Multiomic Approach for Bioprospection: Investigation of Toxins and Peptides of Brazilian Sea Anemone Bunodosoma caissarum
Abstract
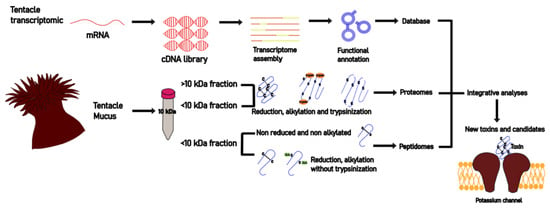
:1. Introduction
2. Results
2.1. Tentacle Transcriptome
2.2. Mucus and Tentacle Proteome
2.3. Mucus and Tentacle Peptidome
Non-Reduced and Non-Alkylated Samples
2.4. Peptidome with Reduced and Alkylated Cysteine Residues
2.5. Analysis of Cleavage Site in the Peptidome
3. Discussion
3.1. Considerations about the Mucus Collection Protocol and Omic Techniques
3.2. Tentacle and Mucus Composition
3.2.1. Toxins
| Toxin | Trinity Code | Transcript (Identity) | E-Value | Toxin Family | Tentacle/Mucus | Omic Approach |
|---|---|---|---|---|---|---|
| * U-actitoxin-Bcs3d | TRINITY_DN72667 | U-actitoxin-Bgr3d (42.3%) | 4.4E−13 | Sea anemone type 3 (BDS) potassium channel toxin | −/+ | Proteome, RA peptidome |
| * U-actitoxin-Bcs3a | TRINITY_DN10015 TRINITY_DN5917 TRINITY_DN38124 | U-actitoxin-Bgr3a (38.1%) | 4.8E−06 | Sea anemone type 3 (BDS) potassium channel toxin | +/+ | Proteome, RA peptidome |
| * U-actitoxin-Bcs2c | TRINITY_DN42790 | U-actitoxin-Ael2c (45.7%) | 1.8E−09 | Sea anemone type 3 (BDS) potassium channel toxin | +/+ | Proteome |
| * Κ-actitoxin-Bcs4m | TRINITY_DN4181 | Kappa-actitoxin-Avd4m (50%) | 1.4E−16 | Sea anemone type 3 (BDS) potassium channel toxin | +/+ | Proteome, RA peptidome |
| U-actitoxin-Bcs2a | TRINITY_DN14686 | U-actitoxin-Bcs2a (100%) | 1.3E−30 | Sea anemone type 3 (BDS) potassium channel toxin | +/+ | Proteome, RA peptidome |
| Bcs Tx3 | TRINITY_DN1024 | type III potassium channel toxin protein, partial (96%) | 1.4E−47 | Sea anemone type 3 (BDS) potassium channel toxin | +/+ | Proteome, RA peptidome |
| * Κ-actitoxin-Bcs2a | TRINITY_DN58216 | Kappa-actitoxin-Ael2a (50%) | 1.2E−08 | Sea anemone type 3 (BDS) potassium channel toxin | −/+ | RA peptidome |
| * ΚΠ-actitoxin-Bcs3e | TRINITY_DN13254 | kappaPI-actitoxin-Avd3e-like (73.5%) | 4.9E−90 | Venom Kunitz type. Sea anemone type 2 potassium channel toxin. | +/+ | Proteome |
| * Π-actitoxin-Bcs2e | TRINITY_DN217 | PI-stichotoxin-Hcr2e (56.4%) | 2.5E−28 | Venom Kunitz type. Sea anemone type 2 potassium channel toxin. | +/− | Proteome |
| * U-actitoxin-Bcs3n | TRINITY_DN6572 | U-actitoxin-Avd3n (74.7%) | 1.6E−51 | Venom Kunitz type. Sea anemone type 2 potassium channel toxin. | +/+ | Proteome, RA peptidome |
| * Π-actitoxin-Bcs3a | TRINITY_DN10770 | PI-actitoxin-Aeq3a-like (85.9%) | 1.2E−59 | Venom Kunitz type. Sea anemone type 2 potassium channel toxin. | −/+ | Proteome |
| * U-actitoxin-Bcs12a | TRINITY_DN7989 | U-actitoxin-Avd12a (74.4%) | 9.4E−38 | EGF-domain peptide | +/+ | Proteome |
| Turripeptide Pal 9.2-like | TRINITY_DN4295 | Turripeptide Pal 9.2 (38.2%) | 1.2E−12 | Kazal-type serine protease inhibitor domain | +/+ | Proteome, RA peptidome |
| * Π-actitoxin-Bcs5a | TRINITY_DN713 | PI-actitoxin-Avd5a (67.4%) | 8E−24 | Kazal-type serine protease inhibitor domain | +/+ | Proteome |
| Basic phospholipase A2 pseudexin A chain-like | TRINITY_DN24551 | Basic phospholipase A2 pseudexin A chain-like (72.8%) | 7.8E−108 | Phospholipase A2 | −/+ | Proteome |
| * Κ-actitoxin-Bcs1a | TRINITY_DN3227 | Kappa-actitoxin-Bgr1a (89.2%) | 1.4E−20 | Sea anemone type 1 potassium channel toxin | −/+ | Proteome |
| * Κ-actitoxin-Bcs3a | TRINITY_DN4127 | Kappa-actitoxin-Aer3a (55.4%) | 1.3E−28 | Sea anemone type 1 potassium channel toxin | +/+ | Proteome |
| * U-actitoxin-Bcs8a | TRINITY_DN1939 | U-actitoxin-Avd8a-like (82.3%) | 1.9E−51 | Sea anemone 8 toxin | +/+ | Proteome |
| * Δ-actitoxin-Bcs2a | TRINITY_DN24411 | Delta-stichotoxin-Sgt2a (48.2%) | 3.1E−15 | Anemone neurotoxin | −/+ | Proteome, RA peptidome |
| Zinc-metalloproteinase nas-13-like | TRINITY_DN24501 | Zinc-metalloproteinase nas-13-like (84.9%) | 3.5E−80 | Astacin | −/+ | Proteome |
| Zinc metalloproteinase nas-4-like | TRINITY_DN2272 | Zinc metalloprotease nas-4-like isoform X2 (82.9%) | 4E−269 | Astacin | +/+ | Proteome |
| Zinc metalloproteinase nas-15-like | TRINITY_DN1535 | Zinc-metalloproteinase nas-15-like (68.2%) | 1.2E−221 | Astacin | +/+ | Proteome |
| Cystatin | TRINITY_DN6968 | Cystatin-like (57.2%) | 7.3E−65 | Cystatin domain | +/+ | Proteome |
| Trinity Code | Transcript (Identity) | E-Value | Toxin Family | Tentacle/Mucus | Omic Approach |
|---|---|---|---|---|---|
| TRINITY_DN4279 | - | - | Sea anemone type 3 (BDS) potassium channel toxin | +/+ | Proteome, RA peptidome |
| TRINITY_DN14881 | Carboxypeptidase inhibitor SmCl-like (64.1%) | 3E−32 | Venom Kunitz type. Sea anemone type 2 potassium channel toxin | −/+ | Proteome |
| TRINITY_DN4129 | Neurogenic locus notch homolog protein 1 (37.1%) | 7.9E−12 | EGF-domain | +/+ | Proteome |
| TRINITY_DN3459 | Agrin-like (42.5%) | 4.1E−71 | Kazal-type serine protease inhibitor domain | −/+ | Proteome |
| TRINITY_DN322 | Fibrilin-2-like isoform X3 (69.5%) | 4.3E−70 | Kazal-type serine protease inhibitor domain | +/+ | Proteome |
| TRINITY_DN14498 | Neurogenic locus notch homolog protein 1-like isoform X6 (81.8%) | 2.4E−55 | Kazal-type serine protease inhibitor domain | +/+ | Proteome |
| TRINITY_DN1312 | Uncharacterized protein ZK643.6-like (47.2%) | 6.4E−52 | ShK domain-like | +/+ | Proteome |
| TRINITY_DN22515 | Uncharacterized protein LOC116291117 (72.1%) | 3.5E−75 | ShK domain-like | +/+ | Proteome |
| TRINITY_DN10788 | Uncharacterized protein LOC116293550 (67.9%) | 2.2E−227 | ShK domain-like | +/− | Proteome |
| TRINITY_DN10521 | Uncharacterized protein LOC116287301 (52.2%) | 1.9E−48 | ShK domain-like | +/+ | Proteome, RA peptidome, NRNA peptidome |
| TRINITY_DN4363 | Uncharacterized protein LOC116301037 (80.4%) | 0 | ShK domain-like | +/− | Proteome |
| TRINITY_DN19291 | Uncharacterized protein LOC116291117 (61.8%) | 1.3E−135 | ShK domain-like | +/+ | Proteome |
| TRINITY_DN2326 | mRNA, partial (76%) | 5.9E−45 | Anemone neurotoxin | −/+ | RA peptidome |
| TRINITY_DN52114 | Uncharacterized protein LOC116291022 (70.8%) | 6.6E−102 | F5/8 C domain | −/+ | Proteome |
| TRINITY_DN16749 | Uncharacterized protein LOC116297803 isoform X6 (67.5%) | 1.4E−50 | F5/8 C domain | −/+ | Proteome |
| TRINITY_DN8245 | Uncharacterized protein LOC116298543 (67.6%) | 5.2E−70 | F5/8 C domain | +/+ | Proteome |
| TRINITY_DN37576 | Uncharacterized protein LOC116297953 (65.3%) | 6.2E−44 | F5/8 C domain | −/+ | RA peptidome, NRNA peptidome |
| TRINITY_DN10809 | Perlucin-like protein (65.7%) | 1.2E−98 | Lectin C-type domain | +/+ | Proteome |
3.2.2. Cysteine Rich Proteins
3.2.3. Neuropeptides
3.2.4. Intracellular Peptides (InPeps)
4. Materials and Methods
4.1. Collection and Maintenance of Specimens
4.2. Transcriptomic
4.2.1. RNA Extraction, mRNA Library Synthesis and Illumina Sequencing
4.2.2. Transcriptome Assembly, Annotation and Functional Enrichment
4.3. Proteomic and Peptidomic
Protein and Peptide Fractions Preparation
4.4. NanoLC and Mass Spectrometry
4.5. Protein and Peptide Identification
4.6. Quantitative Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bakus, G.J.; Targett, N.M.; Schulte, B. Chemical ecology of marine organisms: An overview. J. Chem. Ecol. 1986, 12, 951–987. [Google Scholar] [CrossRef] [PubMed]
- Coll, J.; La Barre, S.; Sammarco, P.; Williams, W.; Bakus, G. Chemical Defences in Soft Corals (Coelenterata: Octocorallia) of the Great Barrier Reef: A Study of Comparative Toxicities. Mar. Ecol. Prog. Ser. 1982, 8, 271–278. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Moran, Y.; Gordon, D.; Gurevitz, M. Sea anemone toxins affecting voltage-gated sodium channels—Molecular and evolutionary features. Toxicon 2009, 54, 1089–1101. [Google Scholar] [CrossRef] [Green Version]
- Diochot, S.; Lazdunski, M. Sea anemone toxins affecting potassium channels. In Marine Toxins as Research Tools. Progress in Molecular and Subcellular Biology; Fusetani, N., Kem, W., Eds.; Springer: Berlin, Germany, 2009; Volume 46, pp. 99–122. [Google Scholar]
- Oliveira, J.S.; Zaharenko, A.J.; Ferreira, W.A.; Konno, K.; Shida, C.S.; Richardson, M.; Lúcio, A.D.; Beirão, P.S.L.; de Freitas, J.C. BcIV, a new paralyzing peptide obtained from the venom of the sea anemone Bunodosoma caissarum. A comparison with the Na+ channel toxin BcIII. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2006, 1764, 1592–1600. [Google Scholar] [CrossRef]
- Chun, H.G.; Davies, B.; Hoth, D.; Suffness, M.; Plowman, J.; Flora, K.; Grieshaber, C.; Leyland-Jones, B.; Didemnin, B. The first marine compound entering clinical trials as an antineoplastic agent. Investig. New Drugs 1986, 4, 279–284. [Google Scholar] [CrossRef]
- Schweikart, K.; Guo, L.; Shuler, Z.; Abrams, R.; Chiao, E.T.; Kolaja, K.L.; Davis, M. The effects of jaspamide on human cardiomyocyte function and cardiac ion channel activity. Toxicol. Vitr. 2012, 27, 745–751. [Google Scholar] [CrossRef] [Green Version]
- Tarcha, E.J.; Olsen, C.M.; Probst, P.; Peckham, D.; Muñoz-Elías, E.J.; Kruger, J.G.; Iadonato, S.P. Safety and pharmacodynamics of dalazatide, a Kv1.3 channel inhibitor, in the treatment of plaque psoriasis: A randomized phase 1b trial. PLoS ONE 2017, 12, e0180762. [Google Scholar] [CrossRef]
- Rinehart, K.L.; Holt, T.G.; Fregeau, N.L.; Keifer, P.A.; Wilson, G.R.; Perun, T.J., Jr.; Sakai, R.; Thompson, A.G.; Stroh, J.G.; Shield, L.S.; et al. Bioactive Compounds from Aquatic and Terrestrial Sources. J. Nat. Prod. 1990, 53, 771–792. [Google Scholar] [CrossRef]
- Castañeda, O.; Sotolongo, V.; Amor, A.M.; Stöcklin, R.; Anderson, A.J.; Harvey, A.L.; Engström, Å.; Wernstedt, C.; Karlsson, E. Characterization of a potassium channel toxin from the Caribbean sea anemone Stichodactyla helianthus. Toxicon 1995, 33, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Beeton, C.; Wulff, H.; Standifer, N.E.; Azam, P.; Mullen, K.M.; Pennington, M.W.; Kolski-Andreaco, A.; Wei, E.; Grino, A.; Counts, D.R.; et al. Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc. Natl. Acad. Sci. USA 2006, 103, 17414–17419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devarajan, P.; Chen, Z. Autoimmune effector memory T cells: The bad and the good. Immunol. Res. 2013, 57, 12–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotton, J.; Crest, M.; Bouet, F.; Alessandri, N.; Gola, M.; Forest, E.; Karlsson, E.; Castaneda, O.; Harvey, A.L.; Vita, C.; et al. A Potassium-Channel Toxin from the Sea Anemone Bunodosoma Granulifera, An Inhibitor for Kv1 Channels—Revision of the Amino Acid Sequence, Disulfide-Bridge Assignment, Chemical Synthesis, and Biological Activity. Eur. J. Biochem. 1997, 244, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Malpezzi, E.L.; Freitas, J.C. Antimitotic effect of an extract of the sea anemone Bunodosoma caissarum on sea urchin egg development. Braz. J. Med. Biol. Res. 1990, 23, 811–814. [Google Scholar]
- Malpezzi, E.L.; Freitas, J.C. Hemolytic activity of the nematocyst venom from the sea anemone Bunodosoma caissarum. Braz. J. Med. Biol. Res. 1991, 24, 1245–1249. [Google Scholar]
- Malpezzi, E.L.A.; Matsui, D.H.; Groote, S.C.T.S.; Freitas, J.C.; Santelli, G.M.; Fernandas, J.B. Antitumoral activity in an organic extract of the sea anemone Bunodosoma caissarum. Toxicon 1993, 33, 291. [Google Scholar]
- Malpezzi, E.L.A. Pharmacologically Active Substances of the Sea Anemone Bunodosoma caissarum, Corrêa, 1964 (Cnidaria, Anthozoa, Actiniidae). J. Venom. Anim. Toxins Incl. Trop. Dis. 1996, 2, 60. [Google Scholar] [CrossRef]
- Malpezzi, E.L.; de Freitas, J.; Muramoto, K.; Kamiya, H. Characterization of peptides in sea anemone venom collected by a novel procedure. Toxicon 1993, 31, 853–864. [Google Scholar] [CrossRef]
- Appeltans, W.; Ahyong, S.T.; Anderson, G.; Angel, M.V.; Artois, T.; Bailly, N.; Bamber, R.; Barber, A.; Bartsch, I.; Berta, A.; et al. The Magnitude of Global Marine Species Diversity. Curr. Biol. 2012, 22, 2189–2202. [Google Scholar] [CrossRef] [Green Version]
- Klompen, A.; Macrander, J.; Reitzel, A.; Stampar, S. Transcriptomic Analysis of Four Cerianthid (Cnidaria, Ceriantharia) Venoms. Mar. Drugs 2020, 18, 413. [Google Scholar] [CrossRef]
- Fu, J.; He, Y.; Peng, C.; Tang, T.; Jin, A.; Liao, Y.; Shi, Q.; Gao, B. Transcriptome Sequencing of the Pale Anemones (Exaiptasia diaphana) Revealed Functional Peptide Gene Resources of Sea Anemone. Front. Mar. Sci. 2022, 9, 856501. [Google Scholar] [CrossRef]
- Warner, J.F.; Röttinger, E. Transcriptomic Analysis in the Sea Anemone Nematostella vectensis. In Developmental Biology of the Sea Urchin and Other Marine Invertebrates; Carroll, D.J., Stricker, S.A., Eds.; Methods in Molecular Biology; Springer Protocols: New York, NY, USA, 2020; Volume 2219, pp. 231–240. [Google Scholar]
- Stefanik, D.J.; Lubinski, T.J.; Granger, B.R.; Byrd, A.L.; Reitzel, A.M.; DeFilippo, L.; Lorenc, A.; Finnerty, J.R. Production of a reference transcriptome and transcriptomic database (EdwardsiellaBase) for the lined sea anemone, Edwardsiella lineata, a parasitic cnidarian. BMC Genom. 2014, 15, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrander, J.; Brugler, M.R.; Daly, M. A RNA-seq approach to identify putative toxins from acrorhagi in aggressive and non-aggressive Anthopleura elegantissima polyps. BMC Genom. 2015, 16, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera-De-Torre, E.; Martínez-Del-Pozo, Á.; Garb, J.E. Stichodactyla helianthus’ de novo transcriptome assembly: Discovery of a new actinoporin isoform. Toxicon 2018, 150, 105–114. [Google Scholar] [CrossRef]
- Madio, B.; Undheim, E.A.B.; King, G.F. Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteom. 2017, 166, 83–92. [Google Scholar] [CrossRef]
- Surm, J.M.; Stewart, Z.K.; Papanicolaou, A.; Pavasovic, A.; Prentis, P.J. The draft genome of Actinia tenebrosa reveals insights into toxin evolution. Ecol. Evol. 2019, 9, 11314–11328. [Google Scholar] [CrossRef] [Green Version]
- Liao, Q.; Feng, Y.; Yang, B.; Lee, S.M.-Y. Cnidarian peptide neurotoxins: A new source of various ion channel modulators or blockers against central nervous systems disease. Drug Discov. Today 2018, 24, 189–197. [Google Scholar] [CrossRef]
- Linial, M.; Rappoport, N.; Ofer, D. Overlooked Short Toxin-Like Proteins: A Shortcut to Drug Design. Toxins 2017, 9, 350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vernon, L.P.; Bell, J.D. Membrane structure, toxins and phospholipase A2 activity. Pharmacol. Ther. 1992, 54, 269–295. [Google Scholar] [CrossRef]
- Rojko, N.; Dalla Serra, M.; Maček, P.; Anderluh, G. Pore formation by actinoporins, cytolysins from sea anemones. Biochimica Biophysica Acta (BBA)—Biomembr. 2016, 1858, 446–456. [Google Scholar] [CrossRef]
- Podobnik, M.; Anderluh, G. Pore-forming toxins in Cnidaria. Semin. Cell Dev. Biol. 2017, 72, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Minagawa, S.; Nagai, H.; Ishida, M.; Nagashima, Y.; Shiomi, K. Novel peptide toxins from acrorhagi, aggressive organs of the sea anemone Actinia equina. Toxicon 2005, 46, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.D.; Alves, R.S.; Martins, A.M.; Barbosa, P.S.F.; Evangelista, J.S.; Evangelista, J.J.F.; Ximenes, R.M.; Toyama, M.H.; Toyama, D.O.; Souza, A.J.F.; et al. Purification and characterization of the biological effects of phospholipase A2 from sea anemone Bunodosoma caissarum. Toxicon 2009, 54, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Orts, D.J.B.; Peigneur, S.; Madio, B.; Cassoli, J.S.; Montandon, G.G.; Pimenta, A.M.C.; Bicudo, J.E.P.W.; Freitas, J.C.; Zaharenko, A.J.; Tytgat, J. Biochemical and Electrophysiological Characterization of Two Sea Anemone Type 1 Potassium Toxins from a Geographically Distant Population of Bunodosoma caissarum. Mar. Drugs 2013, 11, 655–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orts, D.J.B.; Moran, Y.; Cologna, C.T.; Peigneur, S.; Madio, B.; Praher, D.; Quinton, L.; de Pauw, E.; Bicudo, J.E.P.W.; Tytgat, J.; et al. BcsTx3 is a founder of a novel sea anemone toxin family of potassium channel blocker. FEBS J. 2013, 280, 4839–4852. [Google Scholar] [CrossRef]
- Ramírez-Carreto, S.; Vera-Estrella, R.; Portillo-Bobadilla, T.; Licea-Navarro, A.; Bernaldez-Sarabia, J.; Rudiño-Piñera, E.; Verleyen, J.J.; Rodríguez, E.; Rodríguez-Almazán, C. Transcriptomic and Proteomic Analysis of the Tentacles and Mucus of Anthopleura dowii Verrill, 1869. Mar. Drugs 2019, 17, 436. [Google Scholar] [CrossRef] [Green Version]
- Kiyatkin, N.; Dulubova, I.; Grishin, E. Cloning and structural analysis of alpha-latroinsectotoxin cDNA. Abundance of ankyrin-like repeats. Eur. J. Biochem. 1993, 213, 121–127. [Google Scholar] [CrossRef]
- Holz, G.G.; Habener, J.F. Black widow spider α-latrotoxin: A presynaptic neurotoxin that shares structural homology with the glucagon-like peptide-1 family of insulin secretagogic hormones. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1998, 121, 177–184. [Google Scholar] [CrossRef]
- Stabili, L.; Schirosi, R.; Parisi, M.G.; Piraino, S.; Cammarata, M. The Mucus of Actinia equina (Anthozoa, Cnidaria): An Unexplored Resource for Potential Applicative Purposes. Mar. Drugs 2015, 13, 5276–5296. [Google Scholar] [CrossRef] [Green Version]
- Savoca, S.; Di Fresco, D.; Alesci, A.; Capillo, G.; Spanò, N. Mucus secretions in Cnidarian, an ecological, adaptive and evolutive tool. Adv. Oceanogr. Limnol. 2022, 13, 1–24. [Google Scholar] [CrossRef]
- Schweitz, H.; Bruhn, T.; Guillemare, E.; Moinier, D.; Lancelin, J.-M.; Béress, L.; Lazdunski, M. Kalicludines and kaliseptine. J. Biol. Chem. 1995, 270, 25121–25126. [Google Scholar] [CrossRef] [Green Version]
- An, D.; Pinheiro-Junior, E.L.; Béress, L.; Gladkikh, I.; Leychenko, E.; Undheim, E.A.B.; Peigneur, S.; Tytgat, J. AsKC11, a Kunitz Peptide from Anemonia sulcata, Is a Novel Activator of G Protein-Coupled Inward-Rectifier Potassium Channels. Mar. Drugs 2022, 20, 140. [Google Scholar] [CrossRef] [PubMed]
- Eagles, D.A.; Saez, N.J.; Krishnarjuna, B.; Bradford, J.J.; Chin, Y.K.-Y.; Starobova, H.; Mueller, A.; Reichelt, M.E.; Undheim, E.A.B.; Norton, R.S.; et al. A peptide toxin in ant venom mimics vertebrate EGF-like hormones to cause long-lasting hypersensitivity in mammals. Proc. Natl. Acad. Sci. USA 2022, 119, e2112630119. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Morlighem, J.R.; Zhou, H.; Lima, É.P.; Gomes, P.B.; Cai, J.; Lou, I.; Pérez, C.D.; Lee, S.M.; Rádis-Baptista, G. The Transcriptome of the Zoanthid Protopalythoa variabilis (Cnidaria, Anthozoa) Predicts a Basal Repertoire of Toxin-like and Venom-Auxiliary Polypeptides. Genome Biol. Evol. 2016, 8, 3045–3064. [Google Scholar] [CrossRef] [Green Version]
- Liao, Q.; Gong, G.; Poon, T.C.W.; Ang, I.L.; Lei, K.M.K.; Siu, S.W.I.; Wong, C.T.T.; Rádis-Baptista, G.; Lee, S.M.-Y. Combined transcriptomic and proteomic analysis reveals a diversity of venom-related and toxin-like peptides expressed in the mat anemone Zoanthus natalensis (Cnidaria, Hexacorallia). Arch. Toxicol. 2019, 93, 1745–1767. [Google Scholar] [CrossRef]
- Liang, H.; Jiang, G.; Wang, T.; Zhang, J.; Liu, W.; Xu, Z.; Zhang, J.; Xiao, L. An integrated transcriptomic and proteomic analysis reveals toxin arsenal of a novel Antarctic jellyfish Cyanea sp. J. Proteom. 2019, 208, 103483. [Google Scholar] [CrossRef] [PubMed]
- Jaimes-Becerra, A.; Gacesa, R.; Doonan, L.B.; Hartigan, A.; Marques, A.C.; Okamura, B.; Long, P.F. “Beyond primary sequence”—Proteomic data reveal complex toxins in cnidarian venoms. Integr. Comp. Biol. 2019, 59, 777–785. [Google Scholar] [CrossRef]
- Patthy, L.; Nikolics, K. Functions of agrin and agrin-related proteins. Trends Neurosci. 1993, 16, 76–81. [Google Scholar] [CrossRef]
- Lomonte, B.; Križaj, I. Snake venom phospholipase A2 toxins. In Handbook of Venoms and Toxins of Reptiles, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2021; pp. 389–412. [Google Scholar]
- Krayem, N.; Gargouri, Y. Scorpion venom phospholipases A2: A minireview. Toxicon 2020, 184, 48–54. [Google Scholar] [CrossRef]
- Atakuziev, B.U.; Nuritova, F.; Usmanov, P.B. Phospholipase A2 from the venom of the spider Eresus niger. Chem. Nat. Compd. 1991, 27, 487–489. [Google Scholar] [CrossRef]
- Palm, N.W.; Rosenstein, R.K.; Yu, S.; Schenten, D.D.; Florsheim, E.; Medzhitov, R. Bee Venom Phospholipase A2 Induces a Primary Type 2 Response that Is Dependent on the Receptor ST2 and Confers Protective Immunity. Immunity 2013, 39, 976–985. [Google Scholar] [CrossRef] [Green Version]
- Nevalainen, T.J.; Peuravuori, H.J.; Quinn, R.J.; Llewellyn, L.E.; Benzie, J.A.H.; Fenner, P.J.; Winkel, K.D. Phospholipase A2 in cnidaria. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 139, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Razpotnik, A.; Križaj, I.; Šribar, J.; Kordiš, D.; Maček, P.; Frangež, R.; Kem, W.R.; Turk, T. A new phospholipase A2 isolated from the sea anemone Urticina crassicornis—Its primary structure and phylogenetic classification. FEBS J. 2010, 277, 2641–2653. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Honma, T.; Nagai, H.; Ishida, M.; Nagashima, Y.; Shiomi, K. Isolation and cDNA cloning of a potassium channel peptide toxin from the sea anemone Anemonia erythraea. Toxicon 2006, 48, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.S.; Fuentes-Silva, D.; King, G.F. Development of a rational nomenclature for naming peptide and protein toxins from sea anemones. Toxicon 2012, 60, 539–550. [Google Scholar] [CrossRef]
- Ahmmed, M.K.; Bhowmik, S.; Giteru, S.G.; Zilani, N.H.; Adadi, P.; Islam, S.S.; Kanwugu, O.N.; Haq, M.; Ahmmed, F.; Ng, C.C.W.; et al. An Update of Lectins from Marine Organisms: Characterization, Extraction Methodology, and Potential Biofunctional Applications. Mar. Drugs 2022, 20, 430. [Google Scholar] [CrossRef]
- Ojeda, N.; Salazar, C.; Cárdenas, C.; Marshall, S.H. Expression of DC-SIGN-like C-Type Lectin Receptors in Salmo salar. Dev. Comp. Immunol. 2020, 113, 103806. [Google Scholar] [CrossRef]
- Schröder, H.C.; Ushijima, H.; Krasko, A.; Gamulin, V.; Thakur, N.L.; Diehl-Seifert, B.; Müller, I.M.; Müller, W.E.G. Emergence and Disappearance of an Immune Molecule, an Antimicrobial Lectin, in Basal Metazoa. J. Biol. Chem. 2003, 278, 32810–32817. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.-H. SARS-CoV-2 evolutionary adaptation toward host entry and recognition of receptor o-acetyl sialylation in virus–host interaction. Int. J. Mol. Sci. 2020, 21, 4549. [Google Scholar] [CrossRef]
- Rahimi, N. C-Type lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell adhesion molecules turned to pathogen recognition receptors. Biology 2020, 10, 1. [Google Scholar] [CrossRef]
- El-Maradny, Y.A.; El-Fakharany, E.M.; Abu-Serie, M.M.; Hashish, M.H.; Selim, H.S. Lectins purified from medicinal and edible mushrooms: Insights into their antiviral activity against pathogenic viruses. Int. J. Biol. Macromol. 2021, 179, 239–258. [Google Scholar] [CrossRef]
- Baumgartner, S.; Hofmann, K.; Bucher, P.; Chiquet-Ehrismann, R. The discoidin domain family revisited: New members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998, 7, 1626–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarras, M.P., Jr.; Yan, L.; Leontovich, A.; Zhang, J.S. Structure, expression, and developmental function of early divergent forms of metalloproteinases in Hydra. Cell Res. 2002, 12, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Dumermuth, E.; Sterchi, E.; Jiang, W.; Wolz, R.; Bond, J.; Flannery, A.; Beynon, R. The astacin family of metalloendopeptidases. J. Biol. Chem. 1991, 266, 21381–21385. [Google Scholar] [CrossRef] [PubMed]
- Reddi, A.H. BMP-1: Resurrection as Procollagen C-Proteinase. Science 1996, 271, 463. [Google Scholar] [CrossRef]
- Trevisan-Silva, D.; Gremski, L.H.; Chaim, O.M.; da Silveira, R.B.; Meissner, G.O.; Mangili, O.C.; Barbaro, K.C.; Gremski, W.; Veiga, S.S.; Senff-Ribeiro, A. Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 2010, 92, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Möhrlen, F.; Maniura, M.; Plickert, G.; Frohme, M.; Frank, U. Evolution of astacin-like metalloproteases in animals and their function in development. Evol. Dev. 2006, 8, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Zadka, L.; Kulus, M.J.; Piatek, K. ADAM protein family—Its role in tumorigenesis, mechanisms of chemoresistance and potential as diagnostic and prognostic factors. Neoplasma 2018, 65, 823–839. [Google Scholar] [CrossRef] [Green Version]
- Ponce, D.; Brinkman, D.L.; Potriquet, J.; Mulvenna, J. Tentacle Transcriptome and Venom Proteome of the Pacific Sea Nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa). Toxins 2016, 8, 102. [Google Scholar] [CrossRef] [Green Version]
- Neubauer, E.F.; Poole, A.Z.; Weis, V.M.; Davy, S.K. The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ 2016, 4, e2692. [Google Scholar] [CrossRef] [Green Version]
- McFarlane, I.D.; Graff, D.; Grimmelikhuijzen, C.J.P. Excitatory Actions of Antho-Rfamide, an Anthozoan Neuropeptide, on Muscles and Conducting Systems in the Sea Anemone Calliactis parasitica. J. Exp. Biol. 1987, 133, 157–168. [Google Scholar] [CrossRef]
- Tremblay, M.; Henry, J.; Anctil, M. Spawning and gamete follicle rupture in the cnidarian Renilla koellikeri: Effects of putative neurohormones. Gen. Comp. Endocrinol. 2004, 137, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T. Comparative Aspects of Structure and Function of Cnidarian Neuropeptides. Front. Endocrinol. 2020, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Katsukura, Y.; David, C.N.; Grimmelikhuijzen, C.J.P.; Sugiyama, T. Inhibition of metamorphosis by RFamide neuropeptides in planula larvae of Hydractinia echinata. Dev. Genes Evol. 2003, 213, 579–586. [Google Scholar] [CrossRef]
- Leitz, T.; Morand, K.; Mann, M. Metamorphosin A: A Novel Peptide Controlling Development of the Lower Metazoan Hydractinia echinata (Coelenterata, Hydrozoa). Dev. Biol. 1994, 163, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, E.; Watanabe, H.; Menschaert, G.; Holstein, T.W.; Baggerman, G.; Schoofs, L. A combined strategy of neuropeptide prediction and tandem mass spectrometry identifies evolutionarily conserved ancient neuropeptides in the sea anemone Nematostella vectensis. PLoS ONE 2019, 14, e0215185. [Google Scholar] [CrossRef] [Green Version]
- Koch, T.L.; Grimmelikhuijzen, C.J.P. A comparative genomics study of neuropeptide genes in the cnidarian subclasses Hexacorallia and Ceriantharia. BMC Genom. 2020, 21, 666. [Google Scholar] [CrossRef]
- Fricker, L.D. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: Implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol. Biosyst. 2010, 6, 1355–1365. [Google Scholar] [CrossRef]
- Teixeira, C.M.M.; Correa, C.N.; Iwai, L.K.; Ferro, E.S.; de Castro, L.M. Characterization of Intracellular Peptides from Zebrafish (Danio rerio) Brain. Zebrafish 2019, 16, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Gelman, J.S.; Sironi, J.; Castro, L.M.; Ferro, E.S.; Fricker, L.D. Peptidomic Analysis of Human Cell Lines. J. Proteome Res. 2011, 10, 1583–1592. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, S.; Yang, C.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Moir, R.D.; Willis, I.M.; Fricker, L.D. Analysis of the Yeast Peptidome and Comparison with the Human Peptidome. PLoS ONE 2016, 11, e0163312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kmiec, B.; Branca, R.M.; Berkowitz, O.; Li, L.; Wang, Y.; Murcha, M.W.; Whelan, J.; Lehtiö, J.; Glaser, E.; Teixeira, P.F. Accumulation of endogenous peptides triggers a pathogen stress response in Arabidopsis thaliana. Plant J. 2018, 96, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Araujo, C.B.; Heimann, A.S.; Remer, R.A.; Russo, L.C.; Colquhoun, A.; Forti, F.L.; Ferro, E.S. Intracellular Peptides in Cell Biology and Pharmacology. Biomolecules 2019, 9, 150. [Google Scholar] [CrossRef] [Green Version]
- Cunha, F.M.; Berti, D.A.; Ferreira, Z.S.; Klitzke, C.F.; Markus, R.P.; Ferro, E.S. Faculty Opinions recommendation of Intracellular peptides as natural regulators of cell signaling. J. Biol. Chem. 2008, 283, 24448–24459. [Google Scholar] [CrossRef] [Green Version]
- Berti, D.A.; Russo, L.C.; Castro, L.M.; Cruz, L.; Gozzo, F.C.; Heimann, J.C.; Lima, F.B.; Oliveira, A.C.; Andreotti, S.; Prada, P.O.; et al. Identification of intracellular peptides in rat adipose tissue: Insights into insulin resistance. Proteomics 2012, 12, 2668–2681. [Google Scholar] [CrossRef] [PubMed]
- Russo, L.C.; Asega, A.F.; Castro, L.M.; Negraes, P.D.; Cruz, L.; Gozzo, F.C.; Ulrich, H.; Camargo, A.C.; Rioli, V.; Ferro, E.S. Natural intracellular peptides can modulate the interactions of mouse brain proteins and thimet oligopeptidase with 14-3-3epsilon and calmodulin. Proteomics 2012, 12, 2641–2655. [Google Scholar] [CrossRef]
- Monte-Silva, E.R.C.R.C.; Russo, L.C.; Castro, L.M.; Gozzo, F.C.; de Araujo, C.B.; Peron, J.P.S.; Rioli, V.; Ferro, E.S. EL28 is a novel intracellular peptide that activates immune proteasome and CD8+ T-cell response. J. Proteom. 2016, 16, S1874–S3919. [Google Scholar]
- De Araujo, C.B.; Russo, L.C.; Castro, L.M.; Forti, F.L.; do Monte, E.R.; Rioli, V.; Gozzo, F.C.; Colquhoun, A.; Ferro, E.S. A novel intracellular peptide derived from g1/s cyclin d2 induces cell death. J. Biol. Chem. 2014, 289, 16711–16726. [Google Scholar] [CrossRef] [Green Version]
- Russo, L.C.; Araujo, C.B.; Iwai, L.K.; Ferro, E.S.; Forti, F.L. A Cyclin D2-derived peptide acts on specific cell cycle phases by activating ERK1/2 to cause the death of breast cancer cells. J. Proteom. 2016, 151, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Patrzykat, A.; Zhang, L.; Mendoza, V.; Iwama, G.K.; Hancock, R.E.W. Synergy of Histone-Derived Peptides of Coho Salmon with Lysozyme and Flounder Pleurocidin. Antimicrob. Agents Chemother. 2001, 45, 1337–1342. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, J.M.O.; Molle, G.M.; Kemp, G.D.; Smith, V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout Oncorhynchus mykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef]
- Smith, V.J.; Desbois, A.P.; Dyrynda, E.A. Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-algae. Mar. Drugs 2010, 8, 1213–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Café-Mendes, C.; Ferro, E.; Torrão, A.; Crunfli, F.; Rioli, V.; Schmitt, A.; Falkai, P.; Britto, L.; Turck, C.; Martins-De-Souza, D. Peptidomic analysis of the anterior temporal lobe and corpus callosum from schizophrenia patients. J. Proteom. 2017, 151, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Correa, C.N.; Fiametti, L.O.; Mazzi Esquinca, M.E.; Mantovani de Castro, M. Sample Preparation and Relative Quantitation using Reductive Methylation of Amines for Peptidomics Studies. J. Vis. Exp. 2021, 177, e62971. [Google Scholar]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification. Mol. Cell. Proteom. 2012, 11, M111.010587. [Google Scholar] [CrossRef] [Green Version]









| Trinity | TransDecoder | |
|---|---|---|
| Transcripts | 186,978 | 150,493 |
| N50 | 1900 | 501 |
| Genes | 111,386 | 61,465 |
| TRINITY | Name | Peptide | Mucus Sample Area | Tentacle Sample Area |
|---|---|---|---|---|
| TRINITY_DN1213 | - | DCRGKHCQTGPFGD | 9.06E+09 | 4.84E+08 |
| TRINITY_DN58641 | - | TLASSIQCVGKCKIKTSSGQCRTDLRCMLANKGAS | 1.70E+09 | 3.88E+08 |
| TRINITY_DN14686 | U-actitoxin-Bcs2a | GLPCDCHGHTGTYWLNYYSKCPKGYGYTGRCRYLVGSCCYK | 1.32E+09 | - |
| TRINITY_DN3092 | - | MATSCRKCKPGYGCWAVPCPKR | 1.09E+09 | - |
| TRINITY_DN557 | Lamin-A | EELSFKRSVYDKE | 5.92E+08 | - |
| TRINITY_DN1213 | - | DCRGKHCQTGPF | 3.56E+08 | 1.38E+06 |
| TRINITY_DN14686 | U-actitoxin-Bcs2a | GLPCDCHGHTGTY | 2.59E+08 | - |
| TRINITY_DN3092 | - | MATSCRKCKPGYGCWAVPCPKR | 2.16E+08 | - |
| TRINITY_DN3092 | - | ATSCRKCKPGYGCWAVPCPKR | 1.71E+08 | - |
| TRINITY_DN14686 | U-actitoxin-Bcs2a | WLNYYSKCPKGYGYTGRCRYLVGSCCYK | 1.68E+08 | - |
| TRINITY | Name | Peptide | Mucus Sample Area | Tentacle Sample Area |
|---|---|---|---|---|
| TRINITY_DN1213 | - | DCRGKHCQTGPFGD | 9.06E+09 | 4.84E+08 |
| TRINITY_DN282 | Histone H2B | LPGELAKHAVSEGTKAVTKYTSSK | - | 3.99E+08 |
| TRINITY_DN58641 | - | TLASSIQCVGKCKIKTSSGQCRTDLRCMLANKGAS | 1.7E+09 | 3.88E+08 |
| TRINITY_DN3037 | - | NPPYEEILEPAFFHIR | - | 2.61E+08 |
| TRINITY_DN10521 | - | APPDTSILDKLGKL | 7.26E+06 | 2.13E+08 |
| TRINITY_DN12562 | - | DEAGLLKYKTAAGAALVNERLKNLAERY | - | 1.81E+08 |
| TRINITY_DN3037 | - | QPPFLGGPAYFHIR | - | 1.52E+08 |
| TRINITY_DN282 | Histone H2B | LLLPGELAKHAVSEGTKAVTKYTSSK | 8.66E+06 | 1.52E+08 |
| TRINITY_DN25490 | U-actitoxin-Aeq6a | KERCDLLGDPCVKG | - | 1.51E+08 |
| TRINITY_DN1828 | - | AAAYVCDVARNLDCSAH | - | 1.43E+08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazzi Esquinca, M.E.; Correa, C.N.; Marques de Barros, G.; Montenegro, H.; Mantovani de Castro, L. Multiomic Approach for Bioprospection: Investigation of Toxins and Peptides of Brazilian Sea Anemone Bunodosoma caissarum. Mar. Drugs 2023, 21, 197. https://doi.org/10.3390/md21030197
Mazzi Esquinca ME, Correa CN, Marques de Barros G, Montenegro H, Mantovani de Castro L. Multiomic Approach for Bioprospection: Investigation of Toxins and Peptides of Brazilian Sea Anemone Bunodosoma caissarum. Marine Drugs. 2023; 21(3):197. https://doi.org/10.3390/md21030197
Chicago/Turabian StyleMazzi Esquinca, Maria Eduarda, Claudia Neves Correa, Gabriel Marques de Barros, Horácio Montenegro, and Leandro Mantovani de Castro. 2023. "Multiomic Approach for Bioprospection: Investigation of Toxins and Peptides of Brazilian Sea Anemone Bunodosoma caissarum" Marine Drugs 21, no. 3: 197. https://doi.org/10.3390/md21030197
APA StyleMazzi Esquinca, M. E., Correa, C. N., Marques de Barros, G., Montenegro, H., & Mantovani de Castro, L. (2023). Multiomic Approach for Bioprospection: Investigation of Toxins and Peptides of Brazilian Sea Anemone Bunodosoma caissarum. Marine Drugs, 21(3), 197. https://doi.org/10.3390/md21030197