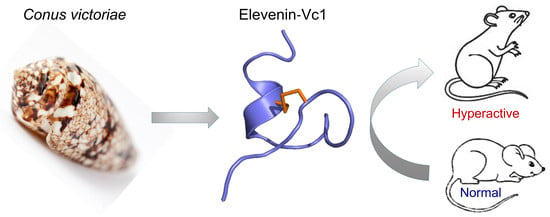
Characterisation of Elevenin-Vc1 from the Venom of Conus victoriae: A Structural Analogue of α-Conotoxins
Abstract
1. Introduction
2. Results and Discussion
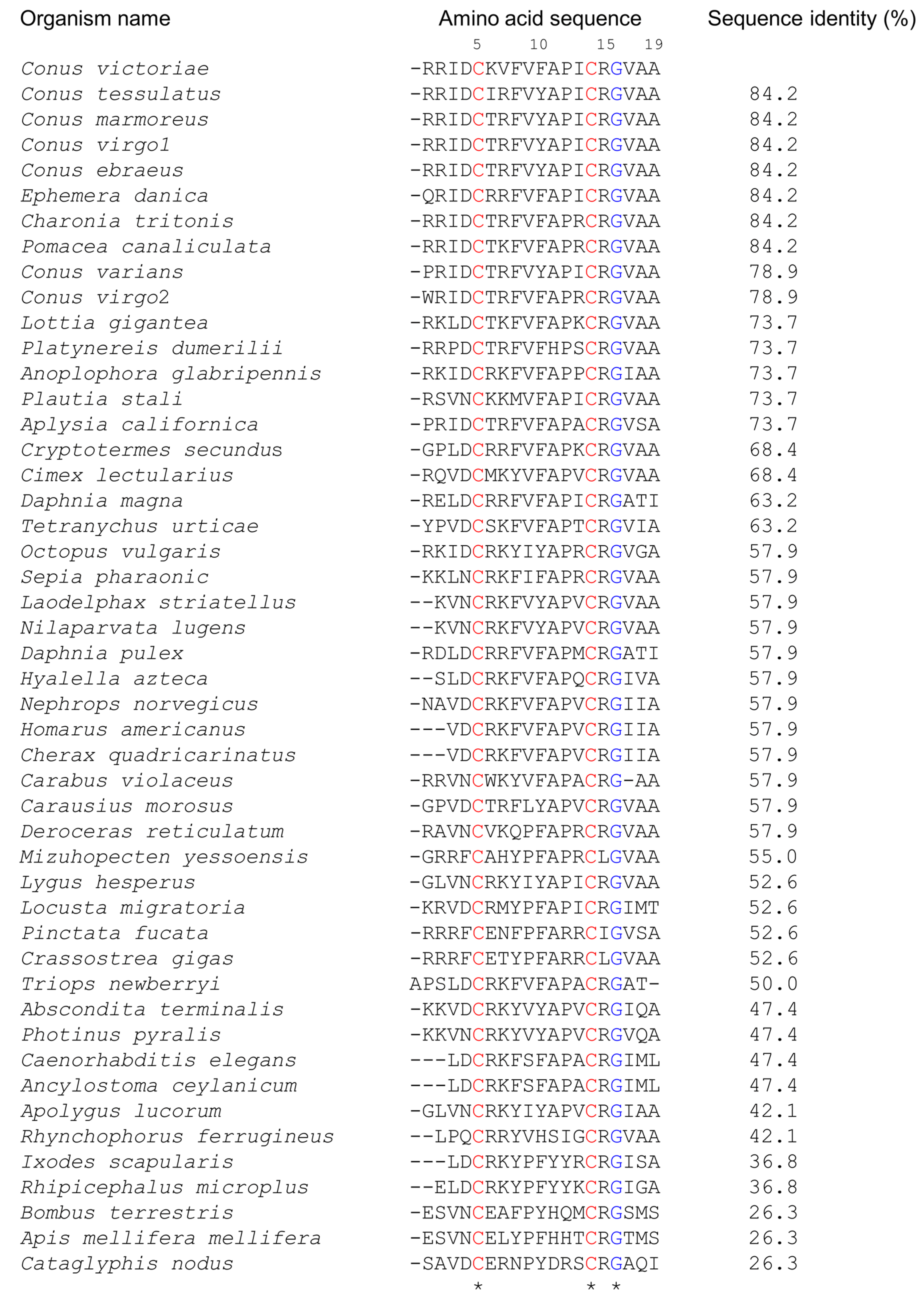
2.1. Elevenin-like Peptides Are Widely Distributed in Protostomes
2.2. Peptide Synthesis, Oxidative Folding, Purification, and Mass Spectroscopy
2.3. Sequential Assignments of NMR Apectra of Elevenin-Vc1
2.4. Solution Structure of Elevenin-Vc1
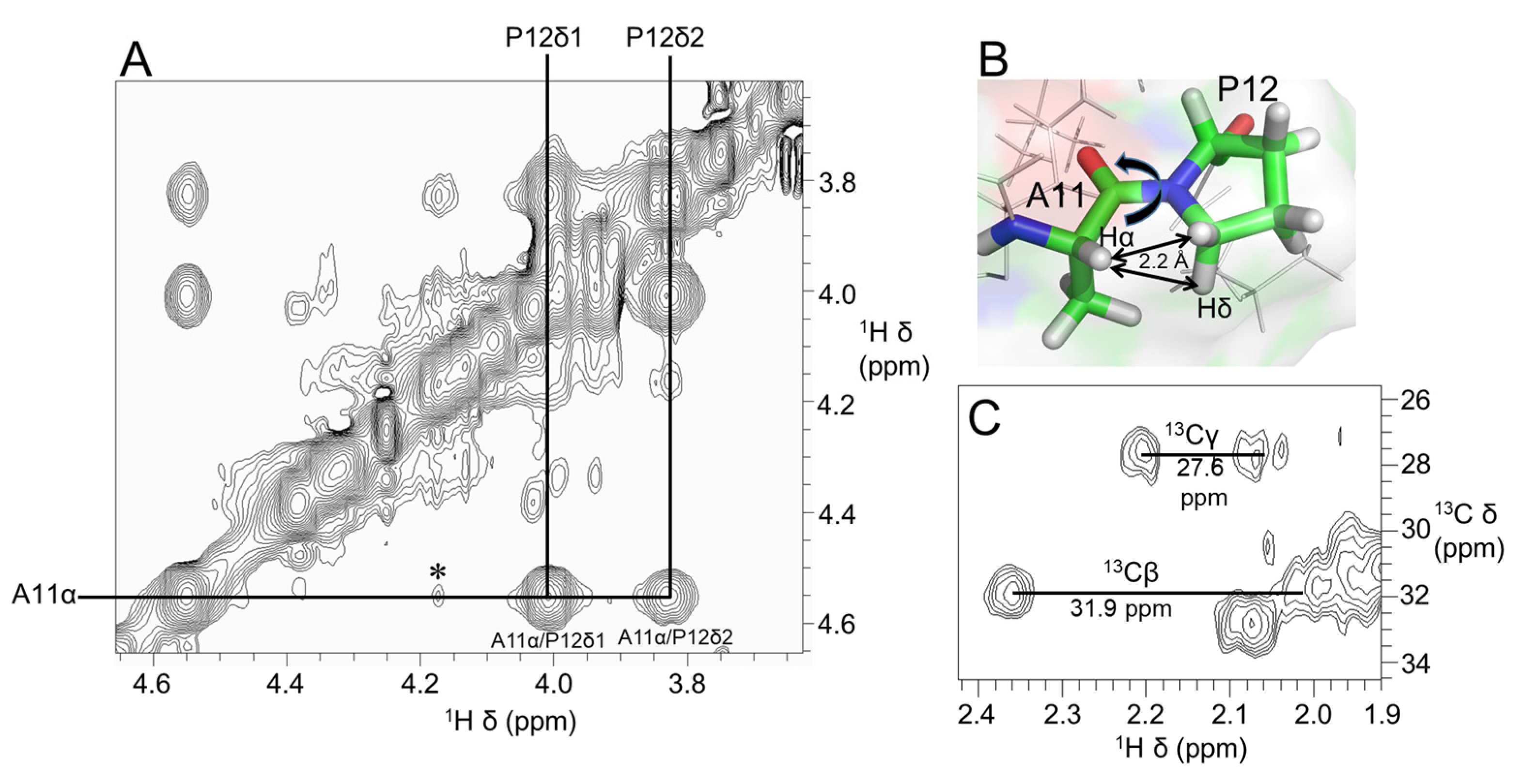
2.5. Valine-Phenylalanine Interactions
2.6. Elevenin-Vc1 Made Mice Hyperactive upon Intracranial Injection
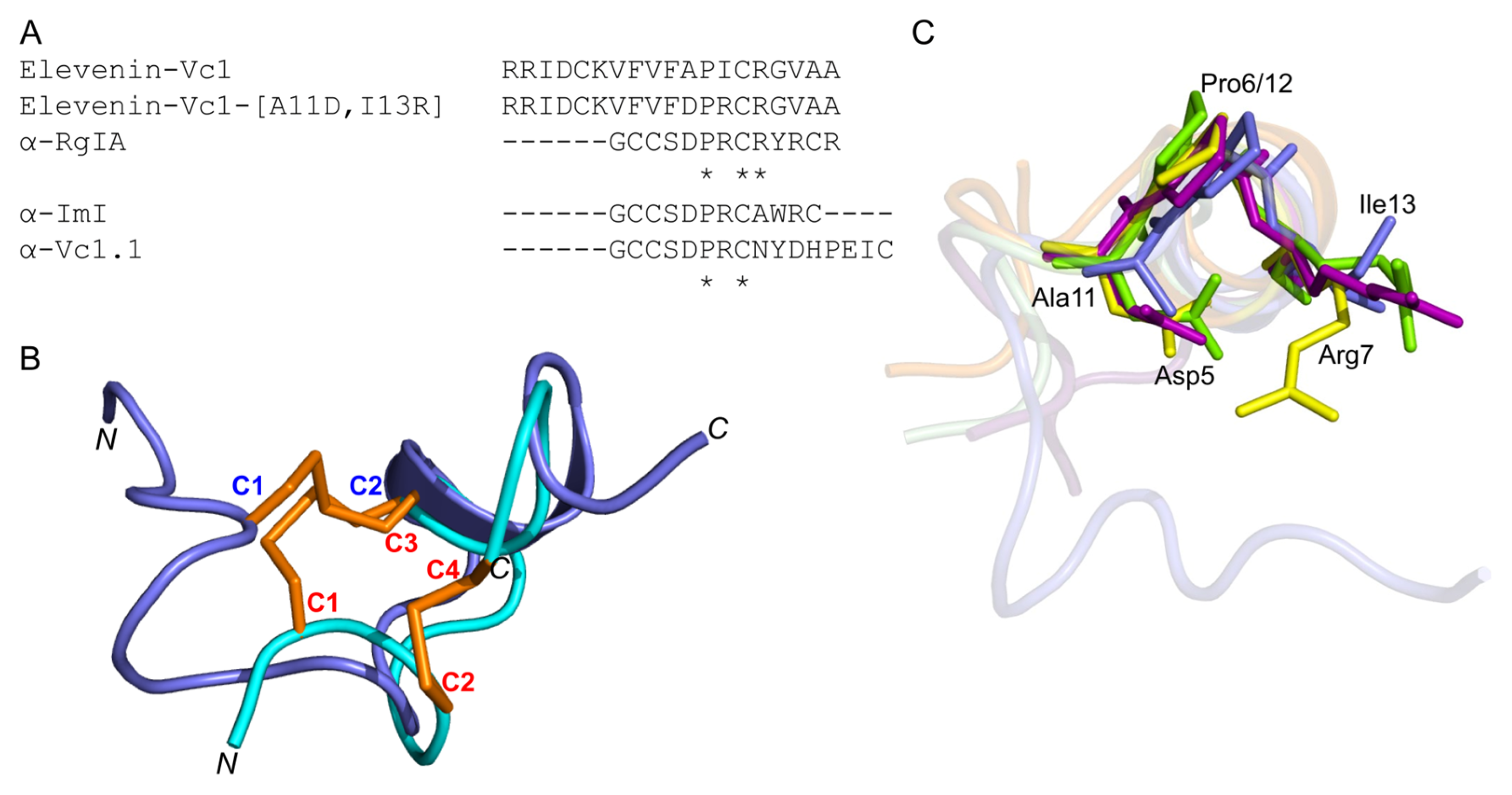
2.7. Elevenin-Vc1 Has a Structural Fold Similar to That of α-Conotoxins
2.8. Elevenin-Vc1 and Elevenin-Vc1[A11D, I13R] (Elevenin-Vc1-DPR) Are not Active against nAChRs
3. Conclusions
4. Materials and Methods
4.1. Peptide Synthesis
4.2. NMR Sample Preparation
4.3. NMR Data Collection, Processing, and Analysis
4.4. Structure Determination
4.5. Mice behavioural Experiments
4.6. Assay against Heterologous Human nAChRs Expressed in Xenopus laevis Oocytes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robinson, S.D.; Safavi-Hemami, H.; McIntosh, L.D.; Purcell, A.W.; Norton, R.S.; Papenfuss, A.T. Diversity of conotoxin gene superfamilies in the venomous snail, Conus victoriae. PLoS ONE 2014, 9, e87648. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, E.K.M.; Tytgat, J. In the picture: Disulfide-poor conopeptides, a class of pharmacologically interesting compounds. J. Venom Anim. Toxins Incl. Trop. Dis. 2016, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Li, Q.; Bandyopadhyay, P.K.; Gajewiak, J.; Yandell, M.; Papenfuss, A.T.; Purcell, A.W.; Norton, R.S.; Safavi-Hemami, H. Hormone-like peptides in the venoms of marine cone snails. Gen. Comp. Endocrinol. 2017, 244, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J. Conotoxins as selective inhibitors of neuronal ion channels, receptors and transporters. IUBMB Life 2004, 56, 89–93. [Google Scholar] [CrossRef]
- Vetter, I.; Lewis, R.J. Therapeutic potential of cone snail venom peptides (conopeptides). Curr. Top. Med. Chem. 2012, 12, 1546–1552. [Google Scholar] [CrossRef]
- Sadeghi, M.; McArthur, J.R.; Finol-Urdaneta, R.K.; Adams, D.J. Analgesic conopeptides targeting G protein-coupled receptors reduce excitability of sensory neurons. Neuropharmacology 2017, 127, 116–123. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Gajewiak, J.; Karanth, S.; Robinson, S.D.; Ueberheide, B.; Douglass, A.D.; Schlegel, A.; Imperial, J.S.; Watkins, M.; Bandyopadhyay, P.K.; et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl. Acad. Sci. USA 2015, 112, 1743–1748. [Google Scholar] [CrossRef]
- Menting, J.G.; Gajewiak, J.; MacRaild, C.A.; Chou, D.H.; Disotuar, M.M.; Smith, N.A.; Miller, C.; Erchegyi, J.; Rivier, J.E.; Olivera, B.M.; et al. A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin. Nat. Struct. Mol. Biol. 2016, 23, 916–920. [Google Scholar] [CrossRef]
- Ahorukomeye, P.; Disotuar, M.M.; Gajewiak, J.; Karanth, S.; Watkins, M.; Robinson, S.D.; Florez Salcedo, P.; Smith, N.A.; Smith, B.J.; Schlegel, A.; et al. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. Elife 2019, 8, e41574. [Google Scholar] [CrossRef]
- Ramiro, I.B.L.; Bjorn-Yoshimoto, W.E.; Imperial, J.S.; Gajewiak, J.; Salcedo, P.F.; Watkins, M.; Taylor, D.; Resager, W.; Ueberheide, B.; Brauner-Osborne, H.; et al. Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads. Sci. Adv. 2022, 8, eabk1410. [Google Scholar] [CrossRef]
- Taussig, R.; Kaldany, R.R.; Scheller, R.H. A cDNA clone encoding neuropeptides isolated from Aplysia neuron L11. Proc. Natl. Acad. Sci. USA 1984, 81, 4988–4992. [Google Scholar] [CrossRef]
- Waldman, J.; Xavier, M.A.; Vieira, L.R.; Logullo, R.; Braz, G.R.C.; Tirloni, L.; Ribeiro, J.M.C.; Veenstra, J.A.; Silva Vaz, I.D. Neuropeptides in Rhipicephalus microplus and other hard ticks. Ticks Tick-Borne Dis. 2022, 13, 101910. [Google Scholar] [CrossRef]
- Taussig, R.; Kaldany, R.-R.; Rothbard, J.B.; Schoolnik, G.; Scheller, R.H. Expression of the L11 neuropeptide gene in the Aplysia central nervous system. J. Comp. Neurol. 1985, 238, 53–64. [Google Scholar] [CrossRef]
- Nagata, S. Chapter 117-Elevenin. In Handbook of Hormones, 2nd ed.; Ando, H., Ukena, K., Nagata, S., Eds.; Academic Press: San Diego, CA, USA, 2021; pp. 879–880. [Google Scholar] [CrossRef]
- Kim, D.; Šimo, L.; Park, Y. Molecular characterization of neuropeptide elevenin and two elevenin receptors, IsElevR1 and IsElevR2, from the blacklegged tick, Ixodes scapularis. Insect Biochem. Mol. Biol. 2018, 101, 66–75. [Google Scholar] [CrossRef]
- Wang, S.L.; Wang, W.W.; Ma, Q.; Shen, Z.F.; Zhang, M.Q.; Zhou, N.M.; Zhang, C.X. Elevenin signaling modulates body color through the tyrosine-mediated cuticle melanism pathway. FASEB J. 2019, 33, 9731–9741. [Google Scholar] [CrossRef]
- Yamada, K.; Hirotsu, T.; Matsuki, M.; Butcher, R.A.; Tomioka, M.; Ishihara, T.; Clardy, J.; Kunitomo, H.; Iino, Y. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science 2010, 329, 1647–1650. [Google Scholar] [CrossRef]
- Conzelmann, M.; Offenburger, S.-L.; Asadulina, A.; Keller, T.; Münch, T.A.; Jékely, G. Neuropeptides regulate swimming depth of Platynereis larvae. Proc. Natl. Acad. Sci. USA 2011, 108, E1174–E1183. [Google Scholar] [CrossRef]
- Bauknecht, P.; Jékely, G. Large-scale combinatorial deorphanization of Platynereis neuropeptide GPCRs. Cell Rep. 2015, 12, 684–693. [Google Scholar] [CrossRef]
- Elphick, M.R.; Mirabeau, O.; Larhammar, D. Evolution of neuropeptide signalling systems. J. Exp. Biol. 2018, 221, jeb151092. [Google Scholar] [CrossRef]
- Uchiyama, H.; Maehara, S.; Ohta, H.; Seki, T.; Tanaka, Y. Elevenin regulates the body color through a G protein-coupled receptor NlA42 in the brown planthopper Nilaparvata lugens. Gen. Comp. Endocrinol. 2018, 258, 33–38. [Google Scholar] [CrossRef]
- Bai, H.; Zhu, F.; Shah, K.; Palli, S.R. Large-scale RNAi screen of G protein-coupled receptors involved in larval growth, molting and metamorphosis in the red flour beetle. BMC Genom. 2011, 12, 388. [Google Scholar] [CrossRef]
- Norton, R.S. Peptide toxin structure and function by NMR. In Modern Magnetic Resonance; Webb, G.A., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 2081–2097. [Google Scholar] [CrossRef]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef]
- Li, S.C.; Goto, N.K.; Williams, K.A.; Deber, C.M. α-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. USA 1996, 93, 6676–6681. [Google Scholar] [CrossRef]
- Krishnarjuna, B.; Ganjiwale, A.; Manjappara, U.; Raghothama, S. NMR structure implications of enhanced efficacy of obestatin fragment analogs. Int. J. Pept. Res. Ther. 2011, 17, 259–270. [Google Scholar] [CrossRef]
- Raghothama, S. NMR of peptides. J. Ind. Inst. Sci. 2010, 90, 145–161. [Google Scholar]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Sunanda, P.; Krishnarjuna, B.; Peigneur, S.; Mitchell, M.L.; Estrada, R.; Villegas-Moreno, J.; Pennington, M.W.; Tytgat, J.; Norton, R.S. Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp. Pept. Sci. 2018, 110, e24073. [Google Scholar] [CrossRef]
- Krishnarjuna, B.; Sunanda, P.; Villegas–Moreno, J.; Csoti, A.; Morales, R.A.V.; Wai, D.C.C.; Panyi, G.; Prentis, P.; Norton, R.S. A disulfide-stabilised helical hairpin fold in acrorhagin I: An emerging structural motif in peptide toxins. J. Struct. Biol. 2021, 213, 107692. [Google Scholar] [CrossRef]
- Mahalakshmi, R.; Raghothama, S.; Balaram, P. NMR analysis of aromatic interactions in designed peptide β-hairpins. J. Am. Chem. Soc. 2006, 128, 1125–1138. [Google Scholar] [CrossRef]
- Olivera, B.M.; Gray, W.R.; Zeikus, R.; McIntosh, J.M.; Varga, J.; Rivier, J.; de Santos, V.; Cruz, L.J. Peptide neurotoxins from fish-hunting cone snails. Science 1985, 230, 1338–1343. [Google Scholar] [CrossRef]
- Ellison, M.; Feng, Z.-P.; Park, A.J.; Zhang, X.; Olivera, B.M.; McIntosh, J.M.; Norton, R.S. α-RgIA, a novel conotoxin that blocks the α9α10 nAChR: Structure and identification of key receptor-binding residues. J. Mol. Biol. 2008, 377, 1216–1227. [Google Scholar] [CrossRef]
- Rogers, J.P.; Luginbühl, P.; Shen, G.S.; McCabe, R.T.; Stevens, R.C.; Wemmer, D.E. NMR solution structure of α-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors. Biochemistry 1999, 38, 3874–3882. [Google Scholar] [CrossRef]
- Clark, R.J.; Fischer, H.; Nevin, S.T.; Adams, D.J.; Craik, D.J. The synthesis, structural characterization, and receptor specificity of the α-conotoxin Vc1.1. J. Biol. Chem. 2006, 281, 23254–23263. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, L.; Zhou, M.; You, Y.; Zhu, X.; Qiang, Y.; Qin, M.; Luo, S.; Ren, Z.; Xu, A. Molecular evolution and diversity of Conus peptide toxins, as revealed by gene structure and intron sequence analyses. PLoS ONE 2013, 8, e82495. [Google Scholar] [CrossRef]
- Pardos-Blas, J.R.; Irisarri, I.; Abalde, S.; Afonso, C.M.L.; Tenorio, M.J.; Zardoya, R. The genome of the venomous snail Lautoconus ventricosus sheds light on the origin of conotoxin diversity. Gigascience 2021, 10, giab037. [Google Scholar] [CrossRef]
- Ellison, M.; Haberlandt, C.; Gomez-Casati, M.E.; Watkins, M.; Elgoyhen, A.B.; McIntosh, J.M.; Olivera, B.M. α-RgIA: A novel conotoxin that specifically and potently blocks the α9α10 nAChR. Biochemistry 2006, 45, 1511–1517. [Google Scholar] [CrossRef]
- Ellison, M.; Gao, F.; Wang, H.-L.; Sine, S.M.; McIntosh, J.M.; Olivera, B.M. α-Conotoxins ImI and ImII target distinct regions of the human α7 nicotinic acetylcholine receptor and distinguish human nicotinic receptor subtypes. Biochemistry 2004, 43, 16019–16026. [Google Scholar] [CrossRef]
- van Lierop, B.J.; Robinson, S.D.; Kompella, S.N.; Belgi, A.; McArthur, J.R.; Hung, A.; MacRaild, C.A.; Adams, D.J.; Norton, R.S.; Robinson, A.J. Dicarba α-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors. ACS Chem. Biol. 2013, 8, 1815–1821. [Google Scholar] [CrossRef]
- Turner, M.W.; Marquart, L.A.; Phillips, P.D.; McDougal, O.M. Mutagenesis of α-conotoxins for enhancing activity and selectivity for nicotinic acetylcholine receptors. Toxins 2019, 11, 113. [Google Scholar] [CrossRef]
- Kennedy, A.C.; Belgi, A.; Husselbee, B.W.; Spanswick, D.; Norton, R.S.; Robinson, A.J. α-conotoxin peptidomimetics: Probing the minimal binding motif for effective analgesia. Toxins 2020, 12, 505. [Google Scholar] [CrossRef]
- Norton, R.S.; Olivera, B.M. Conotoxins down under. Toxicon 2006, 48, 780–798. [Google Scholar] [CrossRef] [PubMed]
- Chittoor, B.; Krishnarjuna, B.; Morales, R.A.V.; Norton, R.S. The single disulfide-directed β-hairpin fold: Role of disulfide bond in folding and effect of an additional disulfide bond on stability. Aust. J. Chem. 2020, 73, 312–320. [Google Scholar] [CrossRef]
- Chittoor, B.; Krishnarjuna, B.; Morales, R.A.V.; MacRaild, C.A.; Sadek, M.; Leung, E.W.W.; Robinson, S.D.; Pennington, M.W.; Norton, R.S. The single disulfide-directed β-hairpin fold. dynamics, stability, and engineering. Biochemistry 2017, 56, 2455–2466. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.D.; Norton, R.S. Conotoxin gene superfamilies. Mar. Drugs 2014, 12, 6058–6101. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Seymour, V.A.; Berecki, G.; Jia, X.; Akcan, M.; Adams, D.J.; Kaas, Q.; Craik, D.J. Less is more: Design of a highly stable disulfide-deleted mutant of analgesic cyclic α-conotoxin Vc1.1. Sci. Rep. 2015, 5, 13264. [Google Scholar] [CrossRef]
- Mohan, M.K.; Abraham, N.; Rajesh, R.P.; Jayaseelan, B.F.; Ragnarsson, L.; Lewis, R.J.; Sarma, S.P. Structure and allosteric activity of a single-disulfide conopeptide from Conus zonatus at human α3β4 and α7 nicotinic acetylcholine receptors. J. Biol. Chem. 2020, 295, 7096–7112. [Google Scholar] [CrossRef]
- Ma, Y.; Cao, Q.; Yang, M.; Gao, Y.; Fu, S.; Du, W.; Adams, D.J.; Jiang, T.; Tae, H.S.; Yu, R. Single-disulfide conopeptide Czon1107, an allosteric antagonist of the human α3β4 nicotinic acetylcholine receptor. Mar. Drugs 2022, 20, 497. [Google Scholar] [CrossRef]
- Joglekar, A.V.; Dehari, D.; Anjum, M.M.; Dulla, N.; Chaudhuri, A.; Singh, S.; Agrawal, A.K. Therapeutic potential of venom peptides: Insights in the nanoparticle-mediated venom formulations. Future J. Pharm. Sci. 2022, 8, 34. [Google Scholar] [CrossRef]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target Ther. 2022, 7, 48. [Google Scholar] [CrossRef]
- Lewis, R.J.; Garcia, M.L. Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2003, 2, 790–802. [Google Scholar] [CrossRef]
- Holford, M.; Daly, M.; King, G.F.; Norton, R.S. Venoms to the rescue. Science 2018, 361, 842–844. [Google Scholar] [CrossRef]
- Pennington, M.W.; Beeton, C.; Galea, C.A.; Smith, B.J.; Chi, V.; Monaghan, K.P.; Garcia, A.; Rangaraju, S.; Giuffrida, A.; Plank, D.; et al. Engineering a stable and selective peptide blocker of the Kv1.3 channel in T lymphocytes. Mol. Pharmacol. 2009, 75, 762–773. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Marius Clore, G. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. [Google Scholar] [CrossRef]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., III; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef]
- Tae, H.-S.; Gao, B.; Jin, A.-H.; Alewood, P.F.; Adams, D.J. Globular and ribbon isomers of Conus geographus α-conotoxins antagonize human nicotinic acetylcholine receptors. Biochem. Pharmacol. 2021, 190, 114638. [Google Scholar] [CrossRef]




| NMR Distance and Dihedral Constraints Used in XPLOR-NIH | |
|---|---|
| Total NOEs | 246 |
| Intra-residue | 68 |
| Inter-residue | 184 |
| Sequential | 98 |
| Medium-range | 74 |
| Long-range | 12 |
| Total dihedral angles | 9 |
| Backbone (ϕ angle) | 7 |
| Structure Statistics | |
| NOE RMSD (Å) | 0.05 |
| Angle (°) | 0.5 |
| Bonds (Å) | 0.006 |
| Improper (°) | 0.18 |
| Number of NOE violations (Å) | 0 |
| Number of angle violations | 0 |
| RMSD between 20 conformers | |
| Average pairwise RMSD for residues 3–16 (Å) | |
| Backbone (Å) (N, Cα, C) | 0.40 |
| All heavy atoms (Å) | 0.80 |
| Ramachandran analysis | |
| Amino acid residues in most favoured regions (%) | 99.4 |
| Amino acid residues in additionally allowed regions (%) | 0.6 |
| Peptide | Dose [nmol] | Observed Behaviour (Time = Approximate Min Post-Injection) |
|---|---|---|
| Saline Control | 0 (n = 4) | Normal: moving around immediately after injection, exploring the cage with intermittent grooming and resting behaviour, no jumping within first 45 min post-injection |
| Elevenin-Vc1 | 10 | 0–1 min: stiff tail, 0–5 min: resting position, 5–60 min: hyperactive, intensive grooming and jumping with no resting |
| 5 | 0–5 min normal behaviour, 5–60 min: hyperactive, intensive grooming and jumping with little resting | |
| 2.5 | No difference from the control mice |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnarjuna, B.; Sunanda, P.; Seow, J.; Tae, H.-S.; Robinson, S.D.; Belgi, A.; Robinson, A.J.; Safavi-Hemami, H.; Adams, D.J.; Norton, R.S. Characterisation of Elevenin-Vc1 from the Venom of Conus victoriae: A Structural Analogue of α-Conotoxins. Mar. Drugs 2023, 21, 81. https://doi.org/10.3390/md21020081
Krishnarjuna B, Sunanda P, Seow J, Tae H-S, Robinson SD, Belgi A, Robinson AJ, Safavi-Hemami H, Adams DJ, Norton RS. Characterisation of Elevenin-Vc1 from the Venom of Conus victoriae: A Structural Analogue of α-Conotoxins. Marine Drugs. 2023; 21(2):81. https://doi.org/10.3390/md21020081
Chicago/Turabian StyleKrishnarjuna, Bankala, Punnepalli Sunanda, Jeffrey Seow, Han-Shen Tae, Samuel D. Robinson, Alessia Belgi, Andrea J. Robinson, Helena Safavi-Hemami, David J. Adams, and Raymond S. Norton. 2023. "Characterisation of Elevenin-Vc1 from the Venom of Conus victoriae: A Structural Analogue of α-Conotoxins" Marine Drugs 21, no. 2: 81. https://doi.org/10.3390/md21020081
APA StyleKrishnarjuna, B., Sunanda, P., Seow, J., Tae, H.-S., Robinson, S. D., Belgi, A., Robinson, A. J., Safavi-Hemami, H., Adams, D. J., & Norton, R. S. (2023). Characterisation of Elevenin-Vc1 from the Venom of Conus victoriae: A Structural Analogue of α-Conotoxins. Marine Drugs, 21(2), 81. https://doi.org/10.3390/md21020081