Coral Lipidome: Molecular Species of Phospholipids, Glycolipids, Betaine Lipids, and Sphingophosphonolipids
Abstract
1. Introduction
2. Coral Host Lipidome
2.1. Phosphatidylcholine and Phosphatidylethanolamine
2.2. Phosphatidylserine and Phosphatidylinositol
2.3. Ceramideaminoethylphosphonate
3. Lipidome of Symbiotic Dinoflagellates
3.1. Plasma Membrane Lipids
3.2. Thylakoid Membrane Lipids
4. Lipidome of Other Members of Coral Holobiont
5. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Imbs, A.B.; Ermolenko, E.V.; Grigorchuk, V.P.; Sikorskaya, T.V.; Velansky, P.V. Current Progress in Lipidomics of Marine Invertebrates. Mar. Drugs 2021, 19, 660. [Google Scholar] [CrossRef]
- Spalding, M.D.; Grenfell, A.M. New estimates of global and regional coral reef areas. Coral Reefs 1997, 16, 225–230. [Google Scholar] [CrossRef]
- Veron, J.E.N.; Stafford-Smith, M.G.; Turak, E.; DeVantier, L.M. Corals of the World. Available online: http://www.coralsoftheworld.org/coral_geographic/interactive_map/ (accessed on 9 May 2023).
- Lewis, J.B. Biology and Ecology of the Hydrocoral Millepora on Coral Reefs. Adv. Mar. Biol. 2006, 50, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Bayer, F.M. Key to the genera of Octocorallia exclusive of Pennatulacea (Coelenterata: Anthozoa), with diagnoses of new taxa. Proc. Biol. Soc. Wash. 1981, 94, 902–947. [Google Scholar]
- Benayahu, Y.; Bridge, T.C.L.; Colin, P.L.; Liberman, R.; McFadden, C.S.; Pizarro, O.; Schleyer, M.H.; Shoham, E.; Reijnen, B.T.; Weis, M.; et al. Octocorals of the Indo-Pacific. In Mesophotic Coral Ecosystems; Loya, Y., Puglise, K.A., Bridge, T.C.L., Eds.; Springer: Cham, Switzerland, 2019; Volume 12, pp. 709–728. [Google Scholar]
- Sanchez, J.A.; Duenas, L.F.; Rowley, S.J.; Gonzalez-Zapata, F.L.; Vergara, D.C.; Montano-Salazar, S.M.; Calixto-Botia, I.; Gomez, C.E.; Abeytia, R.; Colin, P.L.; et al. Gorgonian Corals. In Mesophotic Coral Ecosystems; Loya, Y., Puglise, K.A., Bridge, T.C.L., Eds.; Springer: Cham, Switzerland, 2019; Volume 12, pp. 729–747. [Google Scholar]
- World Register of Marine Species. WoRMS Editorial Board. 2023. Available online: https://www.marinespecies.org (accessed on 12 May 2023).
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the Coral Microbiome: Underpinning the Health and Resilience of Reef Ecosystems. Annu. Rev. Microbiol. 2016, 70, 317–340. [Google Scholar] [CrossRef]
- Muscatine, L. Glycerol Excretion by Symbiotic Algae from Corals and Tridacna and Its Control by the Host. Science 1967, 156, 516–519. [Google Scholar] [CrossRef]
- Hughes, T.P.; Kerry, J.T.; Baird, A.H.; Connolly, S.R.; Chase, T.J.; Dietzel, A.; Hill, T.; Hoey, A.S.; Hoogenboom, M.O.; Jacobson, M.; et al. Global warming impairs stock-recruitment dynamics of corals. Nature 2019, 568, 387–390. [Google Scholar] [CrossRef]
- van Oppen, M.J.H. Coral bleaching: Patterns, processes, causes and consequences. In Coral Bleaching: Patterns, Processes, Causes and Consequences, 2nd ed.; Van Oppen, M.J.H., Lough, J.M., Eds.; Springer International Publishing Ag: Cham, Switzerland, 2018; Volume 233, pp. 1–356. [Google Scholar]
- Wang, Y.-N.; Shao, C.-L.; Zheng, C.-J.; Chen, Y.-Y.; Wang, C.-Y. Diversity and Antibacterial Activities of Fungi Derived from the Gorgonian Echinogorgia rebekka from the South China Sea. Mar. Drugs 2011, 9, 1379–1390. [Google Scholar] [CrossRef]
- Wegley, L.; Edwards, R.; Rodriguez-Brito, B.; Liu, H.; Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007, 9, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Janouskovec, J.; Horak, A.; Barott, K.L.; Rohwer, F.L.; Keeling, P.J. Global analysis of plastid diversity reveals apicomplexan-related lineages in coral reefs. Curr. Biol. 2012, 22, R518–R519. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, H.; Oku, H.; Higa, H.; Chinen, I.; Sakai, K. Composition of lipids, fatty acids and sterols in Okinawan corals. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999, 122, 397–407. [Google Scholar] [CrossRef]
- Lim, C.-S.; Bachok, Z.; Hii, Y.-S. Effects of supplementary polyunsaturated fatty acids on the health of the scleractinian coral Galaxea fascicularis (Linnaeus, 1767). J. Exp. Mar. Biol. Ecol. 2017, 491, 1–8. [Google Scholar] [CrossRef]
- Treignier, C.; Grover, R.; Ferrier-Pagés, C.; Tolosa, I. Effect of light and feeding on the fatty acid and sterol composition of zooxanthellae and host tissue isolated from the scleractinian coral Turbinaria reniformis. Limnol. Oceanogr. 2008, 53, 2702–2710. [Google Scholar] [CrossRef]
- Hamoutene, D.; Puestow, T.; Miller-Banoub, J.; Wareham, V. Main lipid classes in some species of deep-sea corals in the Newfoundland and Labrador region (Northwest Atlantic Ocean). Coral Reefs 2008, 27, 237–246. [Google Scholar] [CrossRef]
- Imbs, A.B. Fatty acids and other lipids of corals: Composition, distribution, and biosynthesis. Russ. J. Mar. Biol. 2013, 39, 153–168. [Google Scholar] [CrossRef]
- Bochkov, V.; Gesslbauer, B.; Mauerhofer, C.; Philippova, M.; Erne, P.; Oskolkova, O.V. Pleiotropic effects of oxidized phos-pholipids. Free. Radic. Biol. Med. 2017, 111, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- O’donnell, V.B.; Aldrovandi, M.; Murphy, R.C.; Krönke, G. Enzymatically oxidized phospholipids assume center stage as essential regulators of innate immunity and cell death. Sci. Signal. 2019, 12, eaau2293. [Google Scholar] [CrossRef]
- Serbulea, V.; DeWeese, D.; Leitinger, N. The effect of oxidized phospholipids on phenotypic polarization and function of macrophages. Free. Radic. Biol. Med. 2017, 111, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Rosic, N.; Ling, E.Y.S.; Chan, C.-K.K.; Lee, H.C.; Kaniewska, P.; Edwards, D.; Dove, S.; Hoegh-Guldberg, O. Unfolding the secrets of coral–algal symbiosis. ISME J. 2015, 9, 844–856. [Google Scholar] [CrossRef]
- Imbs, A.; Latyshev, N.; Dautova, T.; Latypov, Y. Distribution of lipids and fatty acids in corals by their taxonomic position and presence of zooxanthellae. Mar. Ecol. Prog. Ser. 2010, 409, 65–75. [Google Scholar] [CrossRef]
- Joseph, J.D. Lipid composition of marine and estuarine invertebrates: Porifera and cnidaria. Prog. Lipid Res. 1979, 18, 1–30. [Google Scholar] [CrossRef]
- Awai, K.; Matsuoka, R.; Shioi, Y. Lipid and fatty acid compositions of Symbiodinium strains. In Proceedings of the 12th International Coral Reef Symposium, Cairns, Australia, 9–13 July 2012. [Google Scholar]
- Leblond, J.D.; Khadka, M.; Duong, L.; Dahmen, J.L. Squishy lipids: Temperature effects on the betaine and galactolipid profiles of a C-18/C-18 peridinin-containing dinoflagellate, Symbiodinium microadriaticum (Dinophyceae), isolated from the mangrove jellyfish, Cassiopea xamachana. Phycol. Res. 2015, 63, 219–230. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Betaine ether-linked glycerolipids: Chemistry and biology. Prog. Lipid Res. 1996, 35, 1–51. [Google Scholar] [CrossRef] [PubMed]
- Rosset, S.; Koster, G.; Brandsma, J.; Hunt, A.N.; Postle, A.D.; D’angelo, C. Lipidome analysis of Symbiodiniaceae reveals possible mechanisms of heat stress tolerance in reef coral symbionts. Coral Reefs 2019, 38, 1241–1253. [Google Scholar] [CrossRef]
- Sikorskaya, T.V.; Efimova, K.V.; Imbs, A.B. Lipidomes of phylogenetically different symbiotic dinoflagellates of corals. Phytochemistry 2021, 181, 112579. [Google Scholar] [CrossRef] [PubMed]
- Wood, P. Lipidomics; Humana: New York, NY, USA, 2017; p. 248. [Google Scholar]
- Jouhet, J. Importance of the hexagonal lipid phase in biological membrane organization. Front. Plant Sci. 2013, 4, 494. [Google Scholar] [CrossRef] [PubMed]
- Diz-Muñoz, A.; Fletcher, D.A.; Weiner, O.D. Use the force: Membrane tension as an organizer of cell shape and motility. Trends Cell Biol. 2013, 23, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Needham, D. Physical properties of surfactant bilayer membranes: Thermal transitions, elasticity, rigidity, cohesion and colloidal interactions. J. Phys. Chem. 1987, 91, 4219–4228. [Google Scholar] [CrossRef]
- Ermolenko, E.V.; Sikorskaya, T.V. Lipidome of the reef-building coral Acropora cerealis: Changes under thermal stress. Biochem. Syst. Ecol. 2021, 97, 104276. [Google Scholar] [CrossRef]
- Sikorskaya, T.V. Investigation of the total lipidome from a zoantharia Palythoa sp. Chem. Nat. Compd. 2020, 56, 44–49. [Google Scholar] [CrossRef]
- Sikorskaya, T.V.; Ermolenko, E.V.; Boroda, A.V.; Ginanova, T.T. Physiological processes and lipidome dynamics in the soft coral Sinularia heterospiculata under experimental bleaching. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2021, 255, 110609. [Google Scholar] [CrossRef] [PubMed]
- Sikorskaya, T.V.; Ermolenko, E.V.; Imbs, A.B. Effect of experimental thermal stress on lipidomes of the soft coral Sinularia sp. and its symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 2020, 524, 151295. [Google Scholar] [CrossRef]
- Sikorskaya, T.V.; Imbs, A.B. Study of Total Lipidome of the Sinularia siaesensis Soft Coral. Russ. J. Bioorg. Chem. 2018, 44, 712–723. [Google Scholar] [CrossRef]
- Tang, C.-H.; Lin, C.-Y.; Lee, S.-H.; Wang, W.-H. Membrane lipid profiles of coral responded to zinc oxide nanoparticle-induced perturbations on the cellular membrane. Aquat. Toxicol. 2017, 187, 72–81. [Google Scholar] [CrossRef]
- Tang, C.-H.; Shi, S.-H.; Lin, C.-Y.; Li, H.-H.; Wang, W.-H. Using lipidomic methodology to characterize coral response to herbicide contamination and develop an early biomonitoring model. Sci. Total. Environ. 2019, 648, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Imbs, A.B.; Dang, L.T.P. The molecular species of phospholipids of the cold-water soft coral Gersemia rubiformis (Ehrenberg, 1834) (Alcyonacea, Nephtheidae). Russ. J. Mar. Biol. 2017, 43, 239–244. [Google Scholar] [CrossRef]
- Imbs, A.B.; Dang, L.P.T.; Rybin, V.G.; Nguyen, N.T.; Pham, L.Q. Distribution of very-long-chain fatty acids between molecular species of different phospholipid classes of two soft corals. Biochem. Analit. Biochem. 2015, 4, 205. [Google Scholar]
- Imbs, A.B.; Dang, L.P.T.; Rybin, V.G.; Svetashev, V.I. Fatty Acid, Lipid Class, and Phospholipid Molecular Species Composition of the Soft Coral Xenia sp. (Nha Trang Bay, the South China Sea, Vietnam). Lipids 2015, 50, 575–589. [Google Scholar] [CrossRef]
- Sikorskaya, T.V.; Ermolenko, E.V.; Efimova, K.V. Lipids of Indo-Pacific gorgonian corals are modified under the influence of microbial associations. Coral Reefs 2022, 41, 277–291. [Google Scholar] [CrossRef]
- Imbs, A.B.; Dang, L.P.T.; Nguyen, K.B. Comparative lipidomic analysis of phospholipids of hydrocorals and corals from tropical and cold-water regions. PLoS ONE 2019, 14, e0215759. [Google Scholar] [CrossRef]
- Imbs, A.B.; Ermolenko, E.V.; Grigorchuk, V.P.; Dang, L.T.P. Seasonal variation in the lipidome of two species of Millepora hydrocorals from Vietnam coastal waters (the South China Sea). Coral Reefs 2021, 40, 719–734. [Google Scholar] [CrossRef]
- Garrett, T.; Hwang, J.; Schmeitzel, J.L.; Schwarz, J. Lipidomics of Aiptasia pallida and Symbiodinium: A model system for investigating the molecular basis of coral symbiosis. FASEB J. 2011, 25, 938.2. [Google Scholar] [CrossRef]
- Garrett, T.A.; Schmeitzel, J.L.; Klein, J.A.; Hwang, J.J.; Schwarz, J.A. Comparative Lipid Profiling of the Cnidarian Aiptasia pallida and Its Dinoflagellate Symbiont. PLoS ONE 2013, 8, e57975. [Google Scholar] [CrossRef] [PubMed]
- Sikorskaya, T.V.; Ermolenko, E.V.; Efimova, K.V.; Dang, L.T.P. Coral Holobionts Possess Distinct Lipid Profiles That May Be Shaped by Symbiodiniaceae Taxonomy. Mar. Drugs 2022, 20, 485. [Google Scholar] [CrossRef]
- Imbs, A.B.; Ermolenko, E.V.; Grigorchuk, V.P.; Dang, L.T.P. A Lipidomic Approach to the Study of Biodiversity of Symbiotic Dinoflagellates in Millepora Hydrocorals from Vietnam Coral Reefs. Russ. J. Mar. Biol. 2021, 47, 312–317. [Google Scholar] [CrossRef]
- Imbs, A.B.; Rybin, V.G.; Kharlamenko, V.I.; Dang, L.P.T.; Nguyen, N.T.; Pham, K.M.; Pham, L.Q. Polyunsaturated molecular species of galactolipids: Markers of zooxanthellae in a symbiotic association of the soft coral Capnella sp. (Anthozoa: Alcyonacea). Russ. J. Mar. Biol. 2015, 41, 461–467. [Google Scholar] [CrossRef]
- Roach, T.N.F.; Dilworth, J.; H., C.M.; Jones, A.D.; Quinn, R.A.; Drury, C. Metabolomic signatures of coral bleaching history. Nat. Ecol. Evol. 2021, 5, 495–503. [Google Scholar] [CrossRef]
- Botana, T.M.; Chaves-Filho, A.B.; Inague, A.; Güth, A.; Saldanha-Corrêa, F.; Müller, M.N.; Sumida, P.Y.G.; Miyamoto, S.; Kellermann, M.Y.; Valentine, R.C.; et al. Thermal plasticity of coral reef symbionts is linked to major alterations in their lipidome composition. Limnol. Oceanogr. 2022, 67, 1456–1469. [Google Scholar] [CrossRef]
- Dean, J.M.; Lodhi, I.J. Structural and functional roles of ether lipids. Protein Cell 2018, 9, 196–206. [Google Scholar] [CrossRef]
- Vítová, M.; Palyzová, A.; Řezanka, T. Plasmalogens—Ubiquitous molecules occurring widely, from anaerobic bacteria to humans. Prog. Lipid Res. 2021, 83, 101111. [Google Scholar] [CrossRef]
- Zar, J. Biostatistical Analysis; Prentice Hall: Hoboken, NJ, USA, 1999. [Google Scholar]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous Adaptation and the Regulation of Membrane Lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, Y.; Iwao, Y.; Noguchi, S.; Itai, S. Effect of alkyl chain length and unsaturation of the phospholipid on the physi-cochemical properties of lipid nanoparticles. Chem. Pharm. Bull. 2015, 63, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Imbs, A.B. Lipid class and fatty acid compositions of the zoanthid Palythoa caesia (Anthozoa: Hexacorallia: Zoanthidea) and its chemotaxonomic relations with corals. Biochem. Syst. Ecol. 2014, 54, 213–218. [Google Scholar] [CrossRef]
- Kay, J.G.; Fairn, G.D. Distribution, dynamics and functional roles of phosphatidylserine within the cell. Cell Commun. Signal. 2019, 17, 126. [Google Scholar] [CrossRef]
- Vysotskii, M.V.; Svetashev, V.I. Identification, isolation and characterization of tetracosapolyenoic acids in lipids of marine coelenterates. Biochim. Biophys. Acta 1991, 1083, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Imbs, A.B.; Grigorchuk, V.P. Lipidomic study of the influence of dietary fatty acids on structural lipids of cold-water nudi-branch molluscs. Sci. Rep. 2019, 9, 20013. [Google Scholar] [CrossRef]
- Mukhamedova, K.S.; Glushenkova, A.I. Natural phosphonolipids. Chem. Nat. Compd. 2000, 36, 329–341. [Google Scholar] [CrossRef]
- Chintalapati, M.; Truax, R.; Stout, R.; Portier, R.; Losso, J.N. In Vitro and in Vivo Anti-angiogenic Activities and Inhibition of Hormone-Dependent and -Independent Breast Cancer Cells by Ceramide Methylaminoethylphosphonate. J. Agric. Food Chem. 2009, 57, 5201–5210. [Google Scholar] [CrossRef]
- Tomonaga, N.; Tsuduki, T.; Manabe, Y.; Sugawara, T. Sphingoid bases of dietary ceramide 2-aminoethylphosphonate, a marine sphingolipid, absorb into lymph in rats. J. Lipid Res. 2019, 60, 333–340. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Thelen, J.J.; Fedosejevs, E.; Harwood, J.L. The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 2019, 74, 31–68. [Google Scholar] [CrossRef]
- Imbs, A.B.; Yakovleva, I.M.; Latyshev, N.A.; Pham, L.Q. Biosynthesis of polyunsaturated fatty acids in zooxanthellae and polyps of corals. Russ. J. Mar. Biol. 2010, 36, 452–457. [Google Scholar] [CrossRef]
- Chen, H.-K.; Song, S.-N.; Wang, L.-H.; Mayfield, A.; Chen, Y.-J.; Chen, W.-N.U.; Chen, C.-S. A Compartmental Comparison of Major Lipid Species in a Coral-Symbiodinium Endosymbiosis: Evidence that the Coral Host Regulates Lipogenesis of Its Cytosolic Lipid Bodies. PLoS ONE 2015, 10, e0132519. [Google Scholar] [CrossRef]
- Douglas, S.; Zauner, S.; Fraunholz, M.; Beaton, M.; Penny, S.; Deng, L.-T.; Wu, X.; Reith, M.; Cavalier-Smith, T.; Maier, U.-G. The highly reduced genome of an enslaved algal nucleus. Nature 2001, 410, 1091–1096. [Google Scholar] [CrossRef]
- Shimojima, M. Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 2011, 50, 234–239. [Google Scholar] [CrossRef]
- Boudière, L.; Michaud, M.; Petroutsos, D.; Rébeillé, F.; Falconet, D.; Bastien, O.; Roy, S.; Finazzi, G.; Rolland, N.; Jouhet, J.; et al. Glycerolipids in photosynthesis: Composition, synthesis and trafficking. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Dörmann, P.; Benning, C. Galactolipids rule in seed plants. Trends Plant Sci. 2002, 7, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Mansour, J.S.; Pollock, F.J.; Díaz-Almeyda, E.; Iglesias-Prieto, R.; Medina, M. Intra- and interspecific variation and phenotypic plasticity in thylakoid membrane properties across two Symbiodinium clades. Coral Reefs 2018, 37, 841–850. [Google Scholar] [CrossRef]
- Dalsgaard, J.; John, M.S.; Kattner, G.; Muller-Navarra, D.; Hagen, W. Fatty acid trophic markers in the pelagic marine environment. Adv. Mar. Biol. 2003, 46, 225–340. [Google Scholar] [PubMed]
- Kaneda, T. Iso-fatty and anteiso-fatty acids in bacteria—Biosynthesis, function, and taxonomic significance. Microbiol. Rev. 1991, 55, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Řezanka, T.; Sigler, K. Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog. Lipid Res. 2009, 48, 206–238. [Google Scholar] [CrossRef] [PubMed]
- Haubert, D.; Häggblom, M.M.; Langel, R.; Scheu, S.; Ruess, L. Trophic shift of stable isotopes and fatty acids in Collembola on bacterial diets. Soil Biol. Biochem. 2006, 38, 2004–2007. [Google Scholar] [CrossRef]
- Pollierer, M.M.; Scheu, S.; Haubert, D. Taking it to the next level: Trophic transfer of marker fatty acids from basal resource to predators. Soil Biol. Biochem. 2010, 42, 919–925. [Google Scholar] [CrossRef]
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef]
- Ainsworth, T.D.; Krause, L.; Bridge, T.; Torda, G.; Raina, J.-B.; Zakrzewski, M.; Gates, R.D.; Padilla-Gamiño, J.L.; Spalding, H.L.; Smith, C.; et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015, 9, 2261–2274. [Google Scholar] [CrossRef]
- Hester, E.R.; Barott, K.L.; Nulton, J.; Vermeij, M.J.; Rohwer, F.L. Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J. 2016, 10, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Baptista, M.; Lopes, V.M.; Pimentel, M.S.; Bandarra, N.; Narciso, L.; Marques, A.; Rosa, R. Temporal fatty acid dynamics of the octocoral Veretillum cynomorium. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2012, 161, 178–187. [Google Scholar] [CrossRef]
- Imbs, A.B.; Latyshev, N.A.; Zhukova, N.V.; Dautova, T.N. Comparison of fatty acid compositions of azooxanthellate Den-dronephthya and zooxanthellate soft coral species. Comp. Biochem. Physiol. 2007, 148, 314–321. [Google Scholar] [CrossRef]
- Beleneva, I.A.; Dautova, T.I.; Zhukova, N.V. Characterization of communities of heterotrophic bacteria associated with healthy and diseased corals in Nha Trang Bay (Vietnam). Microbiology 2005, 74, 579–587. [Google Scholar] [CrossRef]
- Řezanka, T.; Siristova, L.; Matoulková, D.; Sigler, K. Hydrophilic interaction liquid chromatography: ESI–MS/MS of plasmalogen phospholipids from Pectinatus bacterium. Lipids 2011, 46, 765–780. [Google Scholar] [CrossRef]
- Zuluaga-Montero, A.; Toledo-Hernandez, C.; Rodriguez, J.A.; Sabat, A.M.; Bayman, P. Spatial variation in fungal communities isolated from healthy and diseased sea fans Gorgonia ventalina and seawater. Aquat. Biol. 2010, 8, 151–160. [Google Scholar] [CrossRef]
- Hou, X.M.; Wang, C.Y.; Gu, Y.C.; Shao, C.L. Penimethavone A, a flavone from a gorgonian-derived fungus Penicillium chrysogenum. Nat. Prod. Res. 2016, 30, 2274–2277. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qiu, P.; Liu, H.; Li, J.; Shao, C.; Yan, T.; Cao, W.; She, Z. Identification of anti-inflammatory polyketides from the coral-derived fungus Penicillium sclerotiorin: In vitro approaches and molecular-modeling. Bioorg. Chem. 2019, 88, 102973. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wei, X.; Hu, J.; Wang, S.; Liu, B.; Xie, Z.; Rong, L.; Li, X.; Zhang, C. Researches on the Subergane-Type Sesquiterpenes from the Soft Coral-Derived Fungus Aspergillus sp. EGF15-0-3. Chin. J. Org. Chem. 2020, 40, 1275–1280. [Google Scholar] [CrossRef]
- Khudyakova, Y.V.; Sobolevskaya, M.P.; Smetanina, O.F.; Slinkina, N.N.; Pivkin, M.V.; Moiseenko, O.P.; Kuznetsova, T.A. Fatty-acid composition of certain species of marine mycelial fungi. Chem. Nat. Compd. 2009, 45, 18–20. [Google Scholar] [CrossRef]
- Ostachowska, A.; Stepnowski, P.; Gołębiowski, M. Dicarboxylic acids and hydroxy fatty acids in different species of fungi. Chem. Pap. 2017, 71, 999–1005. [Google Scholar] [CrossRef]
- Liang, Z.-Y.; Shen, N.-X.; Zheng, Y.-Y.; Wu, J.-T.; Miao, L.; Fu, X.-M.; Chen, M.; Wang, C.-Y. Two new unsaturated fatty acids from the mangrove rhizosphere soil-derived fungus Penicillium javanicum HK1-22. Bioorganic Chem. 2019, 93, 103331. [Google Scholar] [CrossRef]
- Wołczańska, A.; Christie, W.W.; Fuchs, B.; Galuska, C.E.; Kowalczyk, B.; Palusińska-Szysz, M. Fatty acid composition and lipid profiles as chemotaxonomic markers of phytopathogenic fungi Puccinia malvacearum and P. glechomatis. Fungal Biol. 2021, 125, 869–878. [Google Scholar] [CrossRef]
- Kotlova, E.R.; Senik, S.V.; Manzhieva, B.S.; Kiyashko, A.A.; Shakhova, N.V.; Puzansky, R.K.; Volobuev, S.V.; Misharev, A.D.; Serebryakov, E.B.; Psurtseva, N.V. Diversity of ESI-MS Based Phosphatidylcholine Profiles in Basidiomycetes. J. Fungi 2022, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Senik, S.V.; Manzhieva, B.S.; Maloshenok, L.G.; Serebryakov, E.B.; Bruskin, S.A.; Kotlova, E.R. Heterogeneous Distribution of Phospholipid Molecular Species in the Surface Culture of Flammulina velutipes: New Facts about Lipids Containing α-Linolenic Fatty Acid. J. Fungi 2023, 9, 102. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef] [PubMed]


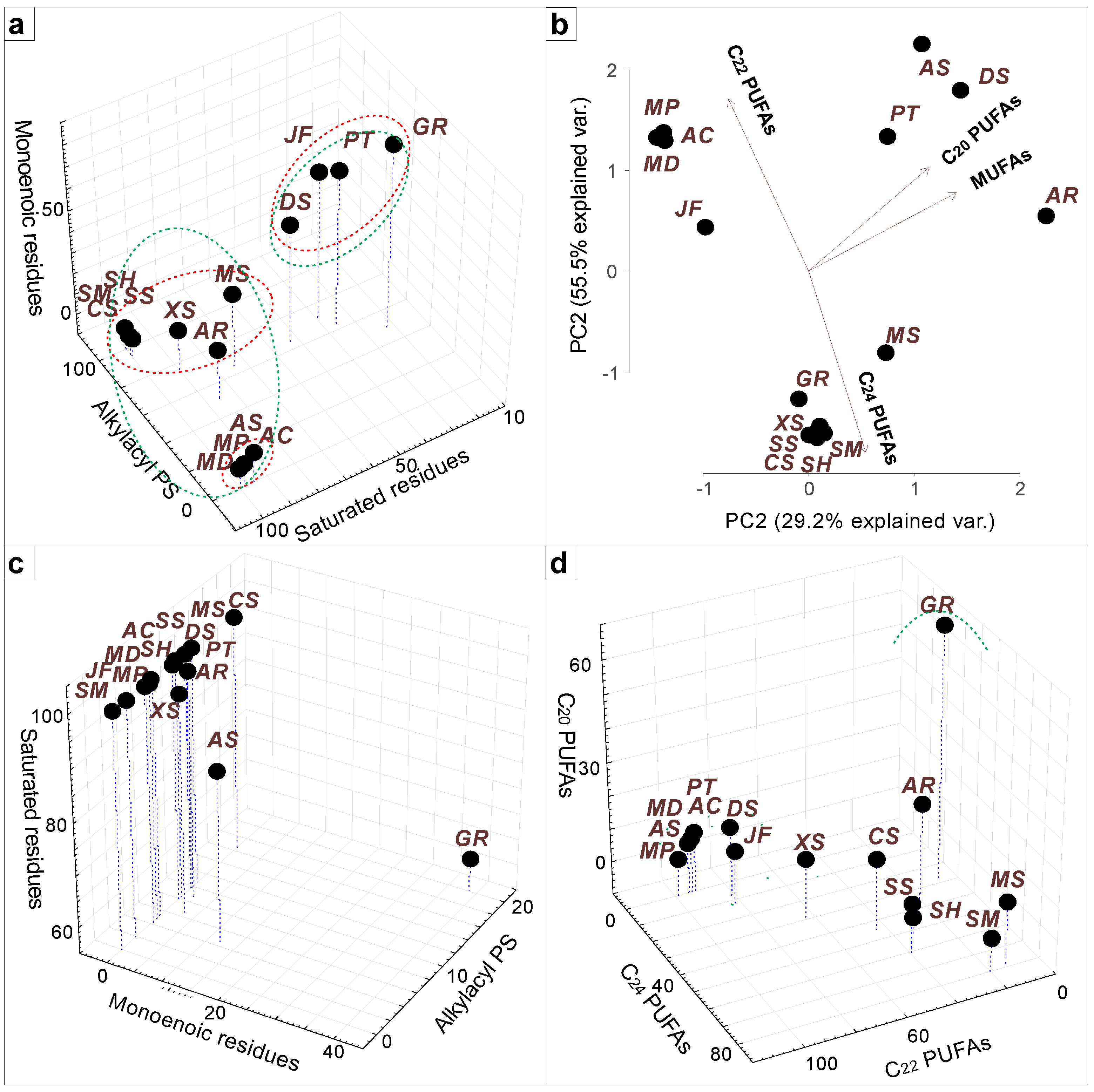
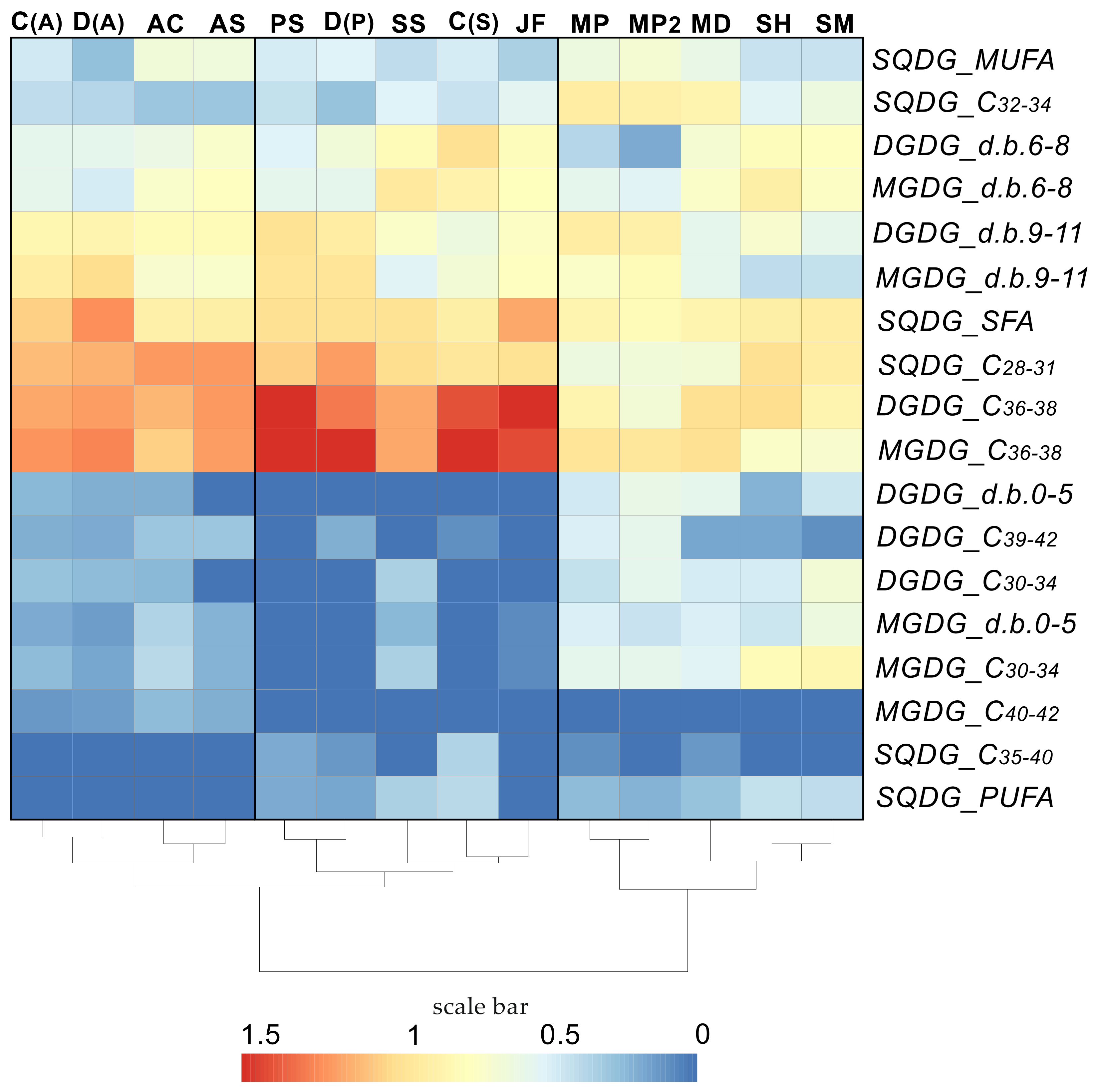
| Class and Subclass of Cnidarians | Coral or Symbionts | Species Name | Membrane Lipid Class of Identified Molecular Species | Reference |
| Anthozoa, Hexacorallia | Reef-building coral | Acropora cerealis | PC, PE, PS, PI, CAEP, GL, BL | [37] |
| Anthozoa, Hexacorallia | Reef-building coral | Acropora sp. | PG, GL, BL | [52] |
| Anthozoa, Hexacorallia | Reef-building coral | Seriatopora caliendrum | PC | [42,43] |
| Anthozoa, Hexacorallia | Zoantharian | Palythoa tuberculosa | PC, PE, PS, PI, CAEP, GL, BL | [38] |
| Anthozoa, Hexacorallia | Sea anemone | Aiptasia pallida | PC, PE, PS, PI, CAEP, SQDG | [50,51] |
| Anthozoa, Octocorallia | Soft coral | Capnella sp. | PC, PE, PS, PI | [45] |
| Anthozoa, Octocorallia | Soft coral | Xenia sp. | PC, PE, PS, PI, CAEP | [46] |
| Anthozoa, Octocorallia | Soft coral | Sinularia heterospiculata | PC, PE, PS, PI, CAEP, GL | [39,40] |
| Anthozoa, Octocorallia | Soft coral | S. siaesensis | PC, PE, PS, PI, CAEP, GL | [41] |
| Anthozoa, Octocorallia | Soft coral | S. macropodia | PC, PE, PS, PI | [45] |
| Anthozoa, Octocorallia | Gorgonian coral | Junceella fragilis | PC, PE, PS, PI, CAEP, GL | [47] |
| Anthozoa, Octocorallia | Gorgonian coral (asymbiotic) | Dichotella sp. | PC, PE, PS, PI, CAEP | [47] |
| Anthozoa, Octocorallia | Gorgonian coral (asymbiotic) | Menella sp. | PC, PE, PS, PI, CAEP | [47] |
| Anthozoa, Octocorallia | Gorgonian coral (asymbiotic) | Astrogorgia rubra | PC, PE, PS, PI, CAEP | [47] |
| Anthozoa, Octocorallia | Soft coral (asymbiotic, cold-water) | Gersemia rubiformis | PC, PE, PS, PI | [44] |
| Hydrozoa | Hydrocoral | Millepora dichotoma | PC, PE, PS, PI, CAEP, GL, BL | [48,49,53] |
| Hydrozoa | Hydrocoral | M. platyphylla | PC, PE, PS, PI, CAEP, GL, BL | [48,49,53] |
| Hydrozoa | Hydrocoral (asymbiotic, cold-water) | Allopora stejnegeri | PC, PE, PS, PI, CAEP | [48] |
| Dinoflagellates of the family Symbiodiniaceae | Symbionts from the jellyfish Cassiopea xamachana | Symbiodinium microadriaticum | GL, BL | [29] |
| Dinoflagellates of the family Symbiodiniaceae | Symbionts from the soft coral S. heterospiculata | Cladocopium C1/C3 | GL, BL | [32] |
| Dinoflagellates of the family Symbiodiniaceae | Symbionts from the soft coral S. heterospiculata | Durusdinium trenchii (D1) | GL, BL | [32] |
| Dinoflagellates of the family Symbiodiniaceae | Symbionts from the reef-building coral A. valida | Cladocopium C1 | GL, BL, PC | [31] |
| Dinoflagellates of the family Symbiodiniaceae | Symbionts from the reef-building coral A. valida | D. trenchii (D1) | GL, BL, PC | [31] |
| Dinoflagellates of the family Symbiodiniaceae | Symbionts from the soft coral Capnella sp. | – | MGDG, DGDG | [54] |
| Dinoflagellates of the family Symbiodiniaceae | Symbionts from the reef-building coral Montipora capitata | Cladocopium sp., D. trenchii (D1) | DGCC | [55] |
| Dinoflagellates of the family Symbiodiniaceae | Cultivated coral symbionts | S. microadriaticum (A1), Cladocopium C1, Breviolum minutum (B1) | GL, BL, PL | [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikorskaya, T.V. Coral Lipidome: Molecular Species of Phospholipids, Glycolipids, Betaine Lipids, and Sphingophosphonolipids. Mar. Drugs 2023, 21, 335. https://doi.org/10.3390/md21060335
Sikorskaya TV. Coral Lipidome: Molecular Species of Phospholipids, Glycolipids, Betaine Lipids, and Sphingophosphonolipids. Marine Drugs. 2023; 21(6):335. https://doi.org/10.3390/md21060335
Chicago/Turabian StyleSikorskaya, Tatyana V. 2023. "Coral Lipidome: Molecular Species of Phospholipids, Glycolipids, Betaine Lipids, and Sphingophosphonolipids" Marine Drugs 21, no. 6: 335. https://doi.org/10.3390/md21060335
APA StyleSikorskaya, T. V. (2023). Coral Lipidome: Molecular Species of Phospholipids, Glycolipids, Betaine Lipids, and Sphingophosphonolipids. Marine Drugs, 21(6), 335. https://doi.org/10.3390/md21060335





