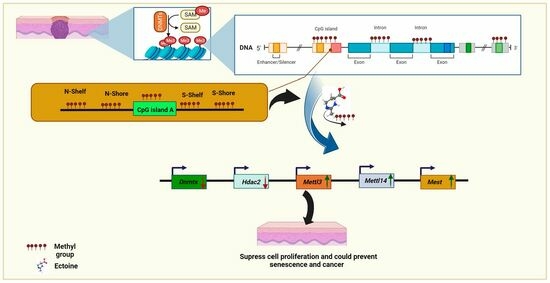
Ectoine Globally Hypomethylates DNA in Skin Cells and Suppresses Cancer Proliferation
Abstract
:1. Introduction
2. Results
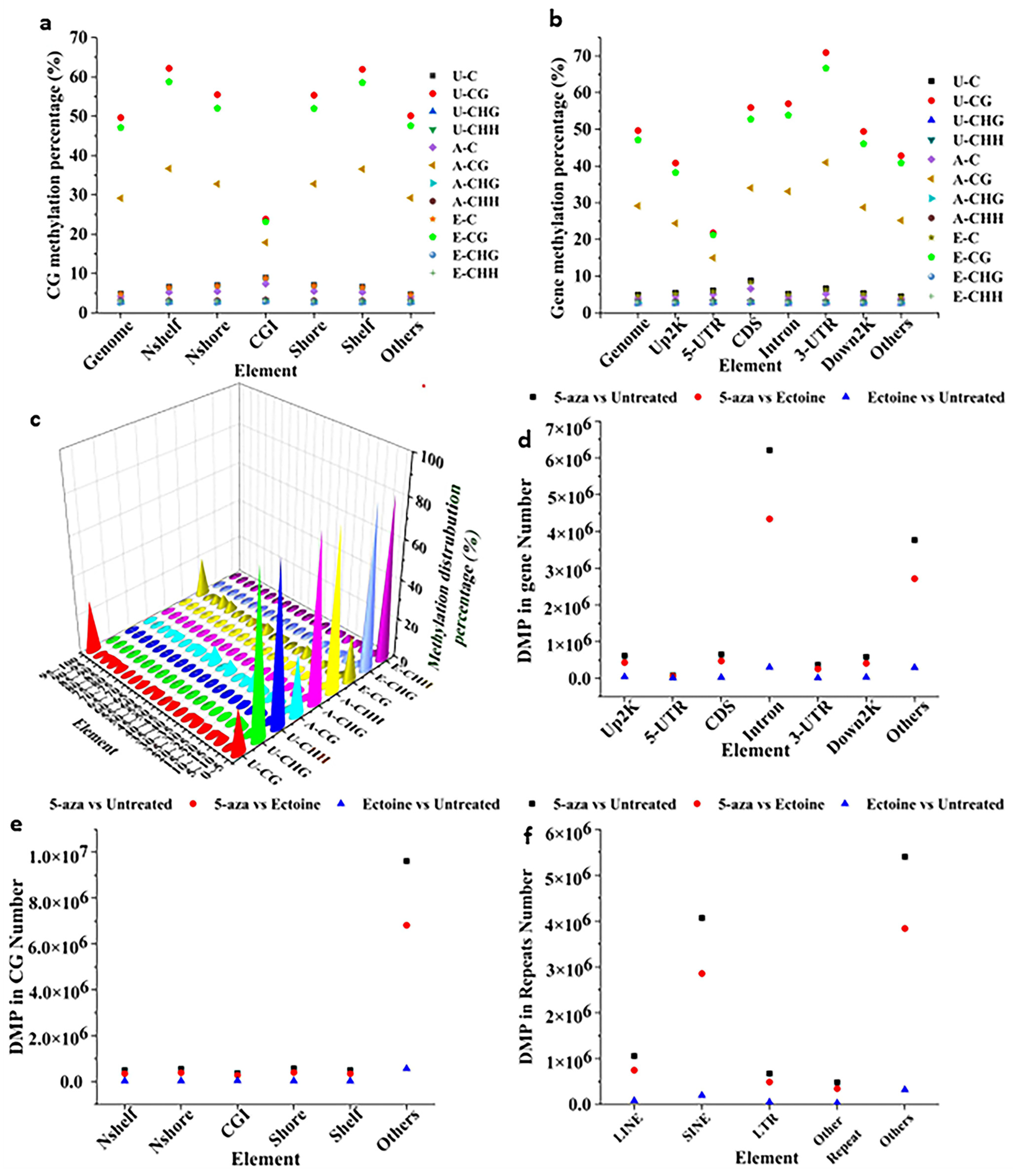
2.1. Global Methylation Sequencing Revealed That Ectoine Hypomethylates DNA and Enriches Nucleotide Binding Ions DEGs
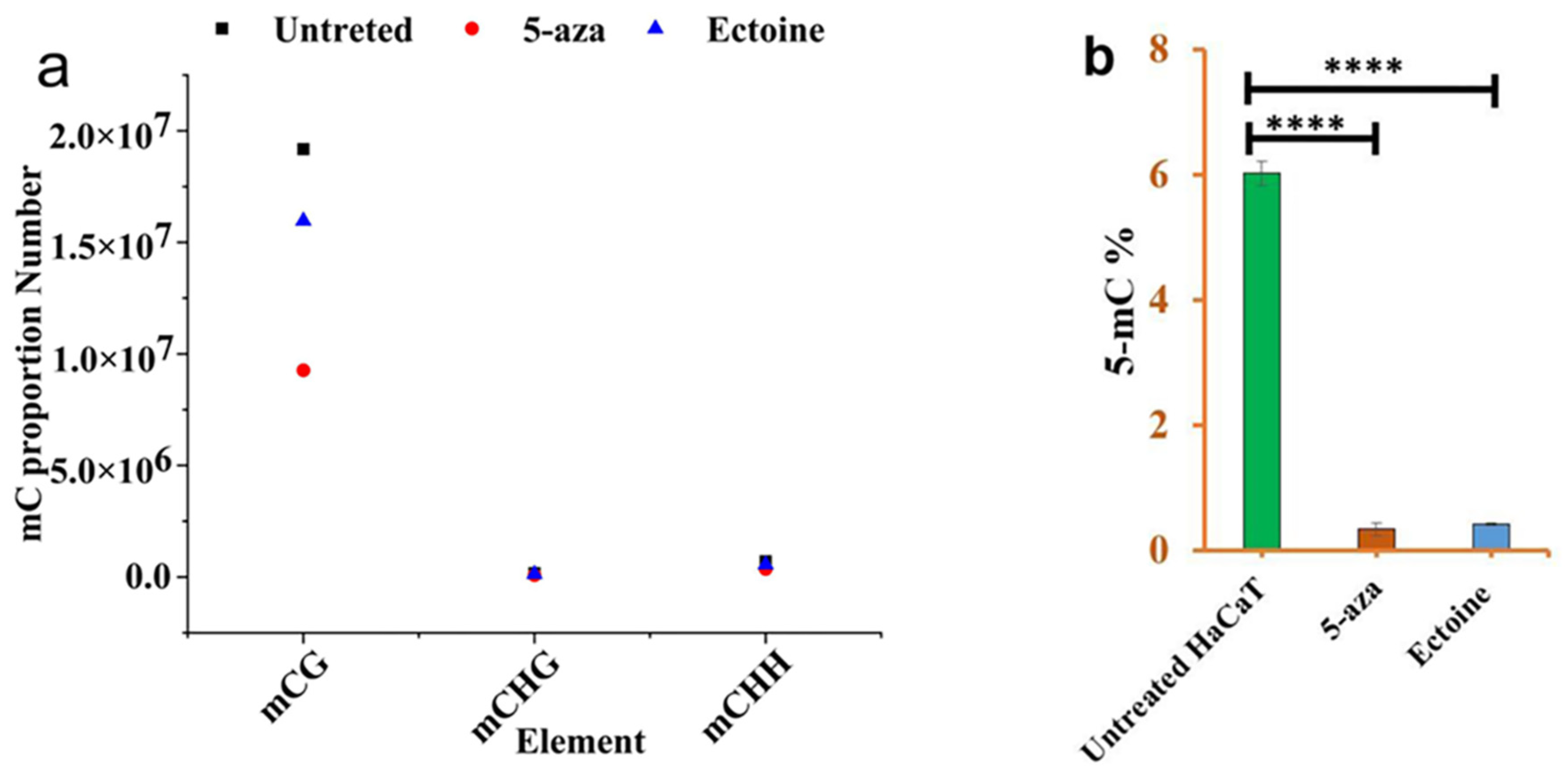
2.2. 5mC Methylation
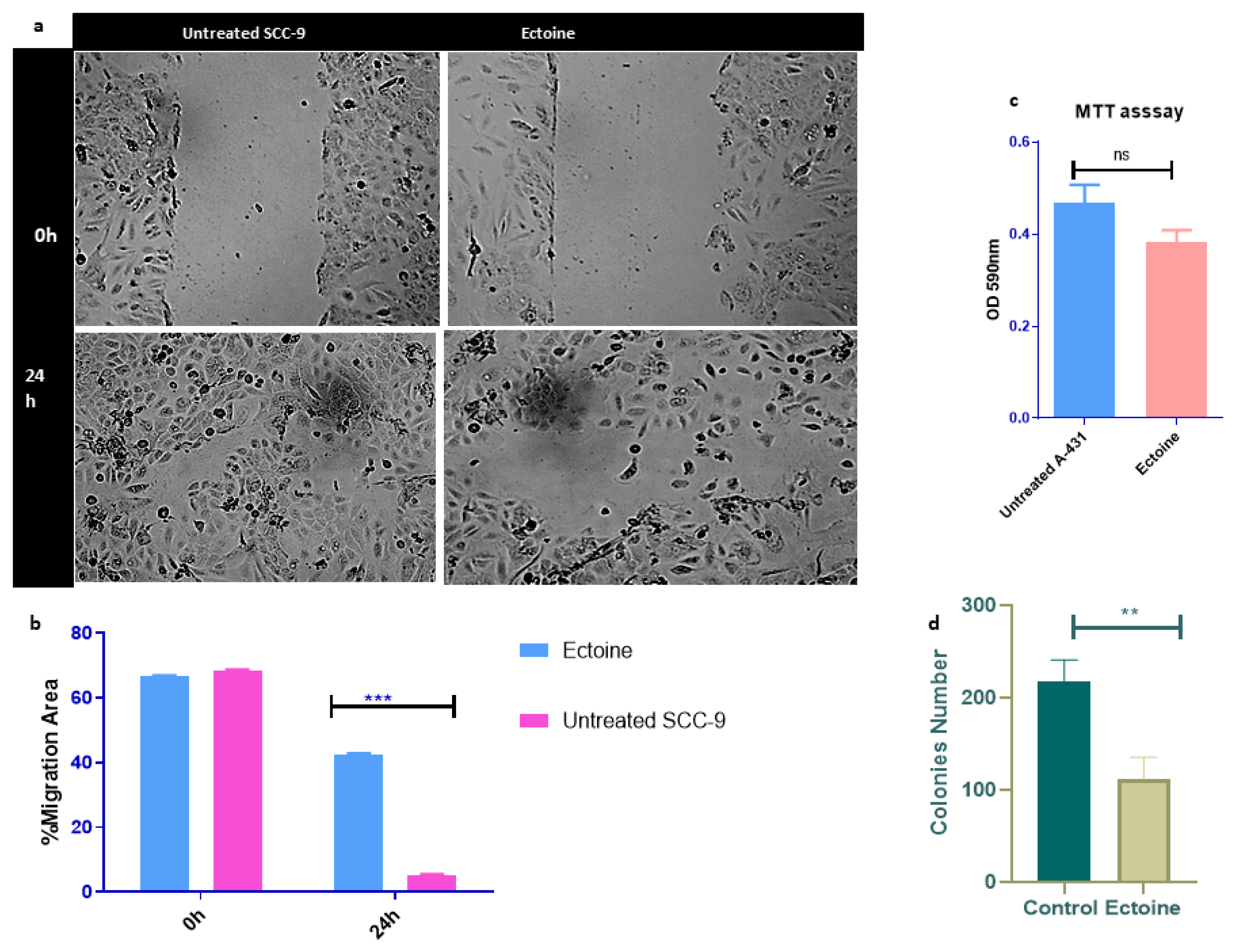
2.3. Ectoine Is a Non-Tumorigenic Molecule and Reduces Cell Proliferation
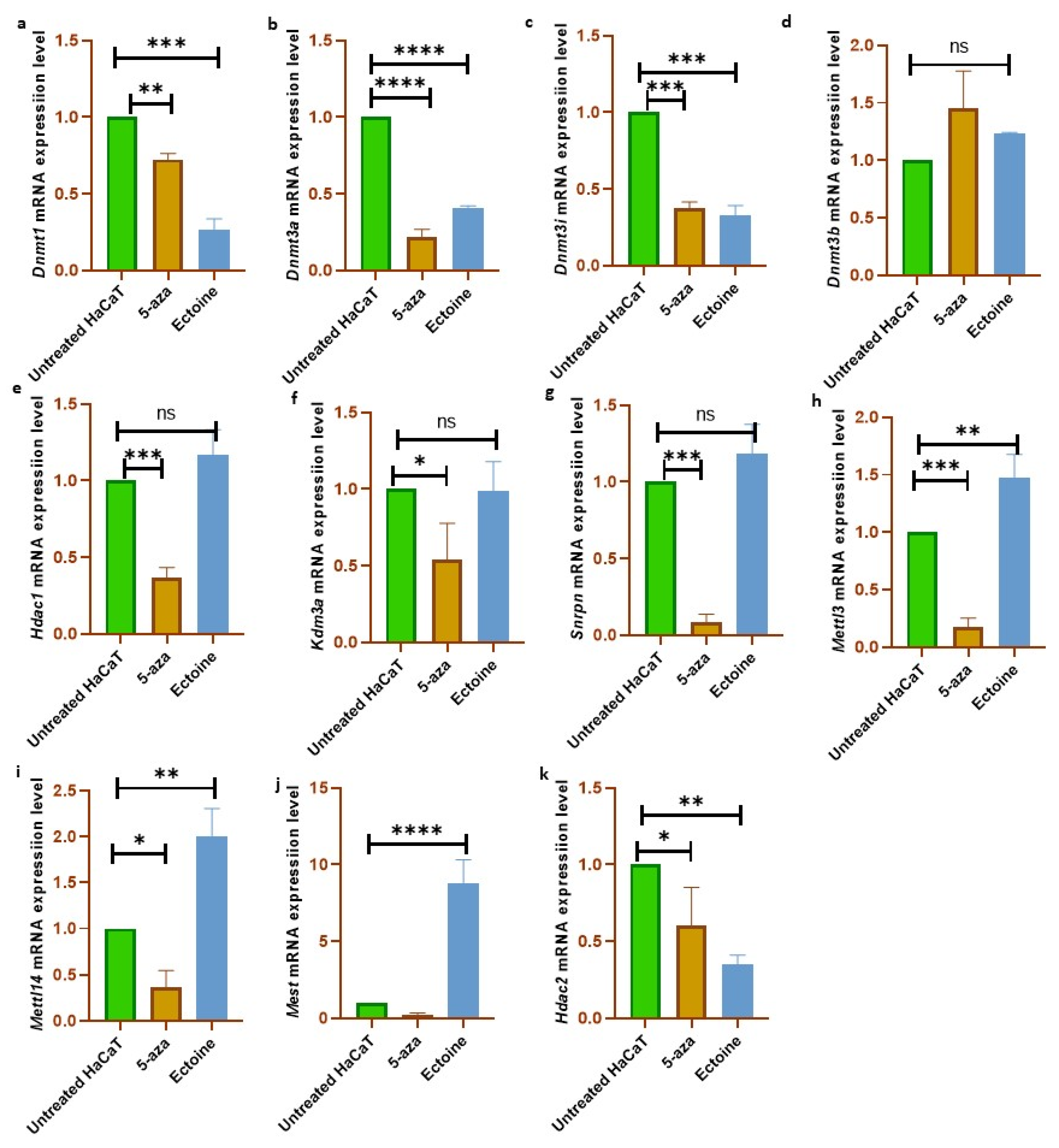
2.4. Ectoine Downregulated Methylation-Related Genes
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Drug Treatments
4.2. Whole-Genome Bisulfite Sequencing and In Silico Methylation Analysis
4.3. Colony Formation Assay
4.4. Wound Healing Assay
4.5. MTT Assay
4.6. Tumorigenicity In Vivo Experiments
4.7. DNA Methylation Analysis
4.8. Quantitative PCR and Gene Expression Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skvortsova, K.; Stirzaker, C.; Taberlay, P. The DNA methylation landscape in cancer. Essays Biochem. 2019, 63, 797–811. [Google Scholar] [PubMed]
- Ghoshal, K.; Datta, J.; Majumder, S.; Bai, S.; Kutay, H.; Motiwala, T.; Jacob, S.T. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 2005, 25, 4727–4741. [Google Scholar] [CrossRef] [PubMed]
- Borthakur, G.; El Ahdab, S.; Ravandi, F.; Faderl, S.; Ferrajoli, A.; Newman, B.; Issa, J.-P.; Kantarjian, H. Activity of decitabine in patients with myelodysplastic syndrome previously treated with azacitidine. Leuk. Lymphoma 2008, 49, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Yang, Y.; Li, W.; Yang, H.; Zhang, Y.; Ge, B.; Zhang, S.; Du, G.; Wang, J. Recent advances in epigenetic anticancer therapeutics and future perspectives. Front. Genet. 2022, 13, 3658. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. Microbial production of extremolytes—High-value active ingredients for nutrition, health care, and well-being. Curr. Opin. Biotechnol. 2020, 65, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Galinski, E.A.; Pfeiffer, H.P.; Trüper, H.G. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid: A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur. J. Biochem. 1985, 149, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Boas Lichty, K.E.; Gregory, G.J.; Boyd, E.F. NhaR, LeuO, and H-NS Are Part of an Expanded Regulatory Network for Ectoine Biosynthesis Expression. Appl. Environ. Microbiol. 2023, 89, e00479-23. [Google Scholar] [CrossRef]
- Kadam, P.; Kajale, S.; Sharma, A.; Dhotre, D.; Barvkar, V.; Shouche, Y.; Zinjarde, S. Whole-Genome Sequencing of the Tropical Marine Bacterium Nocardiopsis dassonvillei NCIM 5124, Containing the Ectoine Biosynthesis Gene Cluster ectABC. Microbiol. Resour. Announc. 2022, 11, e00435-22. [Google Scholar] [CrossRef]
- Kang, J.Y.; Lee, B.; Kim, J.A.; Kim, M.-S.; Kim, C.H. Identification and characterization of an ectoine biosynthesis gene cluster from Aestuariispira ectoiniformans sp. nov. isolated from seawater. Microbiol. Res. 2022, 254, 126898. [Google Scholar] [CrossRef]
- Fenizia, S.; Thume, K.; Wirgenings, M.; Pohnert, G. Ectoine from bacterial and algal origin is a compatible solute in microalgae. Mar. Drugs 2020, 18, 42. [Google Scholar] [CrossRef]
- Azizah, M.; Pohnert, G. Orchestrated Response of Intracellular Zwitterionic Metabolites in Stress Adaptation of the Halophilic Heterotrophic Bacterium Pelagibaca bermudensis. Mar. Drugs 2022, 20, 727. [Google Scholar] [CrossRef] [PubMed]
- Bolen, D.W.; Baskakov, I.V. The osmophobic effect: Natural selection of a thermodynamic force in protein folding. J. Mol. Biol. 2001, 310, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Brands, S.; Schein, P.; Castro-Ochoa, K.F.; Galinski, E.A. Hydroxyl radical scavenging of the compatible solute ectoine generates two N-acetimides. Arch. Biochem. Biophys. 2019, 674, 108097. [Google Scholar] [CrossRef] [PubMed]
- Kanapathipillai, M.; Lentzen, G.; Sierks, M.; Park, C.B. Ectoine and hydroxyectoine inhibit aggregation and neurotoxicity of Alzheimer’s β-amyloid. FEBS Lett. 2005, 579, 4775–4780. [Google Scholar] [CrossRef] [PubMed]
- Unfried, K.; Sydlik, U.; Bierhals, K.; Weissenberg, A.; Abel, J. Carbon nanoparticle-induced lung epithelial cell proliferation is mediated by receptor-dependent Akt activation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 294, L358–L367. [Google Scholar] [CrossRef] [PubMed]
- Eichel, A.; Wittig, J.; Shah-Hosseini, K.; Mösges, R. A prospective, controlled study of SNS01 (ectoine nasal spray) compared to BNO-101 (phytotherapeutic dragées) in patients with acute rhinosinusitis. Curr. Med. Res. Opin. 2013, 29, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Bilstein, A.; Heinrich, A.; Rybachuk, A.; Mösges, R. Ectoine in the Treatment of Irritations and Inflammations of the Eye Surface. BioMed Res. Int. 2021, 2021, 8885032. [Google Scholar] [CrossRef]
- Kauth, M.; Trusova, O.V. Topical Ectoine Application in Children and Adults to Treat Inflammatory Diseases Associated with an Impaired Skin Barrier: A Systematic Review. Dermatol. Ther. 2022, 12, 295–331. [Google Scholar] [CrossRef]
- Wellmerling, J.; Rayner, R.E.; Chang, S.W.; Kairis, E.L.; Kim, S.H.; Sharma, A.; Boyaka, P.N.; Cormet-Boyaka, E. Targeting the EGFR-ERK axis using the compatible solute ectoine to stabilize CFTR mutant F508del. FASEB J. 2022, 36, e22270. [Google Scholar] [CrossRef]
- Wedeking, A.; Hagen-Euteneuer, N.; Gurgui, M.; Broere, R.; Lentzen, G.; Tolba, R.H.; Galinski, E.; van Echten-Deckert, G. A lipid anchor improves the protective effect of ectoine in inflammation. Curr. Med. Chem. 2014, 21, 2565–2572. [Google Scholar] [CrossRef]
- Sydlik, U.; Gallitz, I.; Albrecht, C.; Abel, J.; Krutmann, J.; Unfried, K. The compatible solute ectoine protects against nanoparticle-induced neutrophilic lung inflammation. Am. J. Respir. Crit. Care Med. 2009, 180, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Buenger, J.; Driller, H. Ectoin: An effective natural substance to prevent UVA-induced premature photoaging. Ski. Pharmacol. Appl. Ski. Physiol. 2004, 17, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Sydlik, U.; Peuschel, H.; Paunel-Görgülü, A.; Keymel, S.; Krämer, U.; Weissenberg, A.; Kroker, M.; Seghrouchni, S.; Heiss, C.; Windolf, J. Recovery of neutrophil apoptosis by ectoine: A new strategy against lung inflammation. Eur. Respir. J. 2013, 41, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zang, L.; Shu, Q.; Li, X. From development to diseases: The role of 5hmC in brain. Genomics 2014, 104, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Chedin, F.; Lieber, M.R.; Hsieh, C.-L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 2002, 99, 16916–16921. [Google Scholar] [CrossRef]
- Shetty, M.G.; Pai, P.; Deaver, R.E.; Satyamoorthy, K.; Babitha, K.S. Histone deacetylase 2 selective inhibitors: A versatile therapeutic strategy as next generation drug target in cancer therapy. Pharmacol. Res. 2021, 170, 105695. [Google Scholar] [CrossRef]
- Yeon, M.; Kim, Y.; Jung, H.S.; Jeoung, D. Histone deacetylase inhibitors to overcome resistance to targeted and immuno therapy in metastatic melanoma. Front. Cell Dev. Biol. 2020, 8, 486. [Google Scholar] [CrossRef]
- Xu, Z.; Ma, A.; Ma, Y.C. RNA methylation preserves ES cell identity by chromatin silencing of retrotransposons. Signal Transduct. Target. Ther. 2021, 6, 258. [Google Scholar] [CrossRef]
- Huntriss, J.D.; Hemmings, K.E.; Hinkins, M.; Rutherford, A.J.; Sturmey, R.G.; Elder, K.; Picton, H.M. Variable imprinting of the MEST gene in human preimplantation embryos. Eur. J. Hum. Genet. 2013, 21, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Jeon, Y.H.; Cho, H.Y.; Lee, S.W.; Kim, G.W.; Lee, D.H.; Kwon, S.H. Advances in histone demethylase KDM3A as a cancer therapeutic target. Cancers 2020, 12, 1098. [Google Scholar] [CrossRef]
- Prasad, R.; Singh, T.; Katiyar, S.K. Honokiol inhibits ultraviolet radiation-induced immunosuppression through inhibition of ultraviolet-induced inflammation and DNA hypermethylation in mouse skin. Sci. Rep. 2017, 7, 1657. [Google Scholar] [CrossRef] [PubMed]
- Fontbonne, A.; Teme, B.; Abric, E.; Lecerf, G.; Callejon, S.; Moga, A.; Cadars, B.; Giraud, F.; Chavagnac-Bonneville, M.; Ardiet, N. Positive and ecobiological contribution in skin photoprotection of ectoine and mannitol combined in vivo with UV filters. J. Cosmet. Dermatol. 2023, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schröter, M.A.; Meyer, S.; Hahn, M.B.; Solomun, T.; Sturm, H.; Kunte, H.-J. Ectoine protects DNA from damage by ionizing radiation. Sci. Rep. 2017, 7, 15272. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Schröter, M.A.; Hahn, M.B.; Solomun, T.; Sturm, H.; Kunte, H.-J. Ectoine can enhance structural changes in DNA in vitro. Sci. Rep. 2017, 7, 7170. [Google Scholar] [CrossRef] [PubMed]
- Wittmar, J.; Meyer, S.; Sieling, T.; Kunte, J.R.; Smiatek, J.; Brand, I. What does ectoine do to DNA? A molecular-scale picture of compatible solute–biopolymer interactions. J. Phys. Chem. B 2020, 124, 7999–8011. [Google Scholar] [CrossRef] [PubMed]
- Gombeau, K.; Bonzom, J.-M.; Cavalié, I.; Camilleri, V.; Orjollet, D.; Dubourg, N.; Beaugelin-Seiller, K.; Bourdineaud, J.-P.; Lengagne, T.; Armant, O. Dose-dependent genomic DNA hypermethylation and mitochondrial DNA damage in Japanese tree frogs sampled in the Fukushima Daiichi area. J. Environ. Radioact. 2020, 225, 106429. [Google Scholar] [CrossRef]
- Li, L.; Wang, H.; Chen, X.; Li, X.; Wang, G.; Jie, Z.; Zhao, X.; Sun, X.; Huang, H.; Fan, S. Oxidative stress-induced hypermethylation of KLF5 promoter mediated by DNMT3B impairs osteogenesis by diminishing the interaction with β-catenin. Antioxid. Redox Signal. 2021, 35, 1–20. [Google Scholar] [CrossRef]
- Hahn, M.B.; Solomun, T.; Wellhausen, R.; Hermann, S.; Seitz, H.; Meyer, S.; Kunte, H.-J.R.; Zeman, J.; Uhlig, F.; Smiatek, J. Influence of the compatible solute ectoine on the local water structure: Implications for the binding of the protein G5P to DNA. J. Phys. Chem. B 2015, 119, 15212–15220. [Google Scholar] [CrossRef]
- Hallier, D.C.; Smales, G.J.; Seitz, H.; Hahn, M.B. Bio-SAXS of single-stranded DNA-binding proteins: Radiation protection by the compatible solute ectoine. Phys. Chem. Chem. Phys. 2023, 25, 5372–5382. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Balaguer, F.; Shen, Y.; Lozano, J.J.; Leung, H.-C.E.; Boland, C.R.; Goel, A. Curcumin modulates DNA methylation in colorectal cancer cells. PLoS ONE 2013, 8, e57709. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, V.; Vaid, M.; Tollefsbol, T.O.; Katiyar, S.K. RETRACTED: Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis 2011, 32, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Rieckmann, T.; Gatzemeier, F.; Christiansen, S.; Rothkamm, K.; Münscher, A. The inflammation-reducing compatible solute ectoine does not impair the cytotoxic effect of ionizing radiation on head and neck cancer cells. Sci. Rep. 2019, 9, 6594. [Google Scholar] [CrossRef] [PubMed]
- Boukamp, P.; Popp, S.; Bleuel, K.; Tomakidi, E.; Bürkle, A.; Fusenig, N.E. Tumorigenic conversion of immortal human skin keratinocytes (HaCaT) by elevated temperature. Oncogene 1999, 18, 5638–5645. [Google Scholar] [CrossRef] [PubMed]
- Valerio, H.P.; Ravagnani, F.G.; Ronsein, G.E.; Di Mascio, P. A single dose of Ultraviolet-A induces proteome remodeling and senescence in primary human keratinocytes. Sci. Rep. 2021, 11, 23355. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Dou, G.; Liu, T.; Guo, X.; Bai, Y.; Chu, D.; Liu, S.; Chen, X.; Jin, Y. On-demand manipulation of tumorigenic microenvironments by nano-modulator for synergistic tumor therapy. Biomaterials 2021, 275, 120956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. DNA methyltransferases in cancer: Biology, paradox, aberrations, and targeted therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef]
- Yu, D.; Horton, J.R.; Yang, J.; Hajian, T.; Vedadi, M.; Sagum, C.A.; Bedford, M.T.; Blumenthal, R.M.; Zhang, X.; Cheng, X. Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic Acids Res. 2021, 49, 11629–11642. [Google Scholar] [CrossRef]
- Hseu, Y.-C.; Chen, X.-Z.; Vudhya Gowrisankar, Y.; Yen, H.-R.; Chuang, J.-Y.; Yang, H.-L. The skin-whitening effects of ectoine via the suppression of α-MSH-stimulated melanogenesis and the activation of antioxidant Nrf2 pathways in UVA-irradiated keratinocytes. Antioxidants 2020, 9, 63. [Google Scholar] [CrossRef]
- Poplinski, A.; Tüttelmann, F.; Kanber, D.; Horsthemke, B.; Gromoll, J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int. J. Androl. 2010, 33, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Klinkebiel, D.; Barger, C.J.; Pandey, S.; Guda, C.; Miller, A.; Akers, S.N.; Odunsi, K.; Karpf, A.R. Global DNA hypomethylation in epithelial ovarian cancer: Passive demethylation and association with genomic instability. Cancers 2020, 12, 764. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Van Emburgh, B.O.; Robertson, K.D. DNA methylation in development and human disease. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2008, 647, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Field, A.E.; Robertson, N.A.; Wang, T.; Havas, A.; Ideker, T.; Adams, P.D. DNA methylation clocks in aging: Categories, causes, and consequences. Mol. Cell 2018, 71, 882–895. [Google Scholar] [CrossRef] [PubMed]
- McHugh, D.; Gil, J. Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 2018, 217, 65–77. [Google Scholar] [CrossRef] [PubMed]
- McKinney, B.C.; Lin, C.-W.; Rahman, T.; Oh, H.; Lewis, D.A.; Tseng, G.; Sibille, E. DNA methylation in the human frontal cortex reveals a putative mechanism for age-by-disease interactions. Transl. Psychiatry 2019, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Kang, B.; Petkovich, D.A.; Bhandari, Y.R.; In, J.; Stein-O’Brien, G.; Kong, X.; Xie, W.; Zachos, N.; Maegawa, S. Aging-like spontaneous epigenetic silencing facilitates Wnt activation, stemness, and BrafV600E-induced tumorigenesis. Cancer Cell 2019, 35, 315–328. [Google Scholar] [CrossRef]
- Barajas-Olmos, F.M.; Ortiz-Sanchez, E.; Imaz-Rosshandler, I.; Córdova-Alarcón, E.J.; Martinez-Tovar, A.; Villanueva-Toledo, J.; Morales-Marin, M.E.; Cruz-Colin, J.L.; Rangel, C.; Orozco, L. Analysis of the dynamic aberrant landscape of DNA methylation and gene expression during arsenic-induced cell transformation. Gene 2019, 711, 143941. [Google Scholar] [CrossRef]
- Liang, J.; Liu, L.; Tang, H.; Ma, Q.; Sang, Y.; Kang, X. UVB-induced SFRP1 methylation potentiates skin damage by promoting cell apoptosis and DNA damage. Exp. Dermatol. 2022, 31, 1443–1453. [Google Scholar] [CrossRef]
- Yoshinaga-Sakurai, K.; Rossman, T.G.; Rosen, B.P. Regulation of arsenic methylation: Identification of the transcriptional region of the human AS3MT gene. Cell Biol. Toxicol. 2022, 38, 765–780. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lei, R.; Ding, S.-W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Wu, H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 2016, 32, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Strus, P.; Borensztejn, K.; Szczepankiewicz, A.A.; Lisiecki, K.; Czarnocki, Z.; Nieznanska, H.; Wojcik, C.; Bialy, L.P.; Mlynarczuk-Bialy, I. Novel podophyllotoxin and benzothiazole derivative induces transitional morphological and functional changes in HaCaT cells. Toxicol. Vitr. 2021, 73, 105144. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Alexander, E.T.; Mariner, K.; Borodyanskaya, Y.; Minton, A.; Gilmour, S.K. Polyamine-stimulation of arsenic-transformed keratinocytes. Carcinogenesis 2019, 40, 1042–1051. [Google Scholar] [CrossRef]
- Merckx, C.; Zschüntzsch, J.; Meyer, S.; Raedt, R.; Verschuere, H.; Schmidt, J.; De Paepe, B.; De Bleecker, J.L. Exploring the Therapeutic Potential of Ectoine in Duchenne Muscular Dystrophy: Comparison with Taurine, a Supplement with Known Beneficial Effects in the mdx Mouse. Int. J. Mol. Sci. 2022, 23, 9567. [Google Scholar] [CrossRef]
- Keeter, W.C.; Moriarty, A.K.; Akers, R.; Ma, S.; Mussbacher, M.; Nadler, J.L.; Galkina, E.V. Neutrophil-specific STAT4 deficiency attenuates atherosclerotic burden and improves plaque stability via reduction in neutrophil activation and recruitment into aortas of Ldlr (−/−) mice. bioRxiv 2023, 10, 1175673. [Google Scholar] [CrossRef]
- Yamaga, S.; Murao, A.; Ma, G.; Brenner, M.; Aziz, M.; Wang, P. Radiation upregulates macrophage TREM-1 expression to exacerbate injury in mice. Front. Immunol. 2023, 14, 1151250. [Google Scholar] [CrossRef]
- Schmidt, S.; Luecken, M.D.; Trümbach, D.; Hembach, S.; Niedermeier, K.M.; Wenck, N.; Pflügler, K.; Stautner, C.; Böttcher, A.; Lickert, H.; et al. Primary cilia and SHH signaling impairments in human and mouse models of Parkinson’s disease. Nat. Commun. 2022, 13, 4819. [Google Scholar] [CrossRef] [PubMed]
- Qaria, M.A.; Kumar, N.; Hussain, A.; Qumar, S.; Doddam, S.N.; Sepe, L.P.; Ahmed, N. Roles of Cholesteryl-α-Glucoside Transferase and Cholesteryl Glucosides in Maintenance of Helicobacter pylori Morphology, Cell Wall Integrity, and Resistance to Antibiotics. mBio 2018, 9, e01523-18. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Trinh, E.; Qiang, L.; Xie, L.; Hu, W.-Y.; Prins, G.S.; Pi, J.; He, Y.-Y. Arsenic induces p62 expression to form a positive feedback loop with Nrf2 in human epidermal keratinocytes: Implications for preventing arsenic-induced skin cancer. Molecules 2017, 22, 194. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J. TOPGO: Enrichment Analysis for Gene Ontology. 2.40.0. R Package Version, 2020; 2, 2010. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic. Acids. Res. 2012, 40, 109–114. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qaria, M.A.; Xu, C.; Hu, R.; Alsubki, R.A.; Ali, M.Y.; Sivasamy, S.; Attia, K.A.; Zhu, D. Ectoine Globally Hypomethylates DNA in Skin Cells and Suppresses Cancer Proliferation. Mar. Drugs 2023, 21, 621. https://doi.org/10.3390/md21120621
Qaria MA, Xu C, Hu R, Alsubki RA, Ali MY, Sivasamy S, Attia KA, Zhu D. Ectoine Globally Hypomethylates DNA in Skin Cells and Suppresses Cancer Proliferation. Marine Drugs. 2023; 21(12):621. https://doi.org/10.3390/md21120621
Chicago/Turabian StyleQaria, Majjid A., Chunyan Xu, Ran Hu, Roua A. Alsubki, Mohamed Yassin Ali, Sethupathy Sivasamy, Kotb A. Attia, and Daochen Zhu. 2023. "Ectoine Globally Hypomethylates DNA in Skin Cells and Suppresses Cancer Proliferation" Marine Drugs 21, no. 12: 621. https://doi.org/10.3390/md21120621
APA StyleQaria, M. A., Xu, C., Hu, R., Alsubki, R. A., Ali, M. Y., Sivasamy, S., Attia, K. A., & Zhu, D. (2023). Ectoine Globally Hypomethylates DNA in Skin Cells and Suppresses Cancer Proliferation. Marine Drugs, 21(12), 621. https://doi.org/10.3390/md21120621