Gemmata algarum, a Novel Planctomycete Isolated from an Algal Mat, Displays Antimicrobial Activity
Abstract
:1. Introduction
2. Results and Discussion
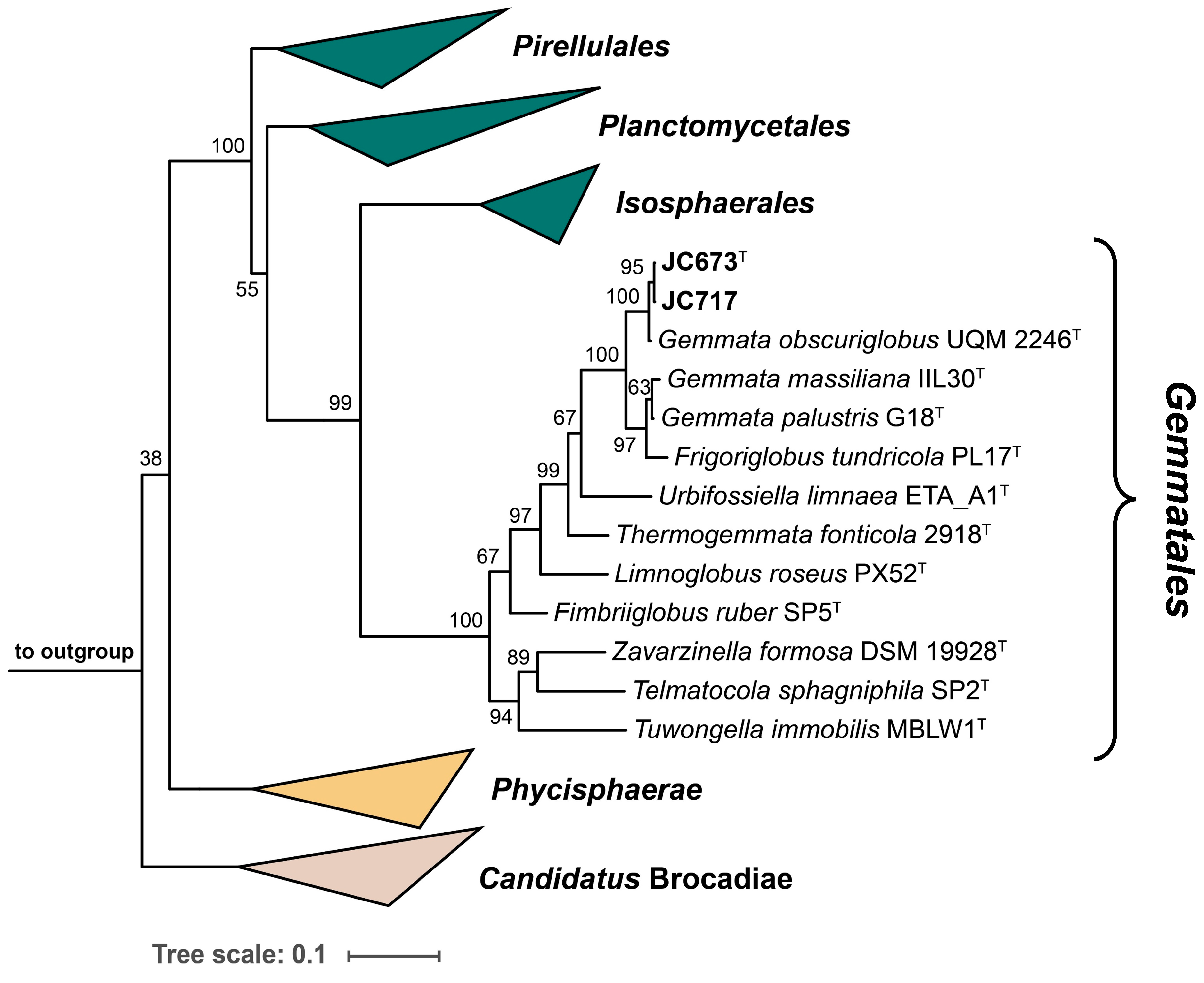
2.1. Phylogenetic Inference
2.2. Genomic Characteristics
2.3. Genome-Based Analysis of Primary and Secondary Metabolic Features
2.4. Morphological and Physiological Analyses
2.5. Chemotaxonomic Characterisation
2.6. Ethyl Acetate Extracts of the Novel Isolates Show Antimicrobial Activities
2.7. Description of Gemmata algarum sp. nov.
3. Conclusions and Outlook
4. Materials and Methods
4.1. Habitat and Isolation
4.2. DNA Isolation, 16S rRNA Gene Sequencing and BLAST Analysis
4.3. Genome Sequencing and In-Silico Analysis of Genome-Encoded Features
4.4. Phylogenetic Analysis
4.5. Physiological Analyses
4.6. Chemotaxonomic Characterization
4.7. Microscopy and Image Analysis
4.8. Metabolite Extraction and Antimicrobial Screening
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Van Teeseling, M.C.F.; Jogler, C. Cultivation of elusive microbes unearthed exciting biology. Nat. Commun. 2021, 12, 75. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.-Y.; Liu, L.; Hua, Z.-S.; Fang, B.-Z.; Zhou, E.-M.; Salam, N.; Hedlund, B.P.; Li, W.-J. Microbial dark matter coming to light: Challenges and opportunities. Natl. Sci. Rev. 2020, 8, nwaa280. [Google Scholar] [CrossRef]
- Boedeker, C.; Schüler, M.; Reintjes, G.; Jeske, O.; van Teeseling, M.C.F.; Jogler, M.; Rast, P.; Borchert, D.; Devos, D.P.; Kucklick, M.; et al. Determining the bacterial cell biology of Planctomycetes. Nat. Commun. 2017, 8, 14853. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Marin, E.; Peeters, S.H.; Claret Fernández, L.; Jogler, C.; van Niftrik, L.; Wiegand, S.; Devos, D.P. Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila. Sci. Rep. 2020, 10, 66. [Google Scholar] [CrossRef]
- Wiegand, S.; Jogler, M.; Boedeker, C.; Pinto, D.; Vollmers, J.; Rivas-Marín, E.; Kohn, T.; Peeters, S.H.; Heuer, A.; Rast, P.; et al. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat. Microbiol. 2020, 5, 126–140. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Jogler, C. The bacterial phylum Planctomycetes as novel source for bioactive small molecules. Biotechnol. Adv. 2021, 53, 107818. [Google Scholar] [CrossRef]
- Belova, S.E.; Saltykova, V.A.; Dedysh, S.N. Antimicrobial Activity of a Novel Freshwater Planctomycete Lacipirellula parvula PX69T. Microbiology 2020, 89, 503–509. [Google Scholar] [CrossRef]
- Vitorino, I.R.; Lage, O.M. The Planctomycetia: An overview of the currently largest class within the phylum Planctomycetes. Antonie Van Leeuwenhoek 2022, 115, 169–201. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.A.; Kulichevskaya, I.S.; Dedysh, S.N. Gemmata palustris sp. nov., a Novel Planctomycete from a Fen in Northwestern Russia. Microbiology 2021, 90, 598–606. [Google Scholar] [CrossRef]
- Kulichevskaya, I.; Serkebaeva, Y.; Kim, Y.; Rijpstra, I.; Sinninghe Damste, J.; Liesack, W.; Dedysh, S. Telmatocola sphagniphila gen. nov., sp. nov., a Novel Dendriform Planctomycete from Northern Wetlands. Front. Microbiol. 2012, 3, 146. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Baulina, O.I.; Bodelier, P.L.E.; Rijpstra, W.I.C.; Damsté, J.S.S.; Dedysh, S.N. Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a Siberian peat bog. Int. J. Syst. Evol. Microbiol. 2009, 59, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaya, I.S.; Ivanova, A.A.; Baulina, O.I.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Dedysh, S.N. Fimbriiglobus ruber gen. nov., sp. nov., a Gemmata-like planctomycete from Sphagnum peat bog and the proposal of Gemmataceae fam. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaya, I.S.; Ivanova, A.A.; Naumoff, D.G.; Beletsky, A.V.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Mardanov, A.V.; Ravin, N.V.; Dedysh, S.N. Frigoriglobus tundricola gen. nov., sp. nov., a psychrotolerant cellulolytic planctomycete of the family Gemmataceae from a littoral tundra wetland. Syst. Appl. Microbiol. 2020, 43, 126129. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaya, I.S.; Naumoff, D.G.; Miroshnikov, K.K.; Ivanova, A.A.; Philippov, D.A.; Hakobyan, A.; Rijpstra, W.I.C.; Damsté, J.S.S.; Liesack, W.; Dedysh, S.N. Limnoglobus roseus gen. nov., sp. nov., a novel freshwater planctomycete with a giant genome from the family Gemmataceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Rast, P.; Jogler, M.; Wiegand, S.; Kohn, T.; Boedeker, C.; Jeske, O.; Heuer, A.; Quast, C.; Glöckner, F.O.; et al. Analysis of bacterial communities in a municipal duck pond during a phytoplankton bloom and isolation of Anatilimnocola aggregata gen. nov., sp. nov., Lacipirellula limnantheis sp. nov. and Urbifossiella limnaea gen. nov., sp. nov. belonging to the phylum Planctomycetes. Environ. Microbiol. 2021, 23, 1379–1396. [Google Scholar] [CrossRef] [PubMed]
- Franzmann, P.D.; Skerman, V.B.D. Gemmata obscuriglobus, a new genus and species of the budding bacteria. Antonie Van Leeuwenhoek 1984, 50, 261–268. [Google Scholar] [CrossRef]
- Seeger, C.; Butler, M.K.; Yee, B.; Mahajan, M.; Fuerst, J.A.; Andersson, S.G.E. Tuwongella immobilis gen. nov., sp. nov., a novel non-motile bacterium within the phylum Planctomycetes. Int. J. Syst. Evol. Microbiol. 2017, 67, 4923–4929. [Google Scholar] [CrossRef]
- Elcheninov, A.G.; Podosokorskaya, O.A.; Kovaleva, O.L.; Novikov, A.A.; Toshchakov, S.V.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Thermogemmata fonticola gen. nov., sp. nov., the first thermophilic planctomycete of the order Gemmatales from a Kamchatka hot spring. Syst. Appl. Microbiol. 2021, 44, 126157. [Google Scholar] [CrossRef] [PubMed]
- Aghnatios, R.; Cayrou, C.; Garibal, M.; Robert, C.; Azza, S.; Raoult, D.; Drancourt, M. Draft genome of Gemmata massiliana sp. nov, a water-borne Planctomycetes species exhibiting two variants. Stand. Genom. Sci. 2015, 10, 120. [Google Scholar] [CrossRef]
- Lhingjakim, K.L.; Smita, N.; Kumar, G.; Jagadeeshwari, U.; Ahamad, S.; Sasikala, C.; Ramana, C.V. Paludisphaera rhizosphaereae sp. nov., a new member of the family Isosphaeraceae, isolated from the rhizosphere soil of Erianthus ravennae. Antonie Van Leeuwenhoek 2022, 115, 1073–1084. [Google Scholar] [CrossRef]
- Kumar, D.; Gaurav, K.; Pk, S.; A, S.; Uppada, J.; Ch, S.; Ch, V.R. Gimesia chilikensis sp. nov., a haloalkali-tolerant planctomycete isolated from Chilika lagoon and emended description of the genus Gimesia. Int. J. Syst. Evol. Microbiol. 2020, 70, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Jagadeeshwari, U.; Sreya, P.; Shabbir, A.; Sasikala, C.; Ramana, C.V. A genomic overview including polyphasic taxonomy of Thalassoroseus pseudoceratinae gen. nov., sp. nov. isolated from a marine sponge, Pseudoceratina sp. Antonie Van Leeuwenhoek 2022, 115, 843–856. [Google Scholar] [CrossRef]
- Kumar, G.; Kumar, D.; Jagadeeshwari, U.; Sreya, P.K.; Shabbir, A.; Sasikala, C.; Ramana, C.V. Crateriforma spongiae sp. nov., isolated from a marine sponge and emended description of the genus “Crateriforma”. Antonie Van Leeuwenhoek 2021, 114, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Lhingjakim, K.L.; Uppada, J.; Ahamad, S.; Kumar, D.; Kashif, G.M.; Sasikala, C.; Ramana, C.V. Aquisphaera insulae sp. nov., a new member in the family Isosphaeraceae, isolated from the floating island of Loktak lake and emended description of the genus Aquisphaera. Antonie Van Leeuwenhoek 2021, 114, 1465–1477. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Klenk, H.-P.; Göker, M. Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.-L.; Xie, B.-B.; Zhang, X.-Y.; Chen, X.-L.; Zhou, B.-C.; Zhou, J.; Oren, A.; Zhang, Y.-Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Rodriguez-R, L.M.; Konstantinidis, K.T. MyTaxa: An advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014, 42, e73. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Stingl, U.; Meuser, K.; Brune, A. Novel lineages of Planctomycetes densely colonize the alkaline gut of soil-feeding termites (Cubitermes spp.). Environ. Microbiol. 2008, 10, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Liesack, W.; Goebel, B.M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993, 7, 232–236. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2020, 49, D1020–D1028. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2, 2.3.1–2.3.22. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Alanjary, M.; Steinke, K.; Ziemert, N. AutoMLST: An automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019, 47, W276–W282. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Tarlachkov, S.; Starodumova, I. TaxonDC: Calculating the similarity value of the 16S rRNA gene sequences of prokaryotes or its regions of fungi. J. Bioinform. Genom. 2017, 3, 1–4. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 2016, 2167–9843. [Google Scholar] [CrossRef]
- Bondoso, J.; Albuquerque, L.; Nobre, M.F.; Lobo-da-Cunha, A.; da Costa, M.S.; Lage, O.M. Aquisphaera giovannonii gen. nov., sp. nov., a planctomycete isolated from a freshwater aquarium. Int. J. Syst. Evol. Microbiol. 2011, 61, 2844–2850. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, R.; Sharma, M.; Gaurav, K.; Jagadeeshwari, U.; Shabbir, A.; Sasikala, C.; Ramana, C.V.; Pandit, M.K. Paludisphaera soli sp. nov., a new member of the family Isosphaeraceae isolated from high altitude soil in the Western Himalaya. Antonie Van Leeuwenhoek 2020, 113, 1663–1674. [Google Scholar] [CrossRef]
- Bondoso, J.; Albuquerque, L.; Nobre, M.F.; Lobo-da-Cunha, A.; da Costa, M.S.; Lage, O.M. Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst. Appl. Microbiol. 2015, 38, 8–15. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; Technical Note 101; Microbial IDentification Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Kates, M. Isolation, analysis and identification of lipids. North-Holland Publishing Comp.: Amsterdam, The Netherlands, 1972; pp. 268–618. [Google Scholar]
- Oren, A.; Duker, S.; Ritter, S. The polar lipid composition of Walsby’s square bacterium. FEMS Microbiol. Lett. 1996, 138, 135–140. [Google Scholar] [CrossRef]
- Imhoff, J.F. Quinones of phototrophic purple bacteria. FEMS Microbiol. Lett. 1984, 25, 85–89. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; van Teeseling, M.C.F.; Thanbichler, M.; Drescher, K. BacStalk: A comprehensive and interactive image analysis software tool for bacterial cell biology. Mol. Microbiol. 2020, 114, 140–150. [Google Scholar] [CrossRef]
- Goedhart, J. SuperPlotsOfData—A web app for the transparent display and quantitative comparison of continuous data from different conditions. Mol. Biol. Cell 2021, 32, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Mujahid, M.; Sasikala, C.; Ramana, C.V. Production of indole-3-acetic acid and related indole derivatives from L-tryptophan by Rubrivivax benzoatilyticus JA2. Appl. Microbiol. Biotechnol. 2011, 89, 1001–1008. [Google Scholar] [CrossRef] [PubMed]






| Characteristics | JC673T | G. obscuriglobus UQM 2246T | G. massiliana IIL30T (#) | G. palustris G18T (#) |
|---|---|---|---|---|
| Cell size (µm) | 1.4–2.2 | 1.4–3.0 | 1.1–2.1 | 1.5–2.8 |
| pH range (optimum) | 6.0–9.0 (8.0) | 7.0–9.0 (7.0) | 6.0–8.0 (8.0) | 6.0–8.0 (7.0) |
| NaCl range (% (w/v)) | 0–4 (2) | 0–1 (0) | 0–0.6 (ND) | 0–1.25 (ND) |
| Temperature range (optimum) | 10–30 (23–25) | 16–35 (26–28) | 25–37 (30) | 4–28 (15–20) |
| Nitrogen source utilization | ||||
| l-Isoleucine | + | − | ND | ND |
| l-Methionine | + | − | ND | ND |
| l-Phenylalanine | + | − | ND | ND |
| l-Proline | + | − | ND | ND |
| dl-Threonine | − | + | ND | ND |
| l-Histidine | − | + | ND | ND |
| Carbon source utilization | ||||
| Ascorbate | + | − | ND | ND |
| Starch | + | − | ND | − |
| Fructose | + | − | ND | ND |
| Galactose | + | − | ND | ND |
| Maltose | − | + | ND | ND |
| Fumarate | − | + | ND | ND |
| Nitrate Reduction | + | − | ND | − |
| Antibiotic resistance | ||||
| Ampicillin | R | R | ND | S |
| Streptomycin | R | R | ND | S |
| Activity of enzymes | ||||
| Valine arylamidase | − | + | + | + |
| β-Galactosidase | − | + | − | − |
| α-Chymotrypsin | + | − | − | + |
| α-Glucosidase | - | + | − | − |
| β-Glucosidase | - | + | − | − |
| Cysteine arylamidase | − | − | − | + |
| Trypsin | − | − | + | + |
| Fatty acid composition | ||||
| iso-C19:0 | + | − | ND | ND |
| C18:0 ω6,9c/ante-C18:0 | + | − | ND | ND |
| iso-C17:0 3-OH | − | − | ND | ND |
| Major Polar lipids | ||||
| Phosphatidylcholine | − | − | + | + |
| Phosphatidylethanolamine | − | − | − | + |
| Major Polyamines | ||||
| Putrescine | + | − | ND | ND |
| Genomic features | ||||
| DNA G+C content (mol%) | 67.6 | 67.4 | 64.0 | 65.0 |
| Genome size (Mb) | 8.20 | 9.03 | 10.14 | 9.23 |
| Number of genes | 6505 | 7408 | 8383 | 7539 |
| RNAs | 88 | 106 | 102 | 97 |
| CRISPRs | 6 | 3 | 4 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, G.; Kallscheuer, N.; Kashif, M.; Ahamad, S.; Jagadeeshwari, U.; Pannikurungottu, S.; Haufschild, T.; Kabuu, M.; Sasikala, C.; Jogler, C.; et al. Gemmata algarum, a Novel Planctomycete Isolated from an Algal Mat, Displays Antimicrobial Activity. Mar. Drugs 2024, 22, 10. https://doi.org/10.3390/md22010010
Kumar G, Kallscheuer N, Kashif M, Ahamad S, Jagadeeshwari U, Pannikurungottu S, Haufschild T, Kabuu M, Sasikala C, Jogler C, et al. Gemmata algarum, a Novel Planctomycete Isolated from an Algal Mat, Displays Antimicrobial Activity. Marine Drugs. 2024; 22(1):10. https://doi.org/10.3390/md22010010
Chicago/Turabian StyleKumar, Gaurav, Nicolai Kallscheuer, Mohammad Kashif, Shabbir Ahamad, Uppada Jagadeeshwari, Sreya Pannikurungottu, Tom Haufschild, Moses Kabuu, Chintalapati Sasikala, Christian Jogler, and et al. 2024. "Gemmata algarum, a Novel Planctomycete Isolated from an Algal Mat, Displays Antimicrobial Activity" Marine Drugs 22, no. 1: 10. https://doi.org/10.3390/md22010010






