Inhibition of Polymicrobial Biofilms of Candida albicans–Staphylococcus aureus/Streptococcus mutans by Fucoidan–Gold Nanoparticles
Abstract
:1. Introduction
2. Results
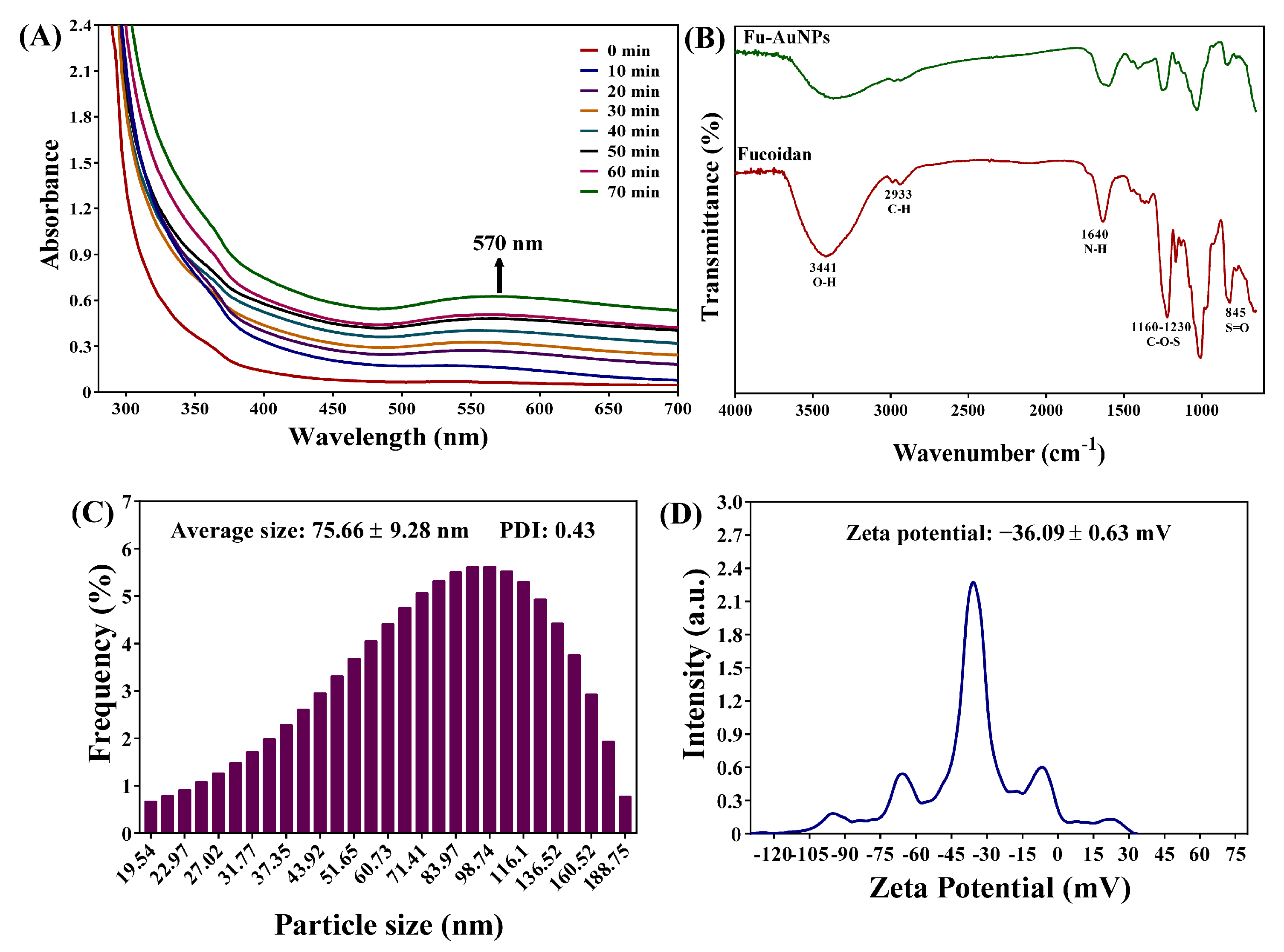
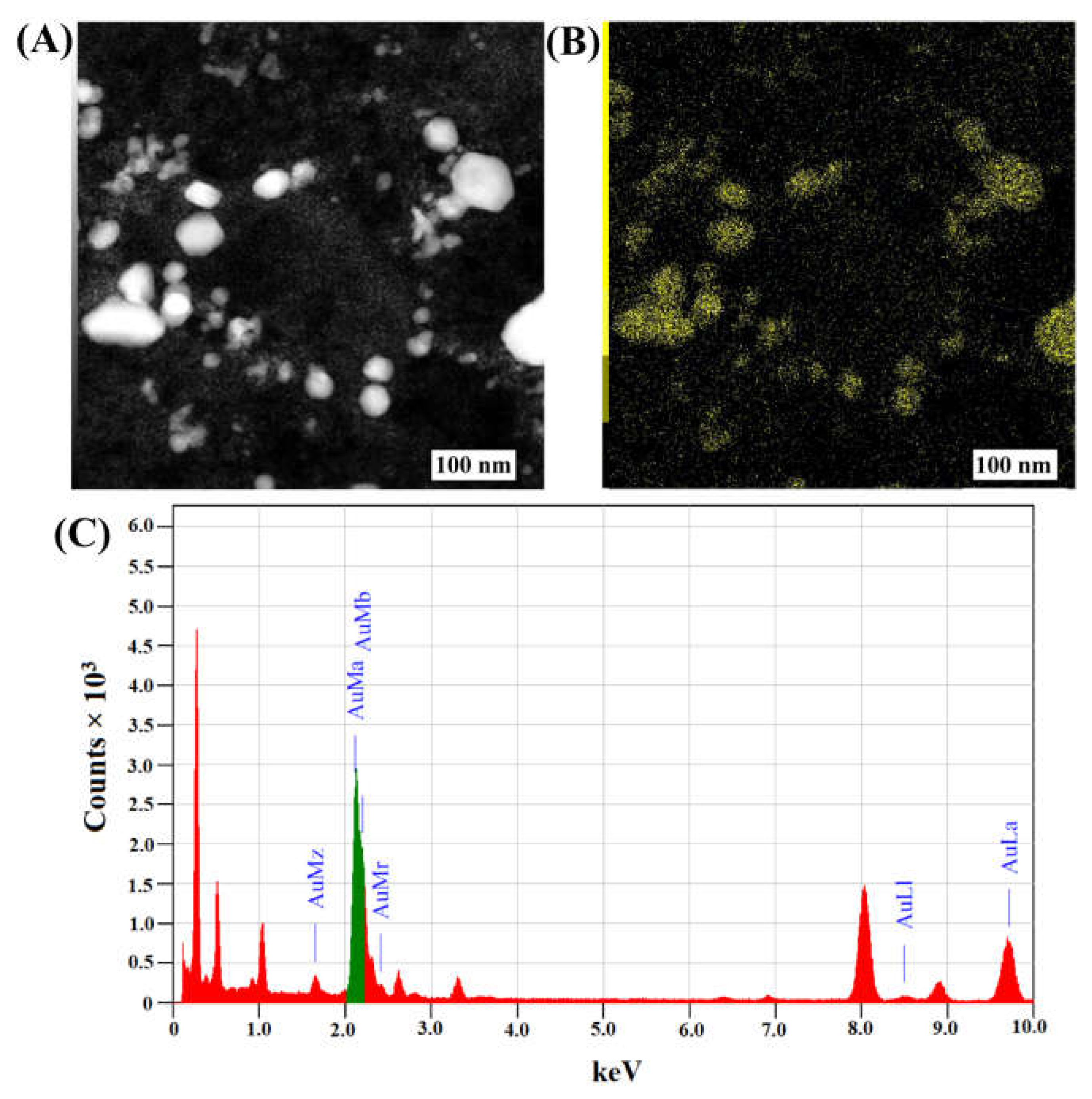
2.1. Synthesis and Characterization of Fu–AuNPs
2.2. MIC and MBC, or MFC, Values of Fu–AuNPs towards Microbial Pathogens
2.3. Inhibitory Effect of Fu–AuNPs towards Single- and Mixed-Species Biofilms
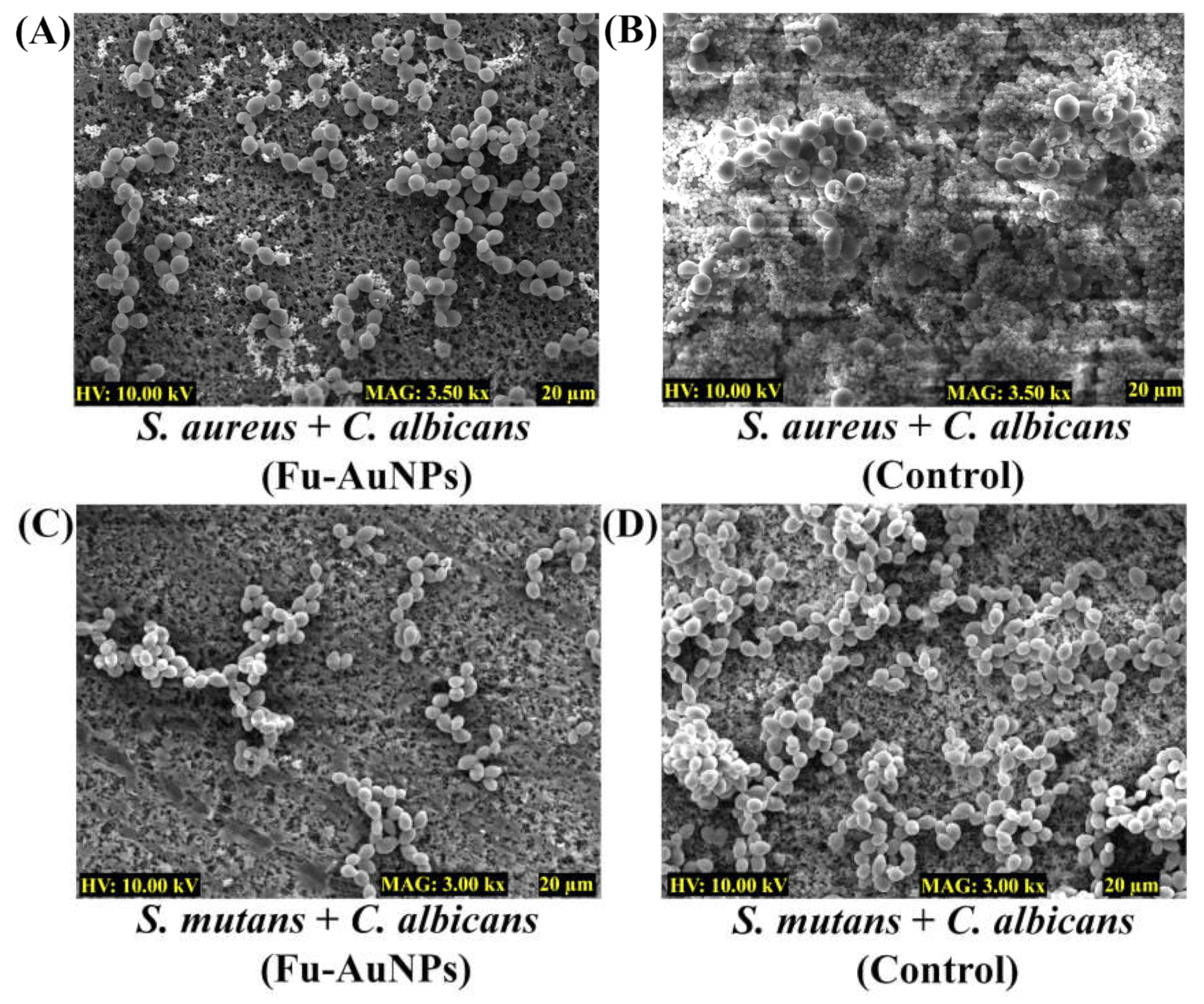
2.4. Microscopic Examination of Single- and Mixed-Species Biofilms Treated with Fu–AuNPs
2.5. Dispersal of Established Mature Biofilms of Single or Mixed Species by Fu–AuNPs
3. Discussion
4. Materials and Methods
4.1. Microbial Strains, Growth Media, and Chemicals
4.2. Synthesis of Fucoidan–Gold Nanoparticles
4.3. Characterization of Fu–AuNPs
4.4. Minimum Inhibitory Concentration (MIC) of Fu–AuNPs
4.5. Biofilm Assays
4.6. Eradication of Established Mature Biofilms
4.7. Examination of Microbial Biofilm Architecture
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Johnson, C.; Berman, J.; Coste, A.T.; Cuomo, C.A.; Perlin, D.S.; Bicanic, T.; Harrison, T.S.; Wiederhold, N.; Bromley, M.; et al. The importance of antimicrobial resistance in medical mycology. Nat. Commun. 2022, 13, 5352. [Google Scholar] [CrossRef]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brüggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Genet. 2022, 20, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K.; on behalf of the CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef]
- Ersoy, S.C.; Heithoff, D.M.; Barnes, L.; Tripp, G.K.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing. Ebiomedicine 2017, 20, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Maset, R.G.; Hapeshi, A.; Hall, S.; Dalgliesh, R.M.; Harrison, F.; Perrier, S. Evaluation of the Antimicrobial Activity in Host-Mimicking Media and In Vivo Toxicity of Antimicrobial Polymers as Functional Mimics of AMPs. ACS Appl. Mater. Interfaces 2022, 14, 32855–32868. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.J.; Figueroa, W.; Welch, M. Decreased efficacy of antimicrobial agents in a polymicrobial environment. ISME J. 2022, 16, 1694–1704. [Google Scholar] [CrossRef]
- Costa, P.d.S.; Prado, A.; Bagon, N.P.; Negri, M.; Svidzinski, T.I.E. Mixed Fungal Biofilms: From Mycobiota to Devices, a New Challenge on Clinical Practice. Microorganisms 2022, 10, 1721. [Google Scholar] [CrossRef] [PubMed]
- Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; De Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcón, K.P.; Melo, W.C.M.A.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal Biofilms and Polymicrobial Diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, F.; Bamunuarachchi, N.I.; Pham, D.T.N.; Tabassum, N.; Khan, M.S.A.; Kim, Y.-M. Mixed biofilms of pathogenic Candida-bacteria: Regulation mechanisms and treatment strategies. Crit. Rev. Microbiol. 2021, 47, 699–727. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Xu, T.; Zeng, L.; Liu, F.; Huang, X.; Liu, Q. Interactions among microorganisms open up a new world for anti-infectious therapy. FEBS J. 2022. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Jeong, G.-J.; Javaid, A.; Pham, D.T.N.; Tabassum, N.; Kim, Y.-M. Surface adherence and vacuolar internalization of bacterial pathogens to the Candida spp. cells: Mechanism of persistence and propagation. J. Adv. Res. 2022. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Krüger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.; Niemiec, M. Fungal-Bacterial Interactions in Health and Disease. Pathogens 2019, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, A.P.; Jorge, P.; Pereira, M.O. Pseudomonas aeruginosa and Staphylococcus aureus communication in biofilm infections: Insights through network and database construction. Crit. Rev. Microbiol. 2019, 45, 712–728. [Google Scholar] [CrossRef]
- Fey, P.D. Modality of bacterial growth presents unique targets: How do we treat biofilm-mediated infections? Curr. Opin. Microbiol. 2010, 13, 610–615. [Google Scholar] [CrossRef]
- Liu, W.; Røder, H.L.; Madsen, J.S.; Bjarnsholt, T.; Sørensen, S.J.; Burmølle, M. Interspecific Bacterial Interactions are Reflected in Multispecies Biofilm Spatial Organization. Front. Microbiol. 2016, 7, 1366. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Moser, D.; Han, F.; Leonhard, M.; Schneider-Stickler, B.; Tan, Y. Preparation and antibiofilm studies of curcumin loaded chitosan nanoparticles against polymicrobial biofilms of Candida albicans and Staphylococcus aureus. Carbohydr. Polym. 2020, 241, 116254. [Google Scholar] [CrossRef]
- Pourhajibagher, M.; Bahador, A. Enhanced reduction of polymicrobial biofilms on the orthodontic brackets and enamel surface remineralization using zeolite-zinc oxide nanoparticles-based antimicrobial photodynamic therapy. BMC Microbiol. 2021, 21, 273. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Xu, L.; Singh, A.K.; Qiu, X.; Liu, M.; Abuzeid, A.; El-Khateib, T.; Bhunia, A.K. Inactivation of Polymicrobial Biofilms of Foodborne Pathogens Using Epsilon Poly-L-Lysin Conjugated Chitosan Nanoparticles. Foods 2022, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef]
- Yetisgin, A.A.; Cetinel, S.; Zuvin, M.; Kosar, A.; Kutlu, O. Therapeutic Nanoparticles and Their Targeted Delivery Applications. Molecules 2020, 25, 2193. [Google Scholar] [CrossRef]
- Zong, T.-X.; Silveira, A.P.; Morais, J.A.V.; Sampaio, M.C.; Muehlmann, L.A.; Zhang, J.; Jiang, C.-S.; Liu, S.-K. Recent Advances in Antimicrobial Nano-Drug Delivery Systems. Nanomaterials 2022, 12, 1855. [Google Scholar] [CrossRef]
- Khan, M.M.; Kim, Y.-M. Recent Progress and Future Perspectives of Antibiofilm Drugs Immobilized on Nanomaterials. Curr. Pharm. Biotechnol. 2018, 19, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Jeong, G.-J.; Singh, P.; Tabassum, N.; Mijakovic, I.; Kim, Y.-M. Retrospective analysis of the key molecules involved in the green synthesis of nanoparticles. Nanoscale 2022, 14, 14824–14857. [Google Scholar] [CrossRef] [PubMed]
- Jeong, G.-J.; Khan, S.; Tabassum, N.; Khan, F.; Kim, Y.-M. Marine-Bioinspired Nanoparticles as Potential Drugs for Multiple Biological Roles. Mar. Drugs 2022, 20, 527. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.-C.; Qin, W.; Lei, C.; Li, Q.-H.; Meng, M.; Fang, M.; Song, W.; Chen, J.-H.; Tay, F.; Niu, L.-N. Biomaterials from the sea: Future building blocks for biomedical applications. Bioact. Mater. 2021, 6, 4255–4285. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Oh, D.; Chandika, P.; Jo, D.-M.; Bamunarachchi, N.I.; Jung, W.-K.; Kim, Y.-M. Inhibitory activities of phloroglucinol-chitosan nanoparticles on mono- and dual-species biofilms of Candida albicans and bacteria. Colloids Surfaces B Biointerfaces 2022, 211, 112307. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Kang, M.-G.; Jo, D.-M.; Chandika, P.; Jung, W.-K.; Kang, H.W.; Kim, Y.-M. Phloroglucinol-Gold and -Zinc Oxide Nanoparticles: Antibiofilm and Antivirulence Activities towards Pseudomonas aeruginosa PAO1. Mar. Drugs 2021, 19, 601. [Google Scholar] [CrossRef]
- Khan, F.; Yu, H.; Kim, Y.-M. Bactericidal Activity of Usnic Acid-Chitosan Nanoparticles against Persister Cells of Biofilm-Forming Pathogenic Bacteria. Mar. Drugs 2020, 18, 270. [Google Scholar] [CrossRef] [PubMed]
- Ale, M.T.; Mikkelsen, J.D.; Meyer, A.S. Important Determinants for Fucoidan Bioactivity: A Critical Review of Structure-Function Relations and Extraction Methods for Fucose-Containing Sulfated Polysaccharides from Brown Seaweeds. Mar. Drugs 2011, 9, 2106–2130. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Li, Y.; Teruya, K.; Katakura, Y.; Ichikawa, A.; Eto, H.; Hosoi, M.; Hosoi, M.; Nishimoto, S.; Shirahata, S. Enzyme-digested Fucoidan Extracts Derived from Seaweed Mozuku of Cladosiphon novae-caledoniae kylin Inhibit Invasion and Angiogenesis of Tumor Cells. Cytotechnology 2005, 47, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lu, F.; Wei, X.J.; Zhao, R.X. Fucoidan: Structure and Bioactivity. Molecules 2008, 13, 1671–1695. [Google Scholar] [CrossRef] [PubMed]
- Luthuli, S.; Wu, S.; Cheng, Y.; Zheng, X.; Wu, M.; Tong, H. Therapeutic Effects of Fucoidan: A Review on Recent Studies. Mar. Drugs 2019, 17, 487. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Nomura, K.; Nagashima, M.; Kamimura, N. Fucoidan alleviates high-fat diet-induced dyslipidemia and atherosclerosis in ApoEshl mice deficient in apolipoprotein E expression. J. Nutr. Biochem. 2016, 32, 46–54. [Google Scholar] [CrossRef]
- Lin, Y.; Qi, X.; Liu, H.; Xue, K.; Xu, S.; Tian, Z. The anti-cancer effects of fucoidan: A review of both in vivo and in vitro investigations. Cancer Cell Int. 2020, 20, 154. [Google Scholar] [CrossRef]
- Duncan, B.; Kim, C.; Rotello, V.M. Gold nanoparticle platforms as drug and biomacromolecule delivery systems. J. Control. Release 2010, 148, 122–127. [Google Scholar] [CrossRef]
- Pechyen, C.; Ponsanti, K.; Tangnorawich, B.; Ngernyuang, N. Waste fruit peel—Mediated green synthesis of biocompatible gold nanoparticles. J. Mater. Res. Technol. 2021, 14, 2982–2991. [Google Scholar] [CrossRef]
- Rossi, O.; van Baarlen, P.; Wells, J.M. Host-recognition of pathogens and commensals in the mammalian intestine. Curr. Top. Microbiol. Immunol. 2013, 358, 291–321. [Google Scholar]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Brogden, K.A.; Guthmiller, J.M.; Taylor, C.E. Human polymicrobial infections. Lancet 2005, 365, 253–255. [Google Scholar] [CrossRef]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced Biofilm Formation and Increased Resistance to Antimicrobial Agents and Bacterial Invasion Are Caused by Synergistic Interactions in Multispecies Biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Ren, B.; Zhou, X.; Zhang, L.; Xu, X. Cross-kingdom interaction between Candida albicans and oral bacteria. Front. Microbiol. 2022, 13, 911623. [Google Scholar] [CrossRef]
- Shirtliff, M.E.; Peters, B.M.; Jabra-Rizk, M.A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 2009, 299, 1–8. [Google Scholar] [CrossRef]
- Pavlaki, M.; Poulakou, G.; Drimousis, P.; Adamis, G.; Apostolidou, E.; Gatselis, N.K.; Kritselis, I.; Mega, A.; Mylona, V.; Papatsoris, A.; et al. Polymicrobial bloodstream infections: Epidemiology and impact on mortality. J. Glob. Antimicrob. Resist. 2013, 1, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Manivasagan, P.; Lee, J.-W.; Pham, D.T.N.; Oh, J.; Kim, Y.-M. Fucoidan-Stabilized Gold Nanoparticle-Mediated Biofilm Inhibition, Attenuation of Virulence and Motility Properties in Pseudomonas aeruginosa PAO1. Mar. Drugs 2019, 17, 208. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-G.; Khan, F.; Jo, D.-M.; Oh, D.; Tabassum, N.; Kim, Y.-M. Antibiofilm and Antivirulence Activities of Gold and Zinc Oxide Nanoparticles Synthesized from Kimchi-Isolated Leuconostoc sp. Strain C2. Antibiotics 2022, 11, 1524. [Google Scholar] [CrossRef]
- Manivasagan, P.; Bharathiraja, S.; Bui, N.Q.; Jang, B.; Oh, Y.-O.; Lim, I.G.; Oh, J. Doxorubicin-loaded fucoidan capped gold nanoparticles for drug delivery and photoacoustic imaging. Int. J. Biol. Macromol. 2016, 91, 578–588. [Google Scholar] [CrossRef]
- Jang, H.; Kang, K.; El-Sayed, M.A. Facile size-controlled synthesis of fucoidan-coated gold nanoparticles and cooperative anticancer effect with doxorubicin. J. Mater. Chem. B 2017, 5, 6147–6153. [Google Scholar] [CrossRef]
- Mikhailova, E.O. Gold Nanoparticles: Biosynthesis and Potential of Biomedical Application. J. Funct. Biomater. 2021, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Shnoudeh, A.J.; Hamad, I.; Abdo, R.W.; Qadumii, L.; Jaber, A.Y.; Surchi, H.S.; Alkelany, S.Z. Chapter 15—Synthesis, Characterization, and Applications of Metal Nanoparticles. In Biomaterials and Bionanotechnology; Tekade, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 527–612. [Google Scholar]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus Form Polymicrobial Biofilms: Effects on Antimicrobial Resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.F.; Tsui, C.; Kucharíková, S.; Andes, D.; Van Dijck, P.; Jabra-Rizk, M.A. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. Mbio 2016, 7, e01365-16. [Google Scholar] [CrossRef]
- Perveen, K.; Husain, F.; Qais, F.; Khan, A.; Razak, S.; Afsar, T.; Alam, P.; Almajwal, A.; Abulmeaty, M. Microwave-Assisted Rapid Green Synthesis of Gold Nanoparticles Using Seed Extract of Trachyspermum ammi: ROS Mediated Biofilm Inhibition and Anticancer Activity. Biomolecules 2021, 11, 197. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.; Wahab, R.; Khan, F.; Mishra, Y.K.; Musarrat, J.; Al-Khedhairy, A.A. Reactive Oxygen Species Mediated Bacterial Biofilm Inhibition via Zinc Oxide Nanoparticles and Their Statistical Determination. PLoS ONE 2014, 9, e111289. [Google Scholar] [CrossRef] [PubMed]
- Anju, V.T.; Busi, S.; Imchen, M.; Kumavath, R.; Mohan, M.S.; Salim, S.A.; Subhaswaraj, P.; Dyavaiah, M. Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics 2022, 11, 1731. [Google Scholar] [CrossRef] [PubMed]
- Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing; CLSI Document M100-S20; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011. [Google Scholar]
- Khan, F.; Lee, J.-W.; Manivasagan, P.; Pham, D.T.N.; Oh, J.; Kim, Y.-M. Synthesis and characterization of chitosan oligosaccharide-capped gold nanoparticles as an effective antibiofilm drug against the Pseudomonas aeruginosa PAO1. Microb. Pathog. 2019, 135, 103623. [Google Scholar] [CrossRef]
- Konaté, K.; Hilou, A.; Mavoungou, J.F.; Lepengué, A.N.; Souza, A.; Barro, N.; Datté, J.Y.; M’Batchi, B.; Nacoulma, O.G. Antimicrobial activity of polyphenol-rich fractions from Sida alba L. (Malvaceae) against co-trimoxazol-resistant bacteria strains. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Microbial Pathogens | Antimicrobial Activities | ||
|---|---|---|---|
| MIC (µg/mL) | MBC or MFC (µg/mL) | MBC or MFC/MIC Ratio | |
| S. aureus | 1024 | 2048 | 2 |
| S. mutans | >512 | 1024 | 2 |
| C. albicans | 1024 | 2048 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabassum, N.; Khan, F.; Kang, M.-G.; Jo, D.-M.; Cho, K.-J.; Kim, Y.-M. Inhibition of Polymicrobial Biofilms of Candida albicans–Staphylococcus aureus/Streptococcus mutans by Fucoidan–Gold Nanoparticles. Mar. Drugs 2023, 21, 123. https://doi.org/10.3390/md21020123
Tabassum N, Khan F, Kang M-G, Jo D-M, Cho K-J, Kim Y-M. Inhibition of Polymicrobial Biofilms of Candida albicans–Staphylococcus aureus/Streptococcus mutans by Fucoidan–Gold Nanoparticles. Marine Drugs. 2023; 21(2):123. https://doi.org/10.3390/md21020123
Chicago/Turabian StyleTabassum, Nazia, Fazlurrahman Khan, Min-Gyun Kang, Du-Min Jo, Kyung-Jin Cho, and Young-Mog Kim. 2023. "Inhibition of Polymicrobial Biofilms of Candida albicans–Staphylococcus aureus/Streptococcus mutans by Fucoidan–Gold Nanoparticles" Marine Drugs 21, no. 2: 123. https://doi.org/10.3390/md21020123





