Description and Genomic Characterization of Oceaniferula flavus sp. nov., a Novel Potential Polysaccharide-Degrading Candidate of the Difficult-to-Cultivate Phylum Verrucomicrobiota Isolated from Seaweed
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Maintenance of Strain 5K15T
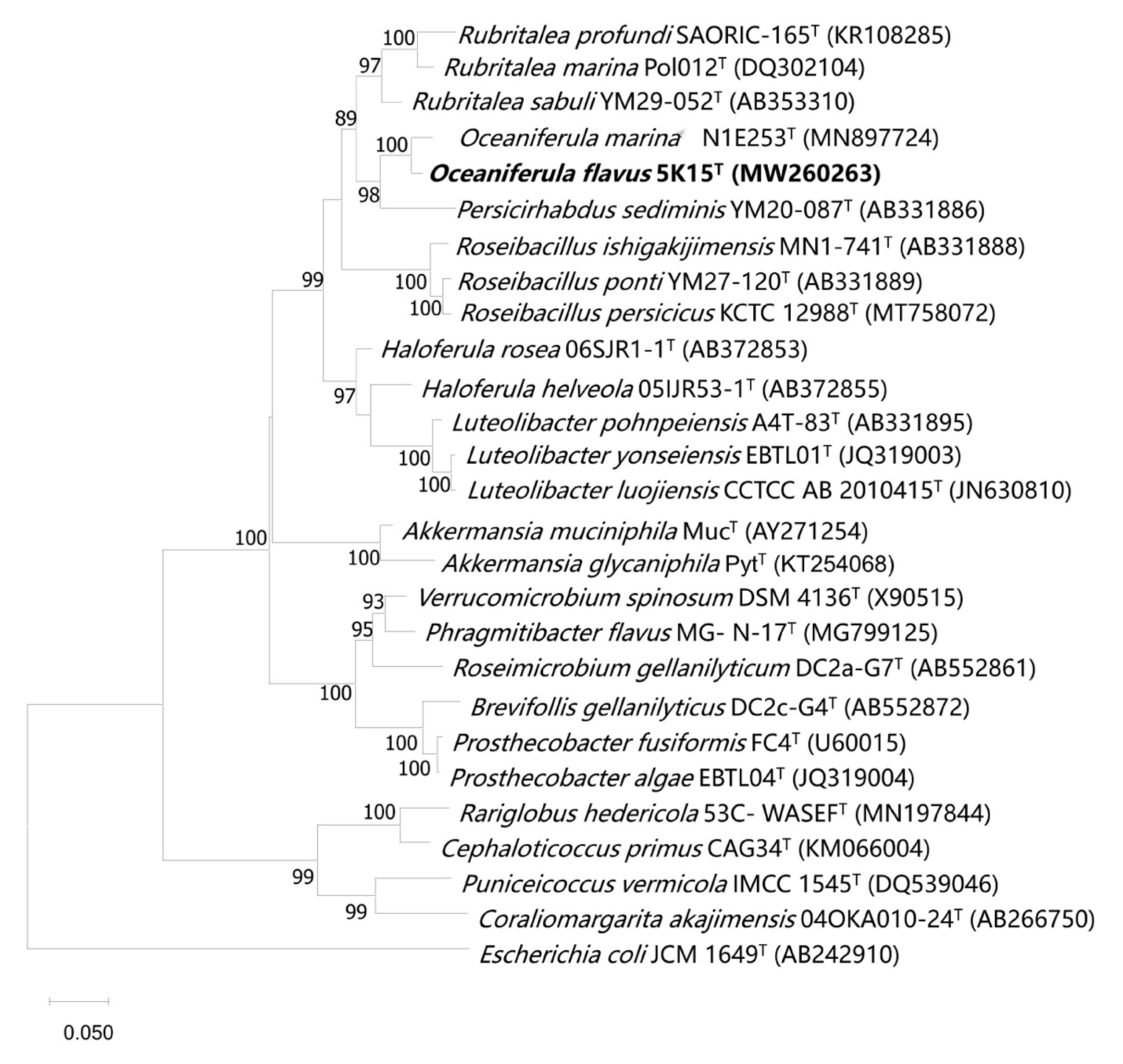
2.2. 16S rRNA Gene Sequence and Phylogenetic Analysis
2.3. Morphological, Physiological, and Biochemical Characteristics
2.4. Chemotaxonomic Characterization
2.5. Genome insight of the Two Strains of Genus Oceaniferula
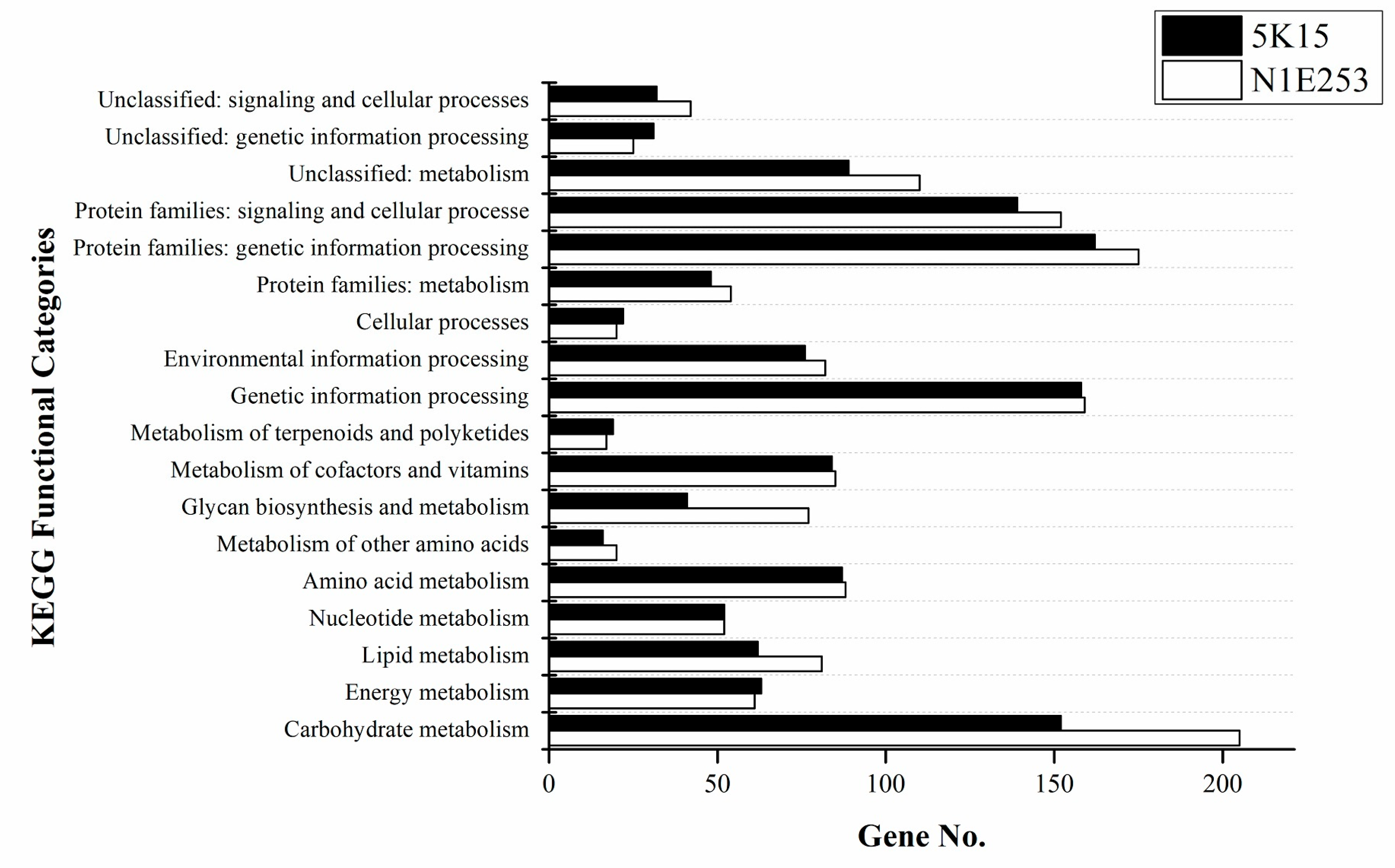
2.5.1. Genome Properties
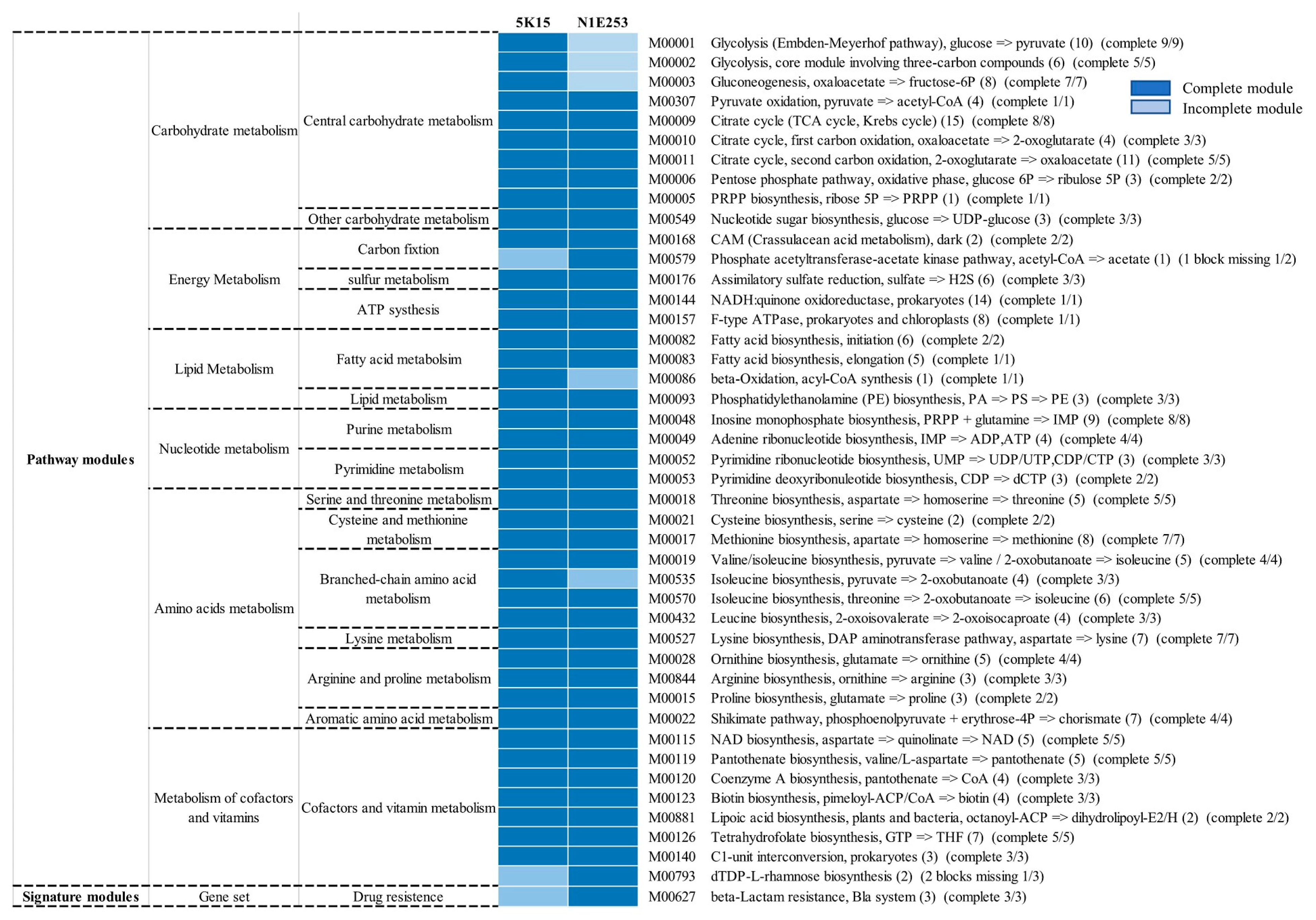
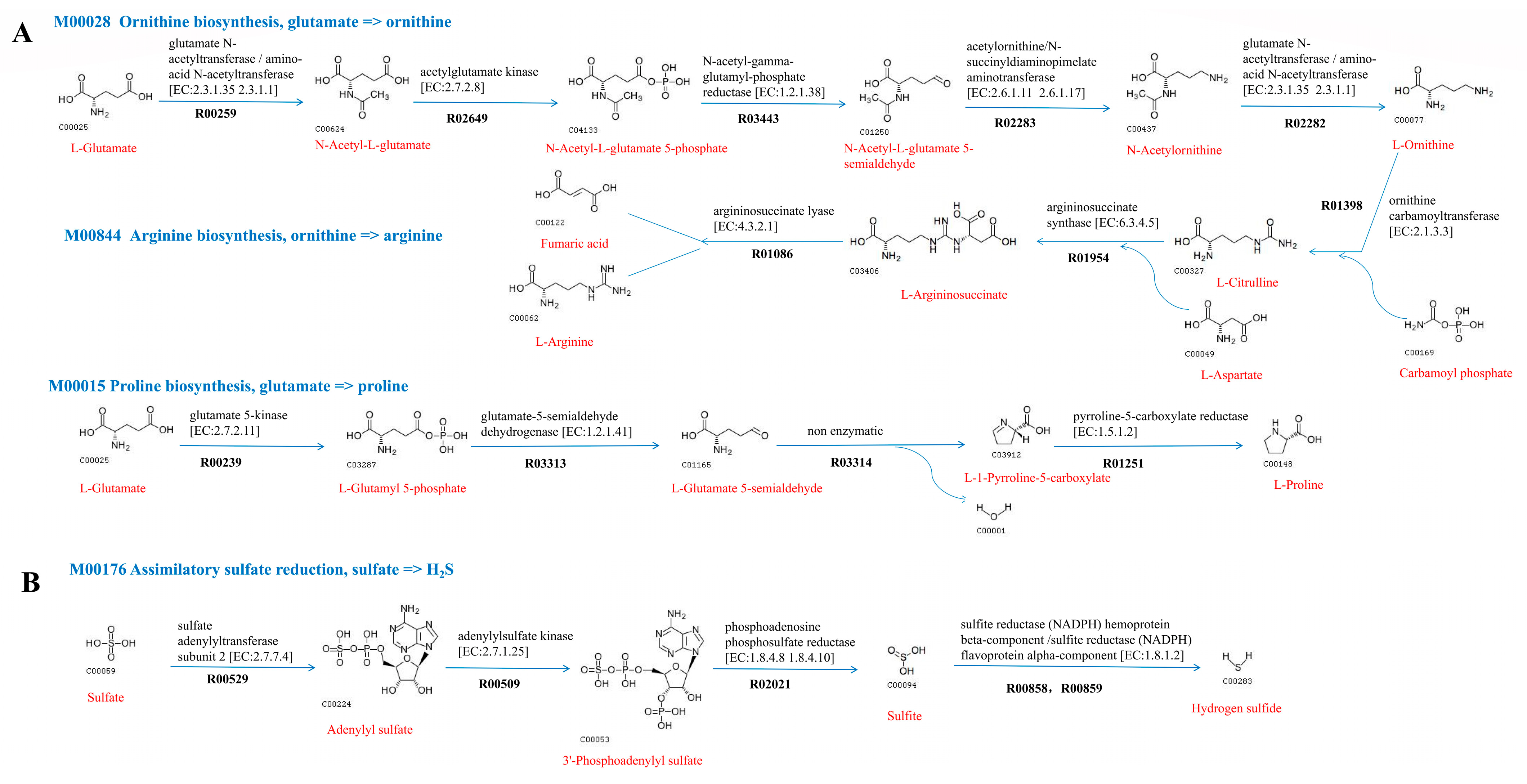
2.5.2. Metabolic Pathways
2.5.3. Prediction of Secondary Metabolites
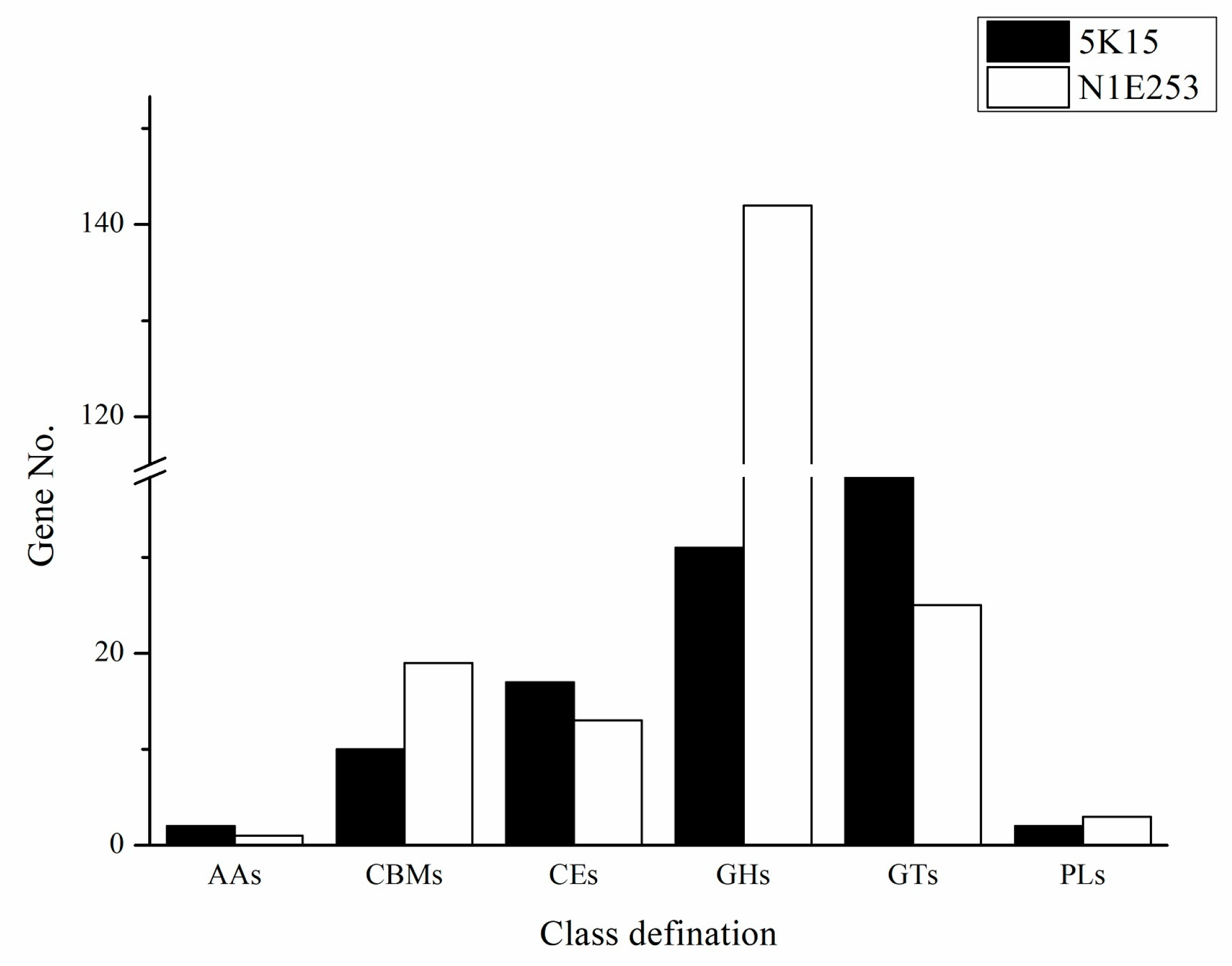
2.5.4. Estimation of Carbohydrate-Active Enzymes (CAZymes)
2.6. Description of Oceaniferula flavus sp. nov. Oceaniferula flavus (L. masc. adj. flavus, Yellow, Referring to the Color of the Colonies)
3. Materials and Methods
3.1. Sample Collection, Isolation, and Preservation
3.2. Physiological and Biochemical Characteristics
3.3. Chemotaxonomy
3.4. DNA Sequencing and Phylogenetic Analyses
3.5. Genomic DNA Sequencing, Assembly, Annotation, and Genome Comparison
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Del Olmo, A.; Picon, A.; Nuñez, M. The microbiota of eight species of dehydrated edible seaweeds from North West Spain. Food Microbiol. 2018, 70, 224–231. [Google Scholar] [CrossRef]
- Xu, S.; Huang, X.; Cheong, K. Recent advances in marine algae polysaccharides: Isolation, structure, and activities. Mar. Drugs 2017, 15, 388. [Google Scholar] [CrossRef] [Green Version]
- Echave, J.; Otero, P.; Garcia-Oliveira, P.; Munekata, P.E.S.; Pateiro, M.; Lorenzo, J.M.; Simal-Gandara, J.; Prieto, M.A. Seaweed-derived proteins and peptides: Promising marine bioactives. Antioxidants 2022, 11, 176. [Google Scholar] [CrossRef]
- Rawiwan, P.; Peng, Y.; Paramayuda, I.G.P.B.; Quek, S.Y. Red seaweed: A promising alternative protein source for global food sustainability. Trends. Food Sci. Technol. 2022, 123, 37–56. [Google Scholar] [CrossRef]
- Brown, E.M.; Allsopp, P.J.; Magee, P.J.; Gill, C.I.; Nitecki, S.; Strain, C.R.; McSorley, E.M. Seaweed and human health. Nutr. Rev. 2014, 72, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Meinita, M.D.N.; Harwanto, D.; Choi, J. Seaweed exhibits therapeutic properties against chronic diseases: An overview. Appl. Sci. 2022, 12, 2638. [Google Scholar] [CrossRef]
- Shah, M.D.; Venmathi Maran, B.A.; Shaleh, S.R.M.; Zuldin, W.H.; Gnanaraj, C.; Yong, Y.S. Therapeutic potential and nutraceutical profiling of north bornean seaweeds: A review. Mar. Drugs 2022, 20, 101. [Google Scholar] [CrossRef]
- Msuya, F.E.; Bolton, J.; Pascal, F.; Narrain, K.; Nyonje, B.; Cottier-Cook, E.J. Seaweed farming in Africa: Current status and future potential. J. Appl. Phycol. 2022, 34, 985–1005. [Google Scholar] [CrossRef]
- Ren, C.G.; Liu, Z.Y.; Wang, X.L.; Qin, S. The seaweed holobiont: From microecology to biotechnological applications. Microb. Biotechnol. 2022, 15, 738–754. [Google Scholar] [CrossRef]
- Ramanan, R.; Kang, Z.; Kim, B.; Cho, D.; Jin, L.; Oh, H.; Kim, H.S. Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats. Algal Res. 2015, 8, 140–144. [Google Scholar] [CrossRef]
- Grant, M.A.; Kazamia, E.; Cicuta, P.; Smith, A.G. Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal–bacterial cocultures. ISME J. 2014, 8, 1418–1427. [Google Scholar] [CrossRef] [Green Version]
- Bengtsson, M.M.; Sjotun, K.; Lanzen, A.; Ovreas, L. Bacterial diversity in relation to secondary production and succession on surfaces of the kelp Laminaria hyperborea. ISME J. 2012, 6, 2188–2198. [Google Scholar] [CrossRef]
- Jin, C.B.; Feng, X.; Zou, Q.H.; Ye, M.Q.; Du, Z.J. Oceaniferula marina gen. nov., sp. nov., an anti-fluoroquinolone bacterium isolated from marine sediment. Antonie Leeuwenhoek 2021, 114, 1855–1865. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.P.; Ward, N.L. Mind the PVCs. Environ. Microbiol. 2014, 16, 1217–1221. [Google Scholar] [CrossRef]
- Fuerst, J.A. Phylum Verrucomicrobia. In Reference Module in Life Sciences; Roitberg, B.D., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 551–563. [Google Scholar]
- Sichert, A.; Corzett, C.H.; Schechter, M.S.; Unfried, F.; Markert, S.; Becher, D.; Fernandez-Guerra, A.; Liebeke, M.; Schweder, T.; Polz, M.F.; et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 2020, 5, 1026–1046. [Google Scholar] [CrossRef]
- Orellana, L.H.; Francis, T.B.; Ferraro, M.; Hehemann, J.; Fuchs, B.M.; Amann, R.I. Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J. 2021, 16, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Qin, Q.L.; Xie, B.B.; Zhang, X.Y.; Chen, X.L.; Zhou, B.C.; Zhou, J.; Oren, A.; Zhang, Y.Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.V.; Jarboe, B.G.; Majer, H.M.; Ma, A.T.; Beld, J. Escherichia coli Nissle 1917 secondary metabolism: Aryl polyene biosynthesis and phosphopantetheinyl transferase crosstalk. Appl. Microbiol. Biotechnol. 2021, 105, 7785–7799. [Google Scholar] [CrossRef]
- Katsuyama, Y.; Ohnishi, Y. Chapter Sixteen—Type III Polyketide Synthases in Microorganisms. In Methods in Enzymolog; Hopwood, D.A., Ed.; Academic Press: Salt Lake City, UT, USA, 2012. [Google Scholar]
- Lim, Y.; Kang, I.; Cho, J. Genome characteristics of Kordia antarctica IMCC3317T and comparative genome analysis of the genus Kordia. Sci. Rep. 2020, 10, 14715. [Google Scholar] [CrossRef]
- Bowman, J.P. Description of Cellulophaga algicola sp. nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Cellulophaga uliginosa comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 1861–1868. [Google Scholar] [CrossRef] [Green Version]
- Bauer, A.; Kirby, W.; Sherris, J.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Cowan, S.T.; Steel, K.J. Bacterial characters and characterization. In Cowan and Steel’s Manual for the Identification of Medical Bacteria, 2nd ed.; Cowan, S.T., Ed.; Cambridge University Press: Cambridge, UK, 1974. [Google Scholar]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, MIDI Technical Note 101; Microbial MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Meth. 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Kroppenstedt, R.M. Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J. Liq. Chromatogr. 1982, 5, 2359–2367. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Liu, Q.Q.; Wang, Y.; Li, J.; Du, Z.J.; Chen, G.J. Saccharicrinis carchari sp. nov., isolated from a shark, and emended descriptions of the genus Saccharicrinis and Saccharicrinis fermentans. Int. J. Syst. Evol. Microbiol. 2014, 64, 2204–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luke, S. Two fast tree-creation algorithms for genetic programming. IEEE Trans. Evol. Comput. 2000, 4, 274–283. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Li, R.Q.; Yu, C.; Li, Y.R.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Valencia, R.; Gonzalez, V.; Undabarrena, A.; Zamora-Leiva, L.; Ugalde, J.A.; Camara, B. An integrative bioinformatic analysis for keratinase detection in marine-derived Streptomyces. Mar. Drugs 2021, 19, 286. [Google Scholar] [CrossRef] [PubMed]
- Konstantinou, D.; Popin, R.V.; Fewer, D.P.; Sivonen, K.; Gkelis, S. Genome reduction and secondary metabolism of the marine sponge-associated Cyanobacterium Leptothoe. Mar. Drugs 2021, 19, 298. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | 5K15T | O. marina N1E253T |
|---|---|---|
| Cell size (μm) | 0.3–0.5 × 0.7–2.0 | 0.5–0.8 × 1.5–2.0 a |
| Temperature range (°C) | 15–37 | 15–37 a |
| Growth pH | 6.0–8.5 | 7.0–9.0 a |
| Tolerance to NaCl (%) | 1.0–5.5 | 1.5–5.0 a |
| Catalase | – | + a |
| Oxidase | – | + a |
| Hydrolysis of starch | – | + a |
| API 20E test: | ||
| Tryptophan deaminase | – | + |
| Voges–Proskauer reaction | + | + |
| Oxidation of (Biolog GEN III): | ||
| d–Maltose | – | + |
| d-Mannose | + | – |
| d-Fructose | + | – |
| d-Raffinose | + | – |
| Enzyme activities (API ZYM): | ||
| Leucine arylamidase | + | + |
| N-acetyl-β-glucosaminidase | – | + |
| Acid production (API 50CHB): | ||
| d-Gentiobiose | – | + |
| d-Trehalose | – | + |
| l-Sorbitose | + | – |
| d-Lyxose | + | – |
| DNA G+C content (mol%) | 55.4 | 52.0 |
| Fatty Acid | 5K15T | O. marina N1E253T |
|---|---|---|
| Branched fatty acids | ||
| Iso-C12:0 | 6.7 | - |
| Anteiso-C13: 0 | 1.4 | - |
| Iso-C14:0 | 28.5 | 33.3 |
| Anteiso-C15: 0 | 11.8 | 4.9 |
| Iso-C16:0 | 9.5 | 7.3 |
| Straight-chain fatty acids | ||
| C12:0 | Tr | 1.2 |
| C14:0 | Tr | 1.4 |
| C16:0 | 10.0 | 12.9 |
| C17:0 | 4.0 | 7.0 |
| C18:0 | Tr | 1.3 |
| Unsaturated fatty acids | ||
| C15: 1ω6c | 2.2 | 1.5 |
| C15: 1ω8c | - | 1.9 |
| C17: 1ω6c | Tr | 4.2 |
| C17: 1ω8c | 1.2 | 6.2 |
| Hydroxy fatty acids | ||
| C12: 0 3-OH | 2.0 | Tr |
| Iso-C12: 0 3-OH | 2.3 | - |
| Iso-C14: 0 3-OH | Tr | 3.8 |
| Summed features * | ||
| Summed feature 3 | 9.3 | 7.0 |
| Characteristic | 5K15T | O. marina N1E253T |
|---|---|---|
| GenBank accession number | JAFBGL000000000 | JACBAZ000000000 |
| Genome size (bp) | 3,953,077 | 5,073,947 |
| N50 Value (bp) | 346,557 | 785,939 |
| Contigs (no.) | 51 | 88 |
| G+C content (mol/%) | 55.4 | 52 |
| Annotation Results by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) | ||
| Genes (no.) | 3354 | 3927 |
| Protein-coding genes | 3293 | 3855 |
| tRNAs (no.) | 45 | 46 |
| rRNAs (no.) | 3 | 7 |
| ncRNAs (no.) | 4 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, M.-Q.; Jin, C.-B.; Liu, X.-J.; Tan, X.-Y.; Ye, Y.-Q.; Du, Z.-J. Description and Genomic Characterization of Oceaniferula flavus sp. nov., a Novel Potential Polysaccharide-Degrading Candidate of the Difficult-to-Cultivate Phylum Verrucomicrobiota Isolated from Seaweed. Mar. Drugs 2023, 21, 31. https://doi.org/10.3390/md21010031
Ye M-Q, Jin C-B, Liu X-J, Tan X-Y, Ye Y-Q, Du Z-J. Description and Genomic Characterization of Oceaniferula flavus sp. nov., a Novel Potential Polysaccharide-Degrading Candidate of the Difficult-to-Cultivate Phylum Verrucomicrobiota Isolated from Seaweed. Marine Drugs. 2023; 21(1):31. https://doi.org/10.3390/md21010031
Chicago/Turabian StyleYe, Meng-Qi, Chuan-Bo Jin, Xin-Jiang Liu, Xin-Yun Tan, Yu-Qi Ye, and Zong-Jun Du. 2023. "Description and Genomic Characterization of Oceaniferula flavus sp. nov., a Novel Potential Polysaccharide-Degrading Candidate of the Difficult-to-Cultivate Phylum Verrucomicrobiota Isolated from Seaweed" Marine Drugs 21, no. 1: 31. https://doi.org/10.3390/md21010031
APA StyleYe, M. -Q., Jin, C. -B., Liu, X. -J., Tan, X. -Y., Ye, Y. -Q., & Du, Z. -J. (2023). Description and Genomic Characterization of Oceaniferula flavus sp. nov., a Novel Potential Polysaccharide-Degrading Candidate of the Difficult-to-Cultivate Phylum Verrucomicrobiota Isolated from Seaweed. Marine Drugs, 21(1), 31. https://doi.org/10.3390/md21010031






