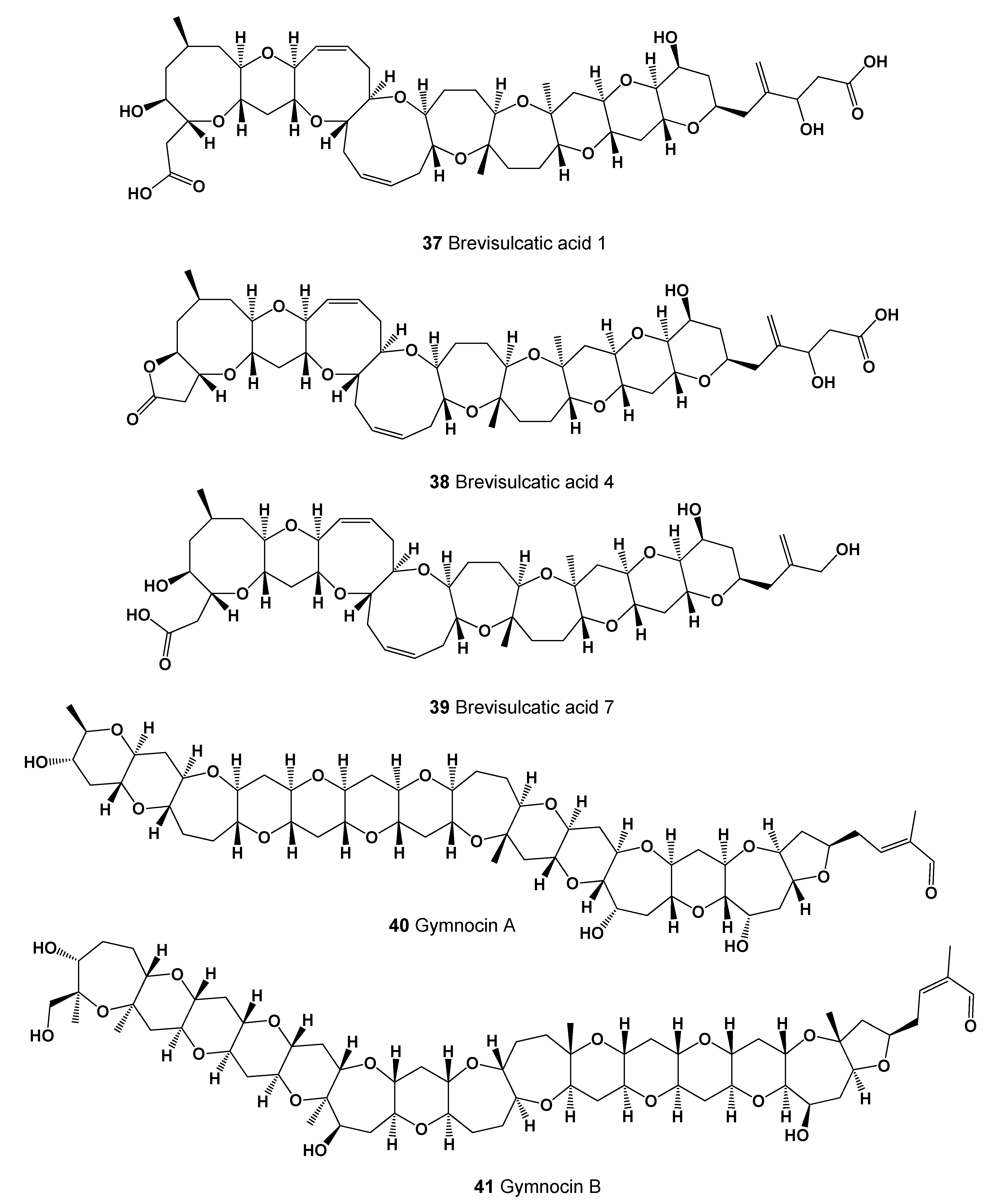
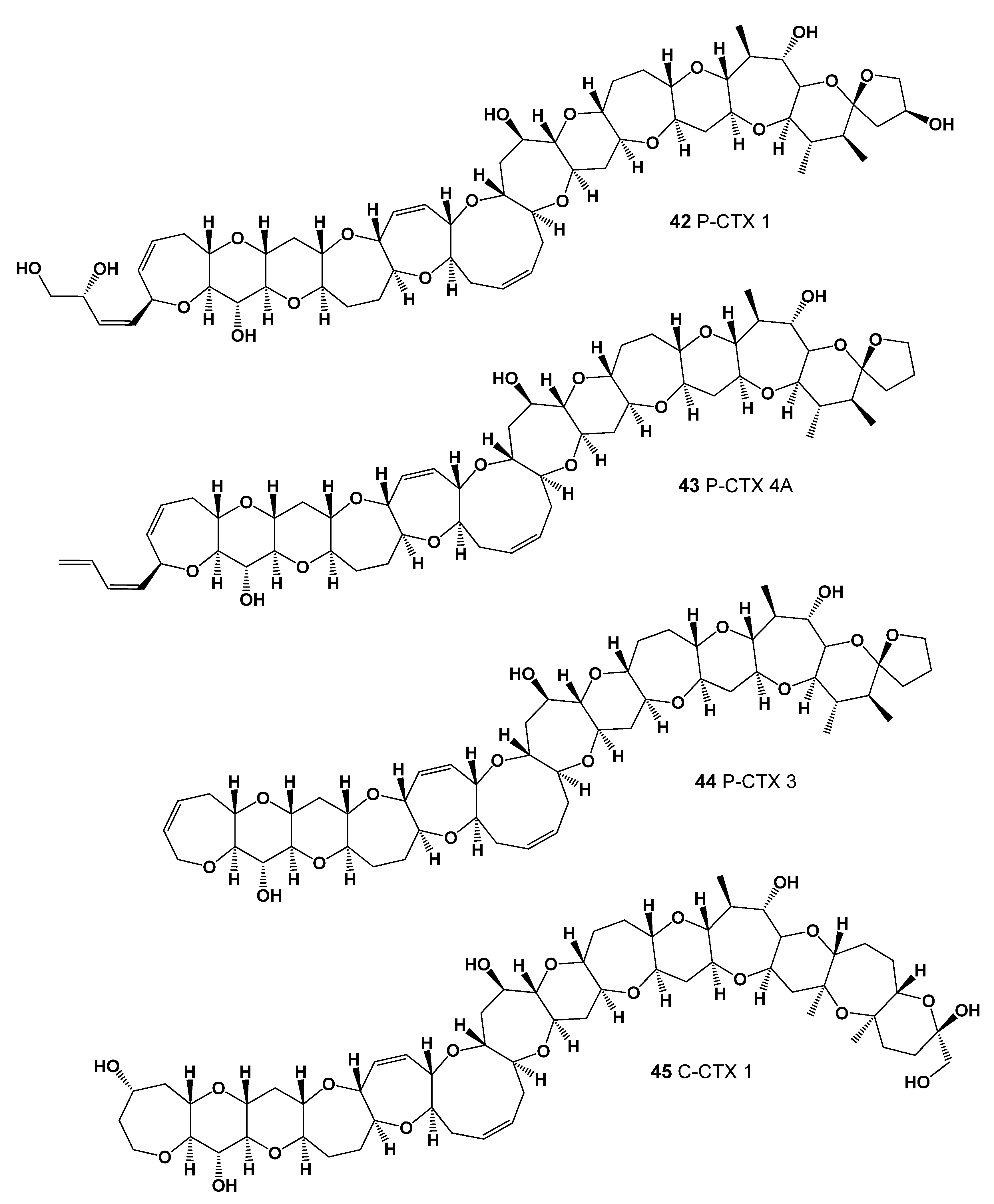
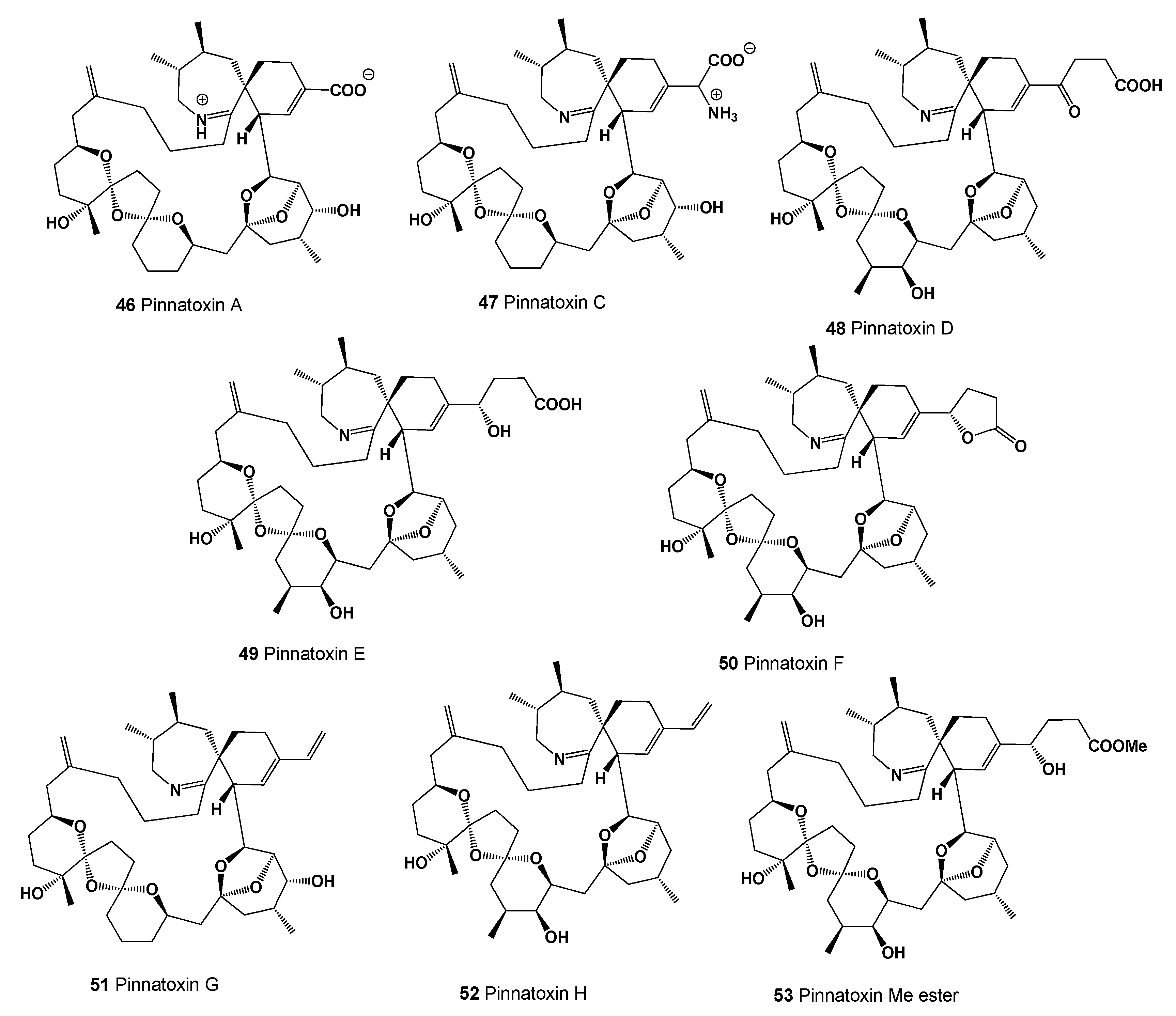

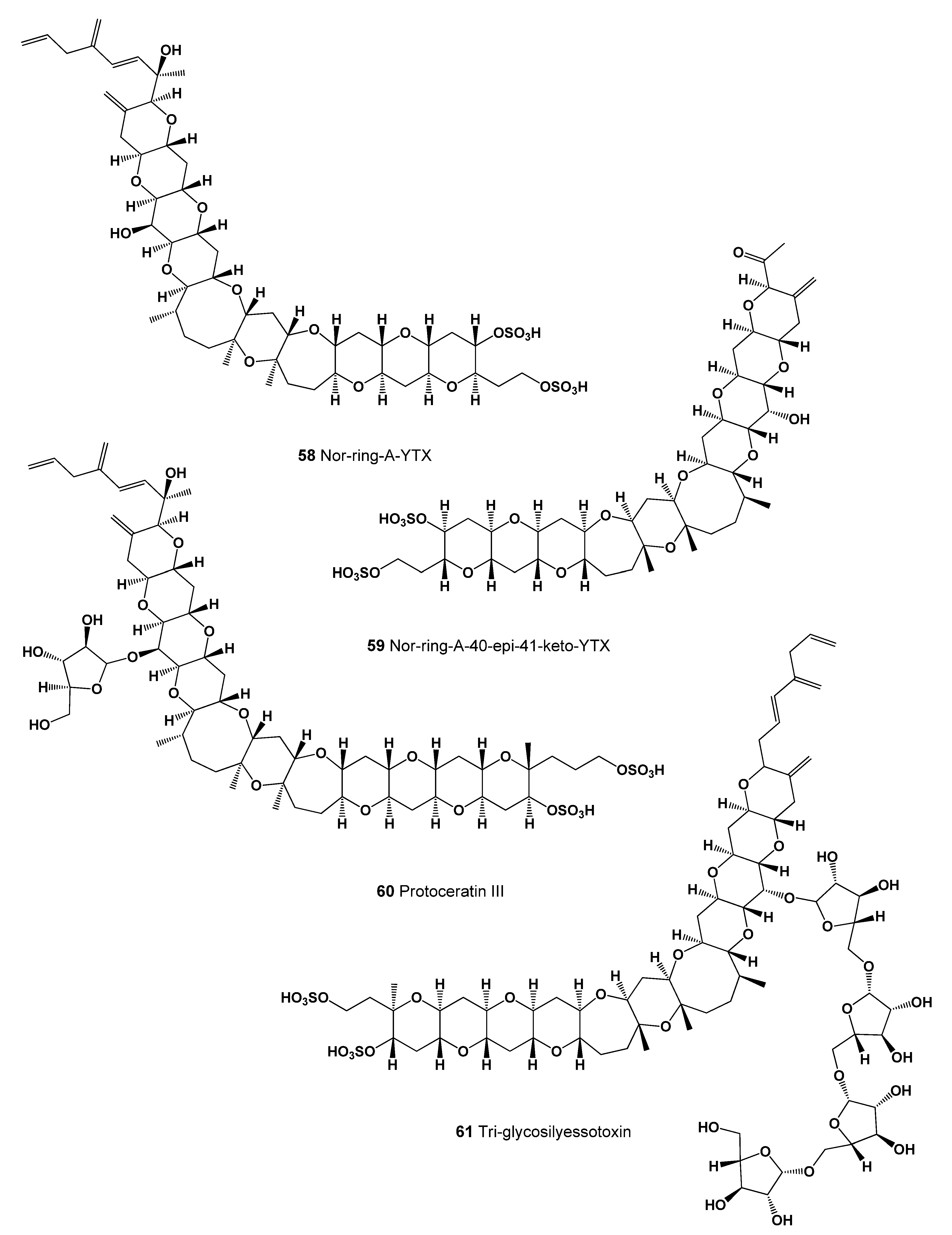


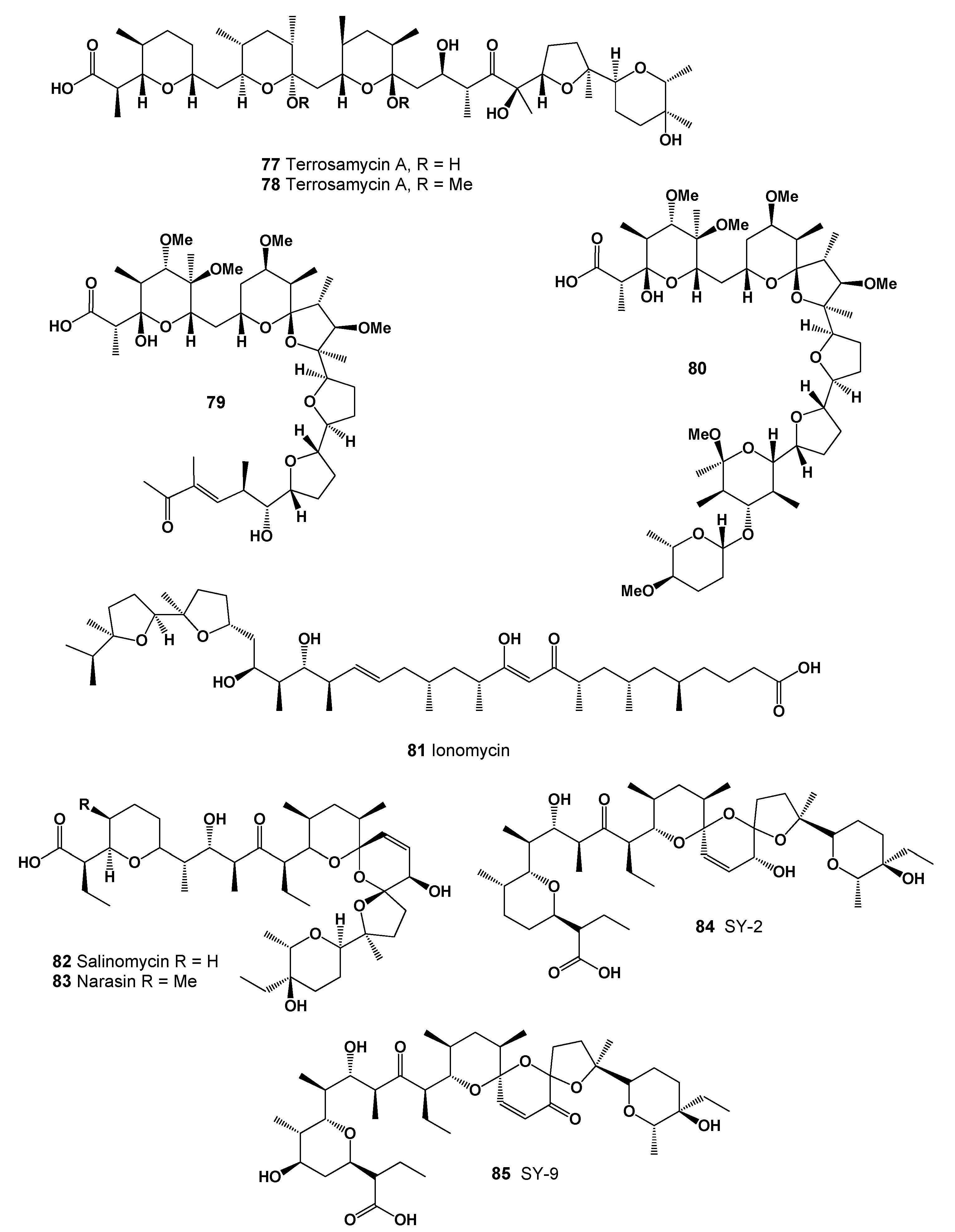





| No. | Predicted Biological Activity, Pa * | Reported Activity [30,31,32,33,34,35,36,37,38,39,40,41,42] |
|---|---|---|
| 1 | Lipid metabolism regulator (0.999) Angiogenesis stimulant (0.995) DNA synthesis inhibitor (0.991) Apoptosis agonist (0.979) Antineoplastic (0.961) Antifungal (0.844) Antiparasitic (0.843) Antibacterial (0.792) Antineoplastic metabolite (0.763) Cardiotonic (0.719) Immunosuppressant (0.719) Antimitotic (0.694) Growth stimulant (0.650) Platelet aggregation inhibitor (0.539) Hypolipemic (0.538) | Angiogenesis inducer Anticancer Antifungal Antimitotic Antiparasitic Apoptosis inducer Immunosuppressive activity Platelet aggregation inhibitor Protein phosphatase inhibitor |
| No. | Predicted Biological Activity, Pa * | Reported Activity [41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63] |
|---|---|---|
| 2 | Lipid metabolism regulator (0.998) Angiogenesis stimulant (0.995) DNA synthesis inhibitor (0.987) Apoptosis agonist (0.984) Antineoplastic (0.964) Antifungal (0.863) Antiparasitic (0.846) Antibacterial (0.818) | Anticancer Apoptosis inducer |
| 3 | Lipid metabolism regulator (0.998) Angiogenesis stimulant (0.995) Apoptosis agonist (0.983) DNA synthesis inhibitor (0.973) Antineoplastic (0.961) | Anticancer Apoptosis inducer |
| 4 | Lipid metabolism regulator (0.999) Angiogenesis stimulant (0.994) DNA synthesis inhibitor (0.983) Apoptosis agonist (0.979) Antineoplastic (0.956) | Anticancer |
| 5 | Lipid metabolism regulator (0.999) Angiogenesis stimulant (0.994) DNA synthesis inhibitor (0.988) Apoptosis agonist (0.974) Antineoplastic (0.959) | Anticancer |
| 6 | Angiogenesis inhibitor (0.979) Antifungal (0.922) Apoptosis agonist (0.906) Antibacterial (0.900) | Antifungal |
| 7 | Apoptosis agonist (0.867) Antineoplastic (0.819) Growth stimulant (0.799) | Neurological activity Anticancer |
| 8 | Antineoplastic (0.846) Apoptosis agonist (0.807) Antifungal (0.796) | Neurological activity Anticancer |
| 9 | Apoptosis agonist (0.879) Antineoplastic (0.822) | Neurological activity Anticancer |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 10 | Antineoplastic (0.945); Apoptosis agonist (0.719); Antineoplastic (sarcoma) (0.664) Antineoplastic (renal cancer) (0.647); T cell inhibitor (0.604); Antineoplastic (pancreatic cancer) (0.594); Antimetastatic (0.590); Antineoplastic (lymphocytic leukemia) (0.581) |
| 11 | Antineoplastic (0.933); Antimitotic (0.747); Apoptosis agonist (0.648) Antineoplastic (sarcoma) (0.639); Antineoplastic (renal cancer) (0.625) Antineoplastic (pancreatic cancer) (0.579); Antimetastatic (0.570) Antineoplastic (lymphocytic leukemia) (0.559); Antineoplastic (multiple myeloma) (0.534) |
| 12 | Antineoplastic (0.936); Apoptosis agonist (0.676); Antineoplastic (sarcoma) (0.656) Antineoplastic (renal cancer) (0.636); Antineoplastic (pancreatic cancer) (0.587) Antineoplastic (lymphocytic leukemia) (0.582); Antineoplastic (myeloid leukemia) (0.564) |
| 13 | Antineoplastic (0.936); Apoptosis agonist (0.717); Antineoplastic (sarcoma) (0.640) Antineoplastic (renal cancer) (0.624); Antineoplastic (pancreatic cancer) (0.579) Antimetastatic (0.575); Antineoplastic (lymphocytic leukemia) (0.551) |
| 14 | Antineoplastic (0.958); Apoptosis agonist (0.835); Antineoplastic (pancreatic cancer) (0.561) |
| 15 | Antineoplastic (0.954); Apoptosis agonist (0.835); Antineoplastic (pancreatic cancer) (0.555) |
| 16 | Antineoplastic (0.917); Antineoplastic (sarcoma) (0.639); Antineoplastic (renal cancer) (0.637) Antineoplastic (lymphocytic leukemia) (0.545); Apoptosis agonist (0.544) |
| 17 | Antineoplastic (0.919); Apoptosis agonist (0.765); Antimitotic (0.739) |
| 18 | Antineoplastic (0.928); Apoptosis agonist (0.748); Antineoplastic (renal cancer) (0.715) Antineoplastic (lymphocytic leukemia) (0.654); Prostate cancer treatment (0.500) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 19 | Antineoplastic (0.899); Apoptosis agonist (0.849); Antimetastatic (0.593) |
| 20 | Antineoplastic (0.901); Apoptosis agonist (0.845); Antimetastatic (0.602) |
| 21 | Antineoplastic (0.940); Apoptosis agonist (0.681); Antineoplastic (renal cancer) (0.664) |
| 22 | Antineoplastic (0.933); Antifungal (0.925); Antibacterial (0.904); Antiparasitic (0.874) Apoptosis agonist (0.679); Antineoplastic (renal cancer) (0.653); Antimetastatic (0.621) |
| 23 | Antifungal (0.933); Antineoplastic (0.933); Antibacterial (0.903); Antiparasitic (0.863) Antimitotic (0.807); Antibiotic (0.791); Apoptosis agonist (0.654) Antineoplastic (renal cancer) (0.650); Antimetastatic (0.633) |
| 24 | Antifungal (0.929); Antineoplastic (0.926); Antibacterial (0.914); Apoptosis agonist (0.652) Antineoplastic (renal cancer) (0.640); Antimetastatic (0.623); Antileukemic (0.591) |
| 25 | Antineoplastic (0.941); Apoptosis agonist (0.773); Antineoplastic (renal cancer) (0.665) |
| 26 | Antineoplastic (0.947); Apoptosis agonist (0.815); Antineoplastic (renal cancer) (0.693) |
| 27 | Antineoplastic (0.951); Apoptosis agonist (0.757); Antineoplastic (renal cancer) (0.732) Alzheimer’s disease treatment (0.631); Antileukemic (0.626); Antimetastatic (0.607) |
| 28 | Antibacterial (0.979); Antifungal (0.965); Antineoplastic (0.931); Antimetastatic (0.565) |
| 29 | Antineoplastic (0.921); Apoptosis agonist (0.664); Antimetastatic (0.651) |
| 30 | Antineoplastic (0.929); Cytostatic (0.922); Apoptosis agonist (0.678) Antineoplastic (lymphocytic leukemia) (0.672); Antineoplastic (renal cancer) (0.606) |
| 31 | Antineoplastic (0.931); Cytostatic (0.899); Antifungal (0.897); Antiparasitic (0.882) Apoptosis agonist (0.664); Antimetastatic (0.655); Antineoplastic (renal cancer) (0.612) |
| 32 | Antineoplastic (0.918); Antifungal (0.884); Cytostatic (0.859); Antibacterial (0.849) Antimitotic (0.817); Antiparasitic (0.793); Antineoplastic (renal cancer) (0.689) Antimetastatic (0.653); Apoptosis agonist (0.616); Prostate cancer treatment (0.500) |
| 33 | Antineoplastic (0.926); Cytostatic (0.907); Antifungal (0.878); Antibacterial (0.850) Antimitotic (0.821); Antineoplastic (renal cancer) (0.698); Antimetastatic (0.654) Antileukemic (0.653); Apoptosis agonist (0.632); Prostate cancer treatment (0.526) |
| 34 | Antineoplastic (0.926); Antifungal (0.885); Cytostatic (0.876); Antibacterial (0.829) Antiparasitic (0.820); Antimitotic (0.815); Antineoplastic (renal cancer) (0.700) Antileukemic (0.684); Antimetastatic (0.655); Apoptosis agonist (0.611) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 35 | Antineoplastic (0.953); Antibacterial (0.907); Antifungal (0.814); Antimitotic (0.721) Apoptosis agonist (0.675); Antiparasitic (0.651); Antimetastatic (0.628) |
| 36 | Antineoplastic (0.936); Antibacterial (0.930); Antifungal (0.848); Apoptosis agonist (0.623) Antimetastatic (0.609); Anti-mycoplasmal (0.577) |
| 37 | Cytostatic (0.912); Antineoplastic (0.912); Antifungal (0.901); Antibacterial (0.860) Antiparasitic (0.849); Antimitotic (0.825); Antineoplastic (renal cancer) (0.656) Antimetastatic (0.655); Apoptosis agonist (0.642) |
| 38 | Cytostatic (0.961); Antineoplastic (0.925); Antifungal (0.910); Antibacterial (0.871) Antiparasitic (0.868); Antimitotic (0.846); Apoptosis agonist (0.694); Antimetastatic (0.672) |
| 39 | Antineoplastic (0.913); Cytostatic (0.910); Antifungal (0.870); Antiparasitic (0.852) Antibacterial (0.842); Antimitotic (0.822); Apoptosis agonist (0.683); Antimetastatic (0.654) Antineoplastic (renal cancer) (0.652); Autoimmune disorders treatment (0.565) |
| 40 | Antineoplastic (0.949); Antineoplastic (liver cancer) (0.905); Antiparasitic (0.869) Cytostatic (0.859); Apoptosis agonist (0.840); Antimetastatic (0.739) |
| 41 | Antineoplastic (0.946); Antimitotic (0.832); Antifungal (0.788); Antiparasitic (0.785) Antineoplastic (renal cancer) (0.750); Antimetastatic (0.691); Prostate cancer treatment (0.521) |
| 42 | Antineoplastic (0.920); Antifungal (0.821); Antibacterial (0.805); Antimetastatic (0.526) |
| 43 | Antineoplastic (0.928); Apoptosis agonist (0.824); Antimetastatic (0.515) |
| 44 | Antineoplastic (0.930); Antimitotic (0.823); Apoptosis agonist (0.762) Alzheimer’s disease treatment (0.741); Antineoplastic (renal cancer) (0.520) |
| 45 | Antineoplastic (0.967); Growth stimulant (0.881); Antiparasitic (0.845); Antibacterial (0.832) Antimitotic (0.821); Antifungal (0.775); Apoptosis agonist (0.699); Cytostatic (0.687) Antimetastatic (0.655); Antineoplastic (non-Hodgkin’s lymphoma) (0.587) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 46 | Antineoplastic (0.871); Apoptosis agonist (0.656); Antibacterial (0.631) |
| 47 | Antineoplastic (0.839); Antibacterial (0.720); Antineoplastic (myeloid leukemia) (0.551) Antileukemic (0.540); Antineoplastic (lymphocytic leukemia) (0.531) |
| 48 | Antineoplastic (0.855); Antifungal (0.793); Antibacterial (0.704); Antimitotic (0.674) Antineoplastic (myeloid leukemia) (0.562); Antineoplastic (lymphocytic leukemia) (0.553) |
| 49 | Antineoplastic (0.858); Antifungal (0.798); Antibacterial (0.743); Apoptosis agonist (0.631) Antineoplastic (myeloid leukemia) (0.573); Antineoplastic (lymphocytic leukemia) (0.550) |
| 50 | Antineoplastic (0.906); Antifungal (0.813); Antibacterial (0.748) Apoptosis agonist (0.692); Alzheimer’s disease treatment (0.625) Antineoplastic (myeloid leukemia) (0.576); Antineoplastic (lymphocytic leukemia) (0.560) |
| 51 | Antineoplastic (0.871); Antifungal (0.736); Antimitotic (0.685); Antibacterial (0.683) Apoptosis agonist (0.670); Antineoplastic (myeloid leukemia) (0.606) Antileukemic (0.593); Antineoplastic (lymphocytic leukemia) (0.568) |
| 52 | Antineoplastic (0.886); Antifungal (0.768); Antimitotic (0.731); Antibacterial (0.716) Apoptosis agonist (0.644); Antineoplastic (myeloid leukemia) (0.581) Antileukemic (0.580); Antineoplastic (lymphocytic leukemia) (0.544) |
| 53 | Antineoplastic (0.851); Antifungal (0.804); Antibacterial (0.735); Antimitotic (0.704) Apoptosis agonist (638); Antineoplastic (myeloid leukemia) (0.566) Antileukemic (0.554); Antineoplastic (lymphocytic leukemia) (0.548) |
| 54 | Antineoplastic (0.928); Antibacterial (0.854); Antifungal (0.827); Antimitotic (0.655) Antimetastatic (0.615); Prostate cancer treatment (0.613); Apoptosis agonist (0.591) |
| 55 | Antineoplastic (0.914); Antifungal (0.820); Antimitotic (0.653); Antimetastatic (0.620) Prostate cancer treatment (0.612); Antineoplastic (lymphocytic leukemia) (0.541) |
| 56 | Antineoplastic (0.915); Antifungal (0.752); Antibacterial (0.746); Antimitotic (0.698) Antineoplastic (myeloid leukemia) (0.677); Antimetastatic (0.613) Prostate cancer treatment (0.592); Antineoplastic (lymphocytic leukemia) (0.550) |
| 57 | Antineoplastic (0.872); Antibacterial (0.793); Antifungal (0.668); Antimitotic (0.662) Antimetastatic (0.656); Antineoplastic (lymphocytic leukemia) (0.576) Apoptosis agonist (0.509); Antineoplastic (non-Hodgkin’s lymphoma) (0.506) |
| 58 | Antineoplastic (0.921); Antibacterial (0.877); Antifungal (0.852); Antimitotic (0.704) Prostate cancer treatment (0.625); Antimetastatic (0.621) Antineoplastic (lymphocytic leukemia) (0.575); Antineoplastic (myeloid leukemia) (0.533) |
| 59 | Antineoplastic (0.895); Antibacterial (0.856); Antifungal (0.796); Antimitotic (0.762) Antineoplastic (lymphocytic leukemia) (0.591); Antineoplastic (myeloid leukemia) (0.545) |
| 60 | Antineoplastic (0.935); Antibacterial (0.912); Antifungal (0.905); Chemopreventive (0.760) Apoptosis agonist (0.638); Anticarcinogenic (0.625); Antimetastatic (0.617) |
| 61 | Antibacterial (0.934); Antineoplastic (0.933); Antifungal (0.913); Chemopreventive (0.758) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 62 | Antifungal (0.884); Lipid metabolism regulator (0.875); Apoptosis agonist (0.851) |
| 63 | Antifungal (0.890); Antibacterial (0.837); Antineoplastic (0.836); Apoptosis agonist (0.782) |
| 64 | Antifungal (0.842); Antineoplastic (0.814); Antibacterial (0.762); Apoptosis agonist (0.738) |
| 65 | Antifungal (0.896); Antineoplastic (0.867); Antibacterial (0.831); Apoptosis agonist (0.806) |
| 66 | Antifungal (0.901); Antibacterial (0.870); Antineoplastic (0.863); Apoptosis agonist (0.718) |
| 67 | Antifungal (0.895); Apoptosis agonist (0.858); Antineoplastic (0.843); Antibacterial (0.826) |
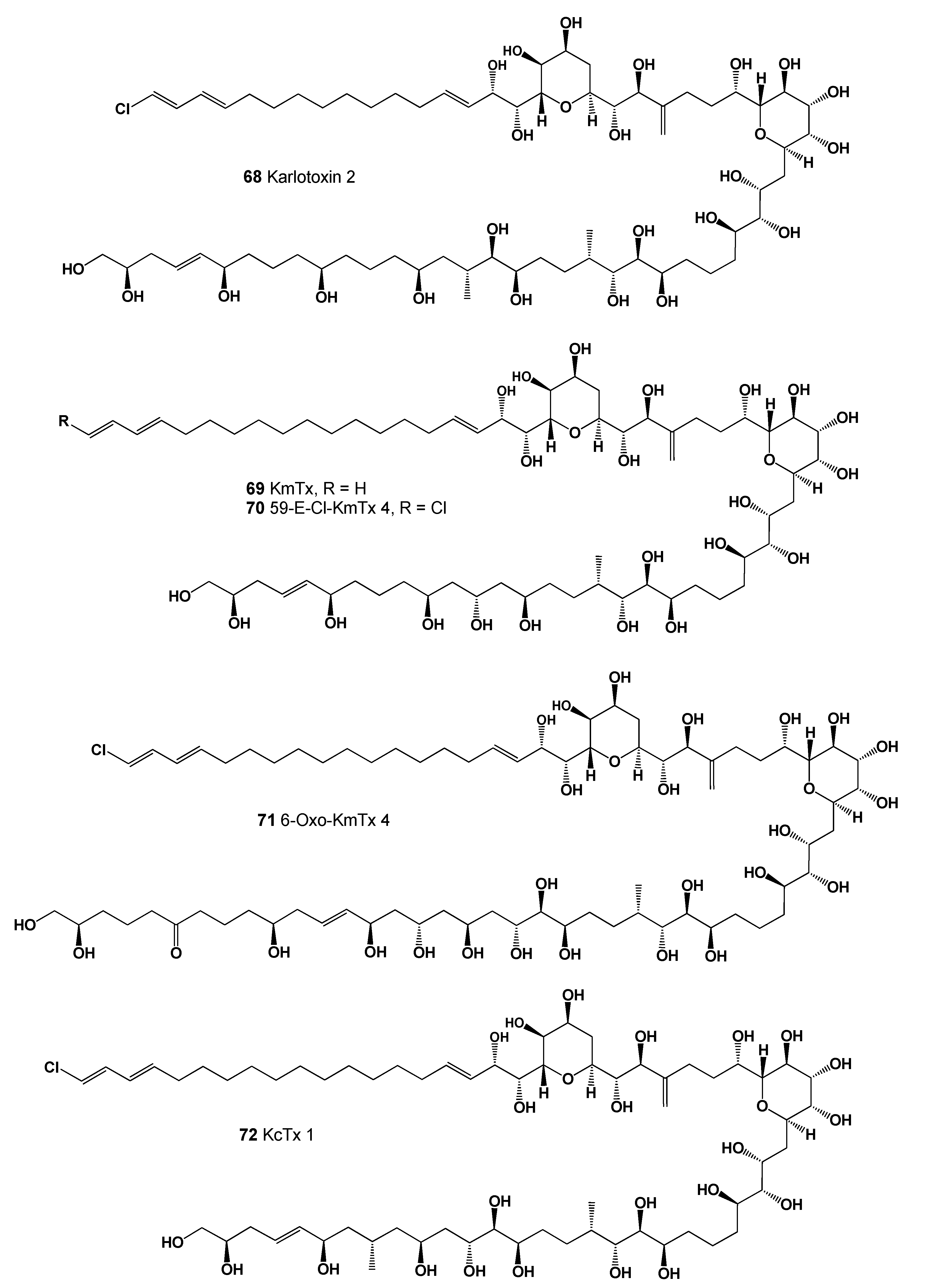
| 68 | Antifungal (0.892); Antineoplastic (0.794); Antibacterial (0.773); Apoptosis agonist (0.772) |
| 69 | Antifungal (0.904); Antineoplastic (0.850); Apoptosis agonist (0.849); Antibacterial (0.821) |
| 70 | Antifungal (0.896); Antineoplastic (0.800); Apoptosis agonist (0.781); Antibacterial (0.779) |
| 71 | Antifungal (0.907); Antineoplastic (0.795); Antibacterial (0.783); Apoptosis agonist (0.777) |
| 72 | Antifungal (0.901); Antineoplastic (0.789); Antibacterial (0.780); Apoptosis agonist (0.772) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
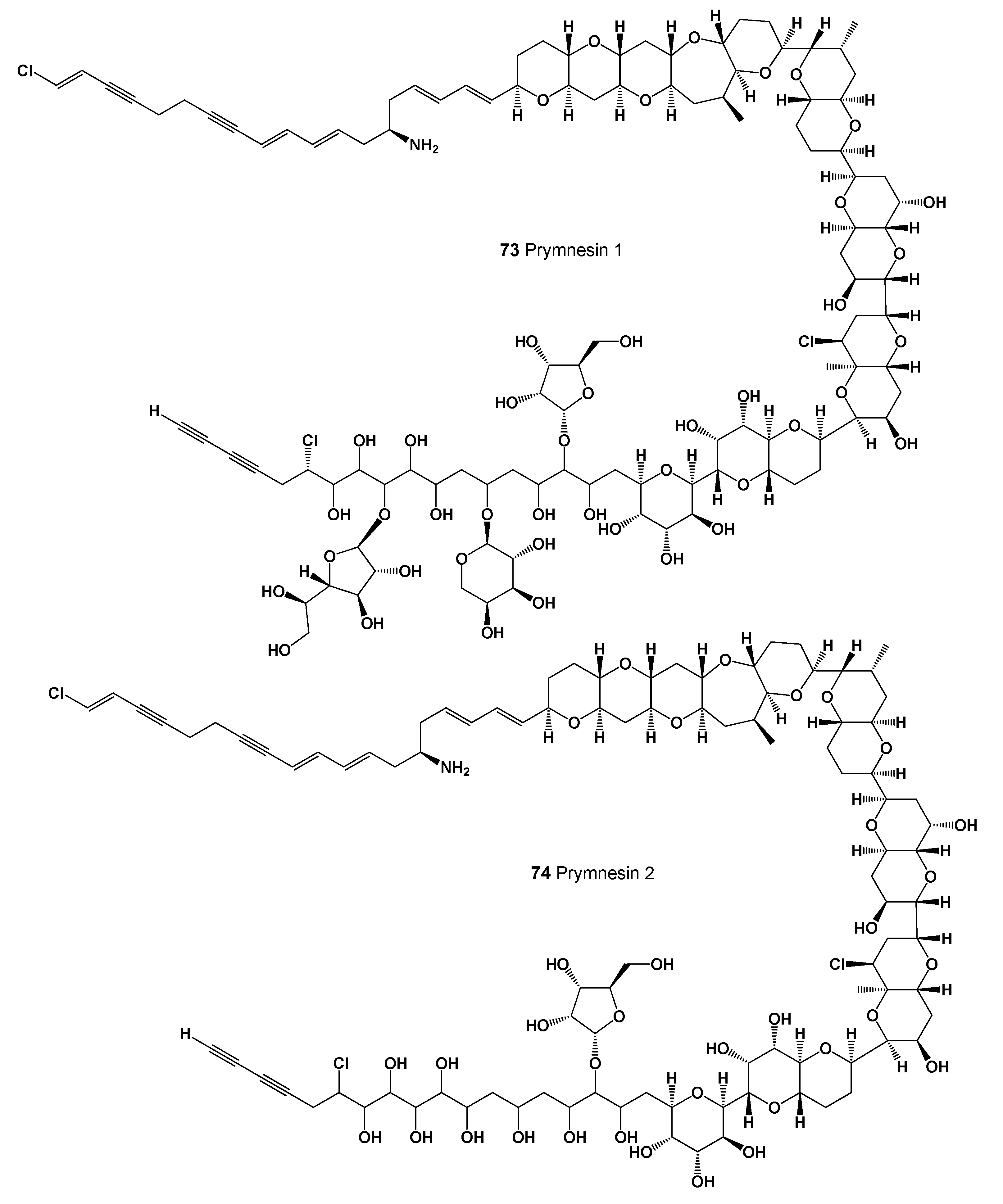
| 73 | Antifungal (0.974); Antibacterial (0.969); Antineoplastic (0.906); Antimetastatic (0.524) |
| 74 | Antifungal (0.979); Antibacterial (0.959); Antineoplastic (0.916); Apoptosis agonist (0.672) |
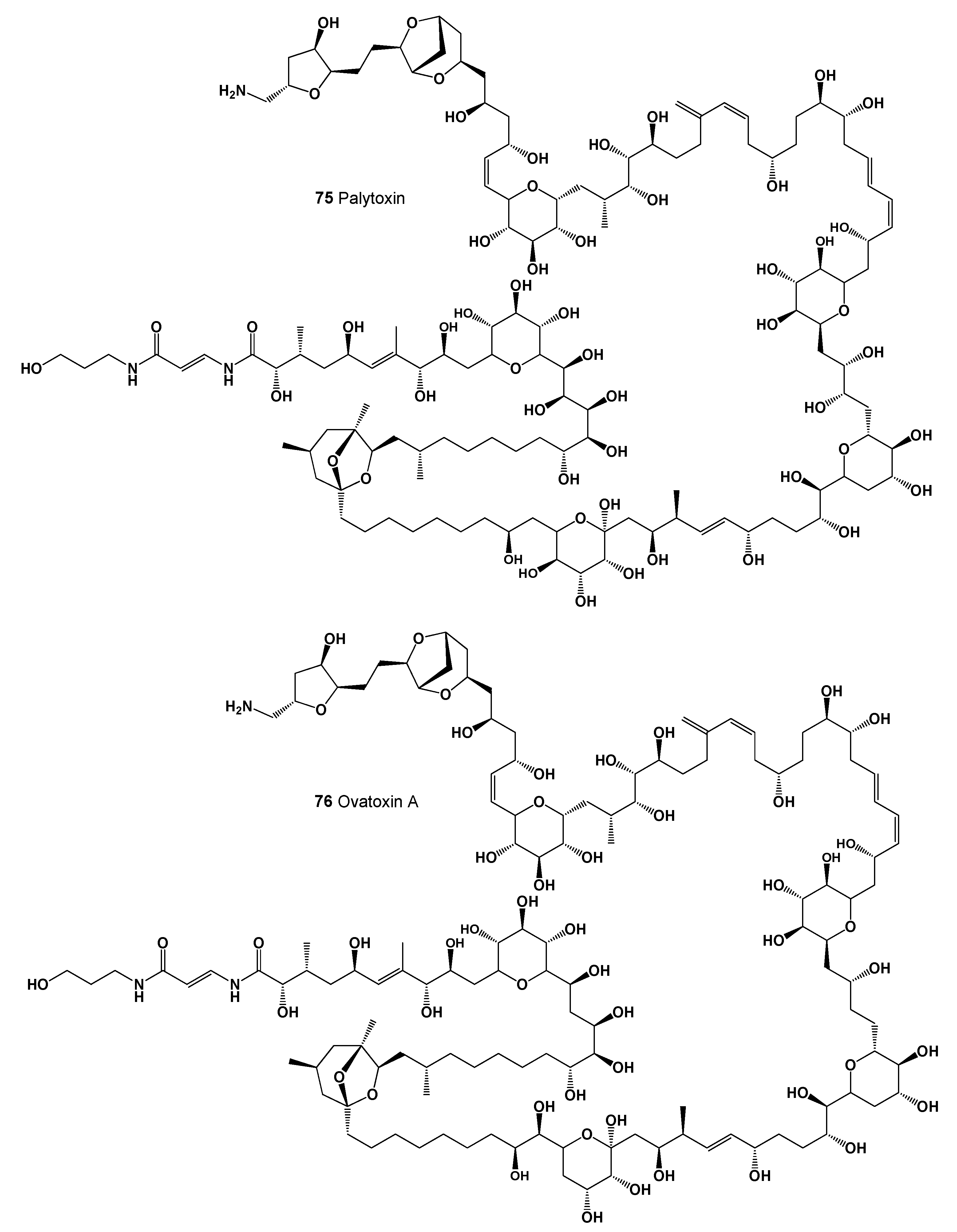
| 75 | Antifungal (0.973); Antibacterial (0.939); Antineoplastic (0.870); Apoptosis agonist (0.628) |
| 76 | Antifungal (0.975); Antibacterial (0.943); Antineoplastic (0.879); Apoptosis agonist (0.638) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 75 | Antifungal (0.973); Antibacterial (0.939); Antineoplastic (0.870); Apoptosis agonist (0.628) |
| 76 | Antifungal (0.975); Antibacterial (0.943); Antineoplastic (0.879); Apoptosis agonist (0.638) |
| 77 | Antiprotozoal (Coccidial) (0.943); Antibacterial (0.939); Antiparasitic (0.867) |
| 78 | Antiprotozoal (Coccidial) (0.928); Antibacterial (0.921); Antiparasitic (0.872) |
| 79 | Antiprotozoal (Coccidial) (0.906); Antibacterial (0.902); Antiparasitic (0.877) |
| 80 | Antibacterial (0.953); Antiprotozoal (Coccidial) (0.924); Antitreponemal (0.906) Antiparasitic (0.898); Antineoplastic (0.867); Antiprotozoal (Plasmodium) (0.686) |
| 81 | Antineoplastic (0.893); Antifungal (0.842); Antibacterial (0.818) Apoptosis agonist (0.638); Antiprotozoal (Coccidial) (0.634); Antimetastatic (0.629) |
| 82 | Growth stimulant (0.974); Antimycobacterial (0.971); Antifungal (0.967) Antiprotozoal (Coccidial) (0.943); Antibacterial (0.942); Antineoplastic (0.937) Antiparasitic (0.919); Antiprotozoal (Plasmodium) (0.699); Antimetastatic (0.602) |
| 83 | Growth stimulant (0.974); Antimycobacterial (0.972); Antifungal (0.971) Antiprotozoal (Coccidial) (0.947); Antibacterial (0.943); Antineoplastic (0.937) Antiparasitic (0.921); Antiprotozoal (Plasmodium) (0.701); Antimetastatic (0.604) Antineoplastic (renal cancer) (0.604); Antineoplastic (sarcoma) (0.590) Apoptosis agonist (0.580); Antineoplastic (lymphocytic leukemia) (0.559) |
| 84 | Growth stimulant (0.974); Antimycobacterial (0.972); Antifungal (0.971) Antiprotozoal (Coccidial) (0.947); Antibacterial (0.943); Antineoplastic (0.937) Antiparasitic (0.921); Antiprotozoal (Plasmodium) (0.701); Antimitotic (0.663) |
| 85 | Antimycobacterial (0.963); Growth stimulant (0.960); Antineoplastic (0.956) Antifungal (0.951); Antibacterial (0.942); Antiprotozoal (Coccidial) (0.937) Antiparasitic (0.917); Apoptosis agonist (0.914); Antiprotozoal (Plasmodium) (0.701) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 86 | Growth stimulant (0.932); Antitreponemal (0.908); Antineoplastic (0.906) Antiprotozoal (Coccidial) (0.900); Antiparasitic (0.891); Antifungal (0.882) |
| 87 | Growth stimulant (0.932); Antitreponemal (0.910); Antineoplastic (0.906) Antiprotozoal (Coccidial) (0.900); Antiparasitic (0.891); Antifungal (0.877) |
| 88 | Antieczematic (0.920); Antibacterial (0.839); Antineoplastic (0.802) |
| 89 | Antieczematic (0.922); Antibacterial (0.834); Antineoplastic (0.820) |
| 90 | Antineoplastic (0.929); Growth stimulant (0.920); Antibacterial (0.918) Antiprotozoal (Coccidial) (0.903); Antifungal (0.898); Antiparasitic (0.877) Antimycobacterial (0.790); Antineoplastic (renal cancer) (0.626); Antimetastatic (0.624) |
| 91 | Antineoplastic (0.928); Growth stimulant (0.919); Antibacterial (0.919) Antiprotozoal (Coccidial) (0.903); Antifungal (0.895); Antiparasitic (0.878) Antimycobacterial (0.660); Antimitotic (0.659); Antineoplastic (renal cancer) (0.623) Antimetastatic (0.622); Antiprotozoal (Plasmodium) (0.547); Chemoprotective (0.523) |
| 92 | Antibacterial (0.896); Growth stimulant (0.886); Antiprotozoal (Coccidial) (0.877) Antineoplastic (0.868); Antifungal (0.854); Antiparasitic (0.747) |
| 93 | Antibacterial (0.945); Antiprotozoal (Coccidial) (0.938); Growth stimulant (0.929) Antiparasitic (0.916); Antitreponemal (0.915); Antiprotozoal (Plasmodium) (0.731) |
| 94 | Antibacterial (0.944); Antiprotozoal (Coccidial) (0.940); Growth stimulant (0.918) Antitreponemal (0.917); Antiparasitic (0.915); Antineoplastic (0.900); Antifungal (0.858) Antiprotozoal (Plasmodium) (0.732); Antineoplastic (renal cancer) (0.677) Antimycoplasmal (0.673); Antineoplastic (lymphocytic leukemia) (0.583) |
| 95 | Antibacterial (0.912); Antineoplastic (0.873); Antiprotozoal (Coccidial) (0.865) Antiparasitic (0.796); Antifungal (0.794); Antineoplastic (renal cancer) (0.633) Antineoplastic (sarcoma) (0.621); Antimetastatic (0.578) Antineoplastic (lymphocytic leukemia) (0.551) |
| 96 | Antibacterial (0.992); Antiprotozoal (Coccidial) (0.964); Antitreponemal (0.954) Antiparasitic (0.948); Antineoplastic (0.901); Antifungal (0.796) Antiprotozoal (Plasmodium) (0.790); Antineoplastic (renal cancer) (0.575) |
| 97 | Antibacterial (0.981); Antiprotozoal (Coccidial) (0.978); Antitreponemal (0.966) Antiparasitic (0.946); Antineoplastic (0.919); Growth stimulant (0.842) Antiprotozoal (Plasmodium) (0.696); Antimycoplasmal (0.597) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 98 | Antibacterial (0.992); Antiprotozoal (Coccidial) (0.977); Antitreponemal (0.959) Antiparasitic (0.950); Antifungal (0.799); Antiprotozoal (Plasmodium) (0.778) |
| 99 | Antibacterial (0.991); Antiprotozoal (Coccidial) (0.964); Antitreponemal (0.953) Antiparasitic (0.950); Antiprotozoal (Plasmodium) (0.803); Antifungal (0.798) |
| 100 | Antimycobacterial (0.957); Antineoplastic (0.956); Antifungal (0.913) Antibacterial (0.910); Antiparasitic (0.876); Antihelmintic (0.804) |
| 101 | Growth stimulant (0.946); Antibacterial (0.921); Antiprotozoal (Coccidial) (0.918) Antiparasitic (0.909); Antineoplastic (0.905); Antifungal (0.881) Antiprotozoal (0.880); Antitreponemal (0.829); Antiprotozoal (Plasmodium) (0.702) |
| 102 | Antiprotozoal (0.944); Antibacterial (0.935); Antiprotozoal (Coccidial) (0.931) Antiparasitic (0.927); Antitreponemal (0.914); Antifungal (0.879) Antiprotozoal (Plasmodium) (0.725); Antimycoplasmal (0.603) |
| 103 | Antiprotozoal (0.948); Antiprotozoal (Coccidial) (0.938); Antibacterial (0.935) Antitreponemal (0.930); Antiparasitic (0.927); Antifungal (0.880); Antihelmintic (0.704) Antiprotozoal (Plasmodium) (0.690); Antimycoplasmal (0.650) |
| 104 | Antibacterial (0.946); Antiprotozoal (Coccidial) (0.943); Antiparasitic (0.932) Antitreponemal (0.924); Antifungal (0.890); Antiprotozoal (Plasmodium) (0.692) |
| 105 | Antibacterial (0.930); Antitreponemal (0.926); Antiprotozoal (Coccidial) (0.922) Antiparasitic (0.914); Antifungal (0.858); Antiprotozoal (Plasmodium) (0.637) |
| 106 | Antineoplastic (0.919); Antifungal (0.894); Antiparasitic (0.872); Antibacterial (0.869) |
| 107 | Antibacterial (0.995); Antiprotozoal (Coccidial) (0.988); Antineoplastic (0.926) Antiparasitic (0.925); Growth stimulant (0.904); Antifungal (0.898) Antitreponemal (0.803); Antimitotic (0.706); Antihelmintic (0.702) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
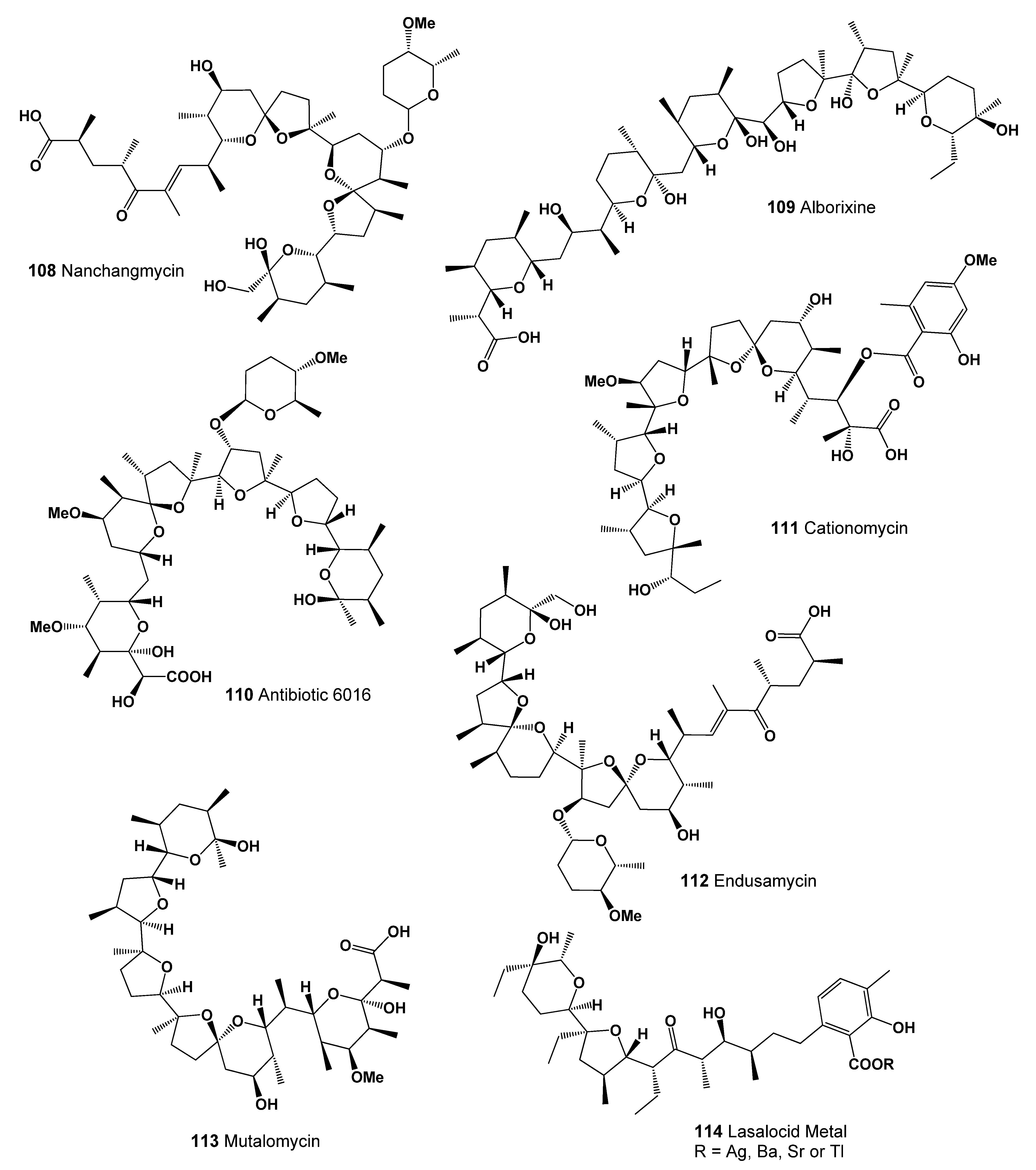
| 108 | Antibacterial (0.996); Antiparasitic (0.994); Antiprotozoal (Coccidial) (0.985) Antineoplastic (0.937); Antifungal (0.935); Antitreponemal (0.919) Antiprotozoal (Plasmodium) (0.769); Antimycoplasmal (0.570) |
| 109 | Antibacterial (0.936); Antiprotozoal (Coccidial) (0.890); Antiparasitic (0.791) Antifungal (0.789); Antiprotozoal (Plasmodium) (0.507); Antimycoplasmal (0.500) |
| 110 | Antibacterial (0.991); Antiprotozoal (Coccidial) (0.990); Antiparasitic (0.924) Antitreponemal (0.921); Antineoplastic (0.915); Antiprotozoal (Plasmodium) (0.746) |
| 111 | Antimycobacterial (0.969); Antibacterial (0.936); Antifungal (0.907) Antiprotozoal (Coccidial) (0.897); Antiparasitic (0.866); Antimycoplasmal (0.533) |
| 112 | Antibacterial (0.994); Antiprotozoal (Coccidial) (0.969); Antiparasitic (0.968) Antineoplastic (0.932); Antifungal (0.924); Antitreponemal (0.904) Antiprotozoal (Plasmodium) (0.703); Antimycoplasmal (0.562) |
| 113 | Antibacterial (0.946); Antiprotozoal (Coccidial) (0.943); Antiparasitic (0.921) Antitreponemal (0.915); Antifungal (0.875); Antiprotozoal (Plasmodium) (0.693) |
| 114 | Growth stimulant (0.932); Antitreponemal (0.908); Antineoplastic (0.906) Antiprotozoal (Coccidial) (0.900); Antiparasitic (0.891); Antifungal (0.882) |
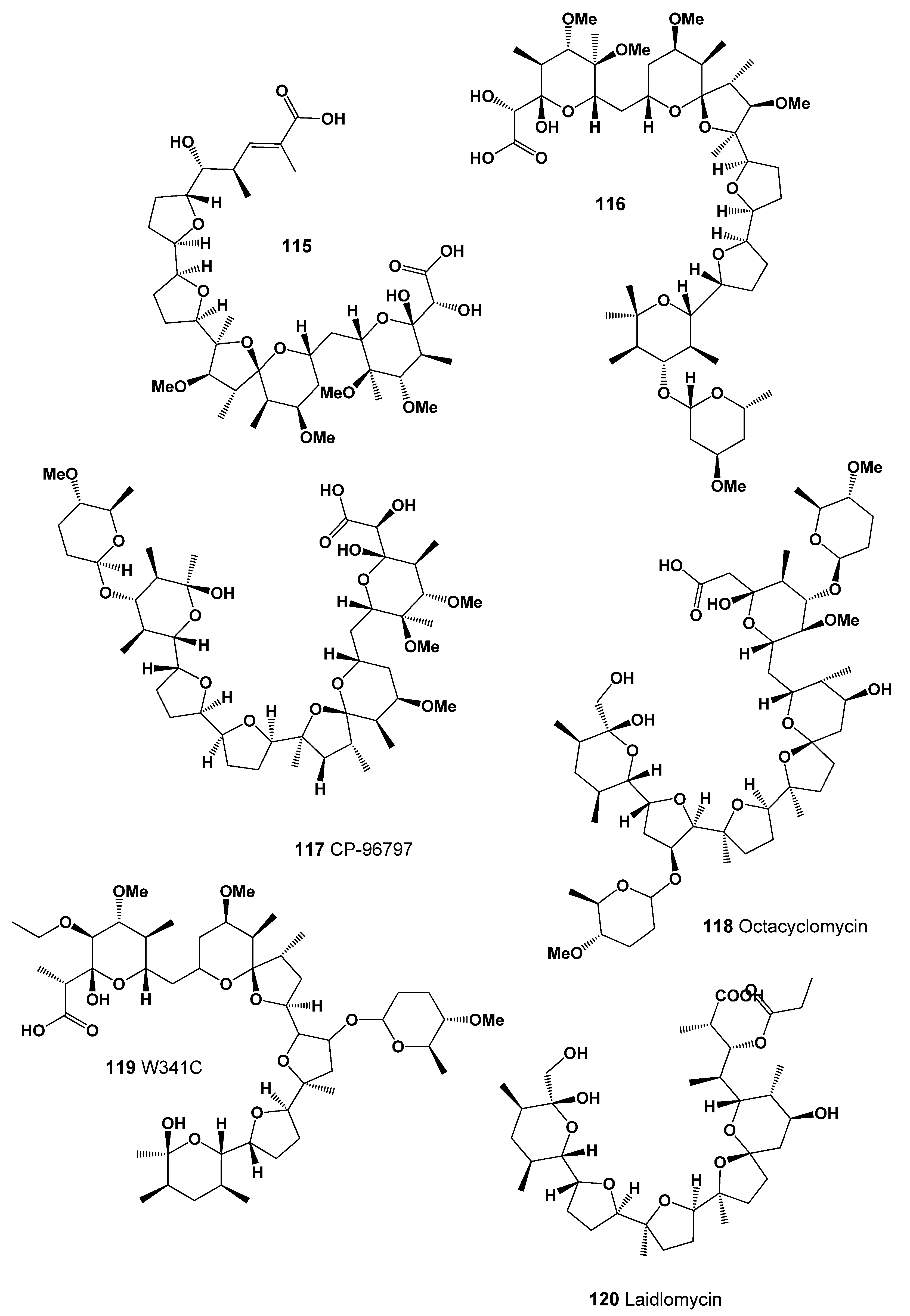
| 115 | Antiprotozoal (Coccidial) (0.916); Antibacterial (0.912); Antiparasitic (0.907) |
| 116 | Antibacterial (0.955); Antiprotozoal (Coccidial) (0.929); Antitreponemal (0.916) |
| 117 | Antibacterial (0.982); Antiprotozoal (Coccidial) (0.977); Antitreponemal (0.952) Antiparasitic (0.941); Antifungal (0.831); Antiprotozoal (Plasmodium) (0.786) |
| 118 | Antibacterial (0.995); Antiprotozoal (Coccidial) (0.986); Antineoplastic (0.936) Antiparasitic (0.921); Growth stimulant (0.907); Antifungal (0.884) Antitreponemal (0.843); Antiprotozoal (Plasmodium) (0.708); Antimycoplasmal (0.647) |
| 119 | Antibacterial (0.995); Antiprotozoal (Coccidial) (0.984); Antiparasitic (0.950) Antineoplastic (0.930); Growth stimulant (0.889); Antitreponemal (0.886) Antifungal (0.842); Antiprotozoal (Plasmodium) (0.723); Antimycoplasmal (0.582) |
| 120 | Growth stimulant (0.945); Antiprotozoal (Coccidial) (0.922); Antibacterial (0.921) Antiparasitic (0.916); Antitreponemal (0.775); Antiprotozoal (Plasmodium) (0.729) |
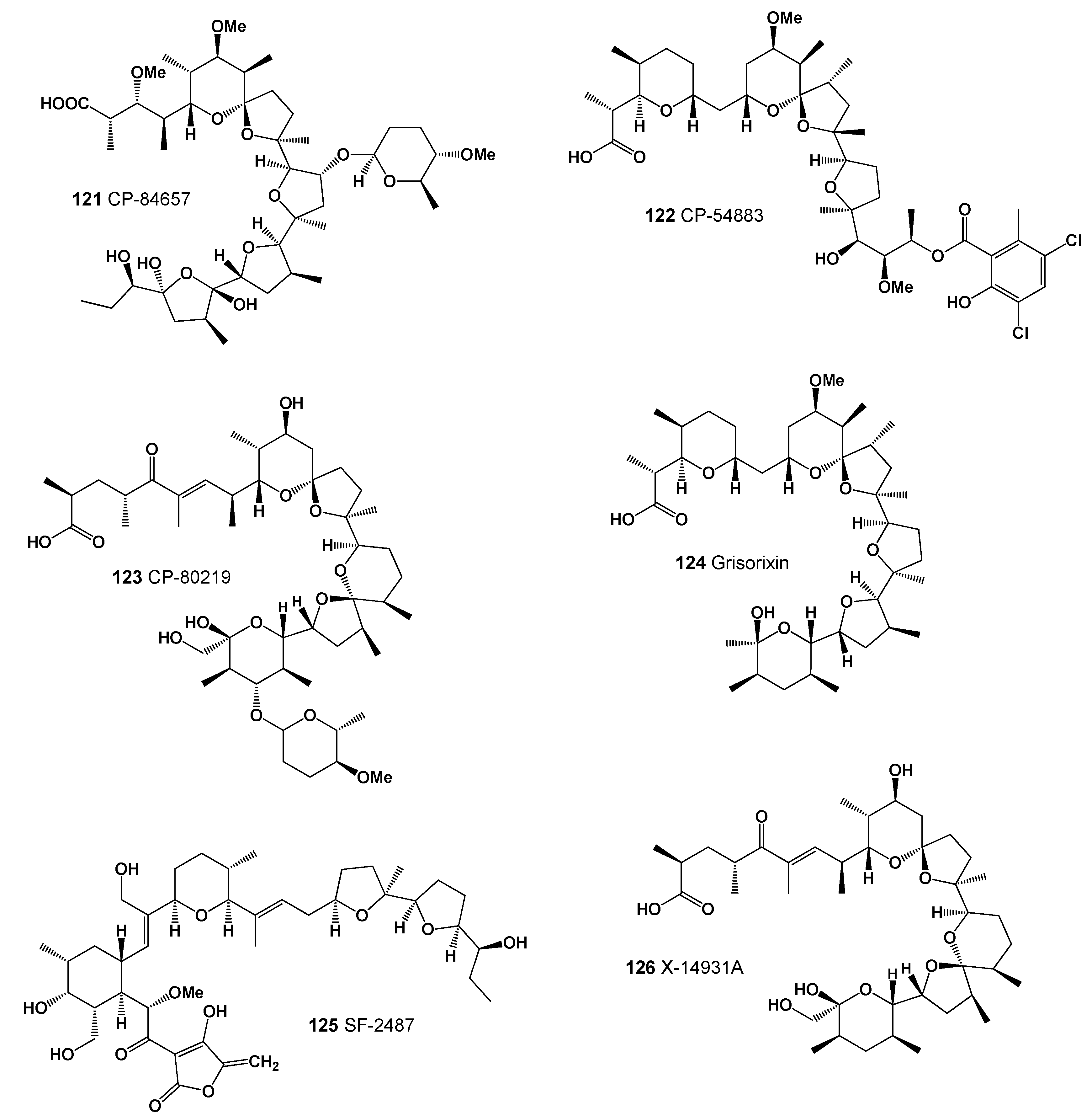
| 121 | Antibacterial (0.987); Antiprotozoal (Coccidial) (0.962); Antineoplastic (0.947) Antifungal (0.907); Antiparasitic (0.889); Antiprotozoal (Plasmodium) (0.686) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 122 | Antibacterial (0.944); Antiprotozoal (Coccidial) (0.940); Antitreponemal (0.917) Antiparasitic (0.915); Antifungal (0.858); Antiprotozoal (Plasmodium) (0.732) |
| 123 | Antiprotozoal (Coccidial) (0.894); Antiparasitic (0.893); Antibacterial (0.876) |
| 124 | Antineoplastic (0.907); Antibacterial (0.893); Antifungal (0.863); Apoptosis agonist (0.799) |
| 125 | Antibacterial (0.944); Antiparasitic (0.941); Antiprotozoal (Coccidial) (0.935) Antitreponemal (0.925); Antifungal (0.905); Antiprotozoal (Plasmodium) (0.764) |
| 126 | Antiparasitic (0.995); Antibacterial (0.995); Antiprotozoal (Coccidial) (0.986) Antifungal (0.935); Antitreponemal (0.933); Antiprotozoal (Plasmodium) (0.776) |
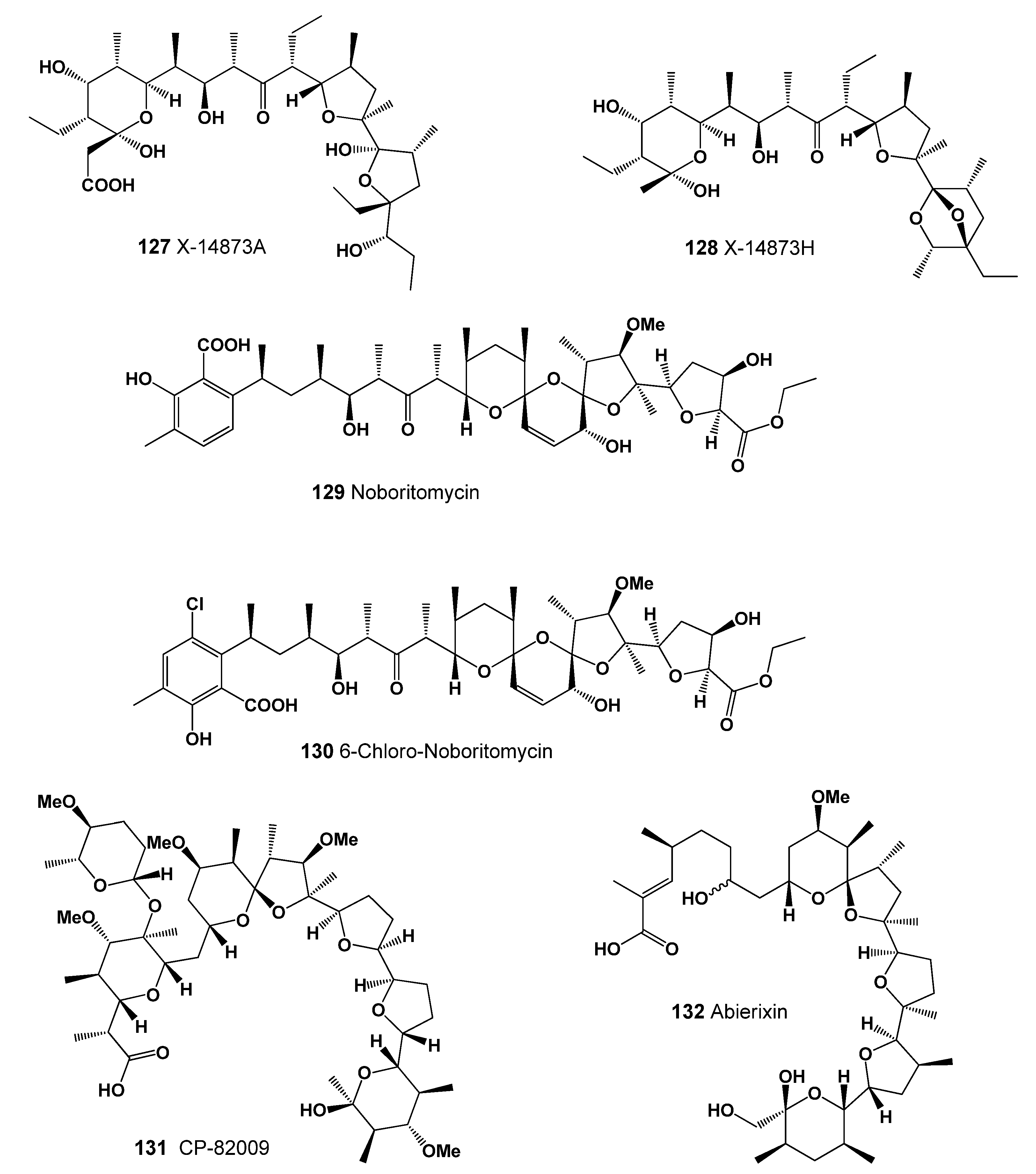
| 127 | Antibacterial (0.925); Antineoplastic (0.907); Antiprotozoal (Coccidial) (0.899) |
| 128 | Antineoplastic (0.985); Chemoprotective (0.964); Growth stimulant (0.934) Antitreponemal (0.913); Antiparasitic (0.902); Antifungal (0.902); Antiprotozoal (Coccidial) (0.895) Antibacterial (0.874); Antihelmintic (0.660); Antiprotozoal (Plasmodium) (0.602) |
| 129 | Antineoplastic (0.932); Growth stimulant (0.926); Antibacterial (0.912) Antiprotozoal (Coccidial) (0.910); Antifungal (0.897); Antiparasitic (0.878) Antimycobacterial (0.775); Antitreponemal (0.676); Antimetastatic (0.629) |
| 130 | Antineoplastic (0.924); Growth stimulant (0.922); Antiprotozoal (Coccidial) (0.910) Antibacterial (0.904); Antifungal (0.898); Antiparasitic (0.880); Antimycobacterial (0.800) |
| 131 | Antibacterial (0.981); Antiprotozoal (Coccidial) (0.978); Antitreponemal (0.961) Antiparasitic (0.950); Antineoplastic (0.930); Antiprotozoal (Plasmodium) (0.646) |
| 132 | Antibacterial (0.949); Antiprotozoal (Coccidial) (0.911); Growth stimulant (0.909) Antiparasitic (0.899); Antifungal (0.894); Antitreponemal (0.841) |
| No. | Predicted Biological Activity, Pa * |
|---|---|
| 133 | Antibacterial (0.975); Antineoplastic (0.948); Antiprotozoal (Coccidial) (0.935) Antiparasitic (0.909); Antifungal (0.904); Antihelmintic (0.821); Antitreponemal (0.724) Antimycoplasmal (0.633); Antiprotozoal (Plasmodium) (0.628) |
| 134 | Antibacterial (0.994); Antiparasitic (0.979); Antiprotozoal (Coccidial) (0.973) Antineoplastic (0.935); Antifungal (0.922); Antitreponemal (0.911) Antiprotozoal (Plasmodium) (0.738); Antimycoplasmal (0.525) |
| 135 | Antibacterial (0.969); Antiprotozoal (Coccidial) (0.933); Antineoplastic (0.917) Antiparasitic (0.904); Antitreponemal (0.897); Antiprotozoal (Plasmodium) (0.716) |
| 136 | Growth stimulant (0.965); Antimycobacterial (0.965); Antifungal (0.945) Antineoplastic (0.940); Antiprotozoal (Coccidial) (0.935); Antibacterial (0.935) Antiparasitic (0.921); Antitreponemal (0.861); Antihelmintic (0.855) |
| 137 | Growth stimulant (0.968); Antimycobacterial (0.967); Antifungal (0.949) Antineoplastic (0.944); Antiprotozoal (Coccidial) (0.933); Antibacterial (0.930) Antiparasitic (0.922); Antiprotozoal (Plasmodium) (0.788) |
| 138 | Antibacterial (0.940); Antiprotozoal (Coccidial) (0.935); Antineoplastic (0.891) Antifungal (0.880); Antiparasitic (0.848); Antimycoplasmal (0.579) |
| 139 | Growth stimulant (0.883); Antitreponemal (0.837); Antiprotozoal (Coccidial) (0.829) Antibacterial (0.823); Antiparasitic (0.814); Antiprotozoal (Plasmodium) (0.546) |
| 140 | Growth stimulant (0.883); Antitreponemal (0.837); Antiprotozoal (Coccidial) (0.829) Antibacterial (0.823); Antiparasitic (0.814); Antiprotozoal (Plasmodium) (0.546) |
| 141 | Antimycobacterial (0.965); Antibacterial (0.932); Antifungal (0.905) Antiprotozoal (Coccidial) (0.894); Antiparasitic (0.861) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dembitsky, V.M. Natural Polyether Ionophores and Their Pharmacological Profile. Mar. Drugs 2022, 20, 292. https://doi.org/10.3390/md20050292
Dembitsky VM. Natural Polyether Ionophores and Their Pharmacological Profile. Marine Drugs. 2022; 20(5):292. https://doi.org/10.3390/md20050292
Chicago/Turabian StyleDembitsky, Valery M. 2022. "Natural Polyether Ionophores and Their Pharmacological Profile" Marine Drugs 20, no. 5: 292. https://doi.org/10.3390/md20050292
APA StyleDembitsky, V. M. (2022). Natural Polyether Ionophores and Their Pharmacological Profile. Marine Drugs, 20(5), 292. https://doi.org/10.3390/md20050292








