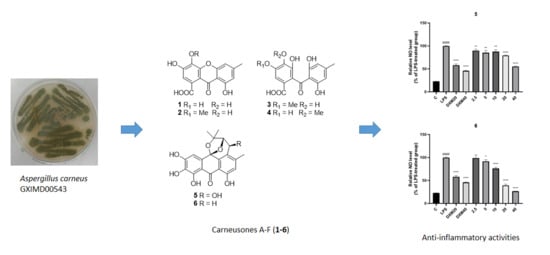
Carneusones A-F, Benzophenone Derivatives from Sponge-Derived Fungus Aspergillus carneus GXIMD00543
Abstract
1. Introduction
2. Results and Discussion
2.1. Strain Isolation and Species Identification
2.2. Elucidation of Chemical Structures
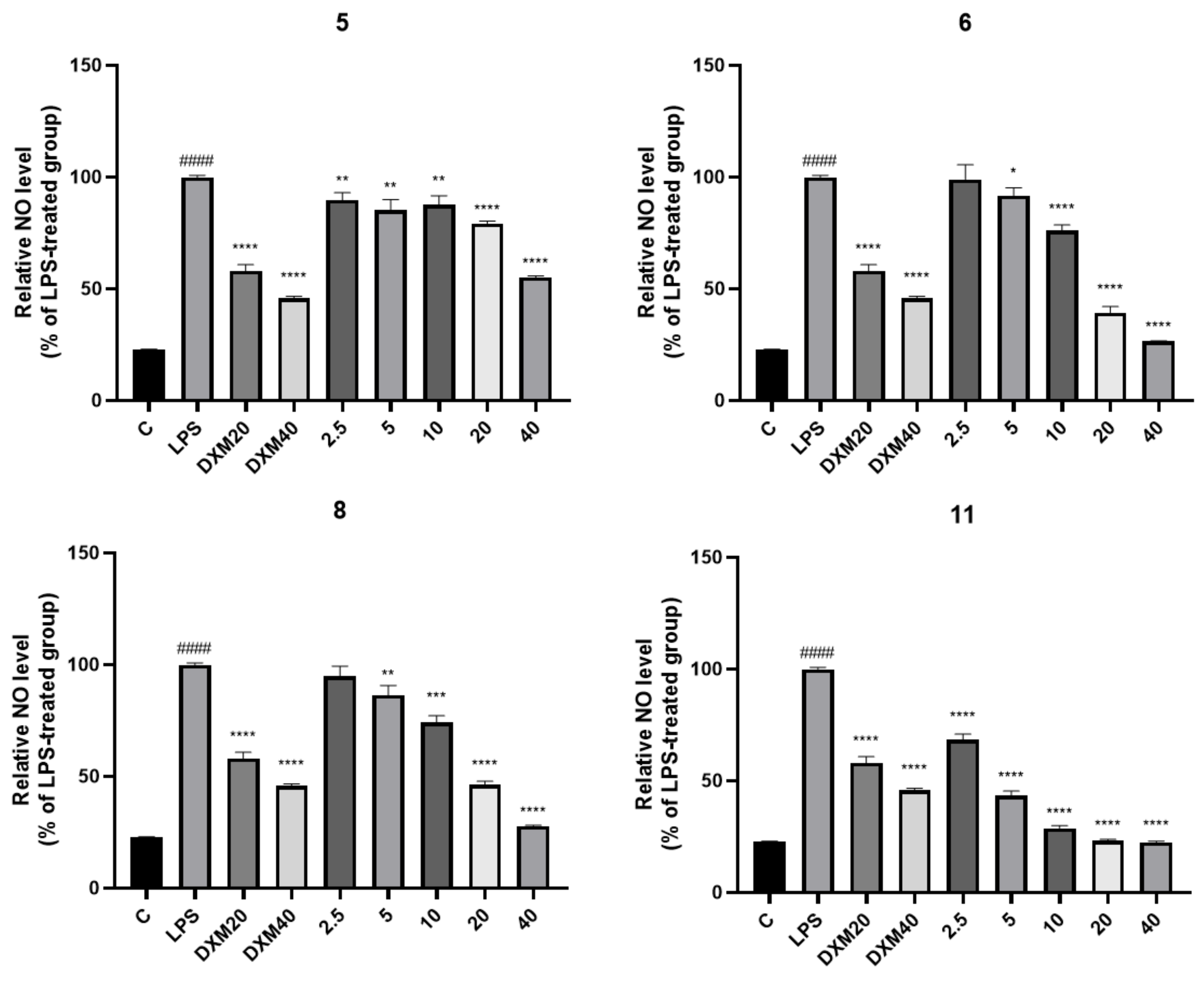
2.3. Anti-Inflammatory Activity Test
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Isolation and Species Identification of Marine Fungus
3.3. Fungal Fermentation
3.4. Extration and Isolation
3.5. Spectroscopic and Spectrometric Data
3.6. Computational Methods
3.7. Anti-Inflammatory Activity Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gonçalves, M.F.M.; Esteves, A.C.; Alves, A. Marine fungi: Opportunities and challenges. Encyclopedia 2022, 2, 559–577. [Google Scholar] [CrossRef]
- Sebak, M.; Molham, F.; Greco, C.; Tammam, M.A.; Sobeh, M.; El-Demerdash, A. Chemical diversity, medicinal potentialities, biosynthesis, and pharmacokinetics of anthraquinones and their congeners derived from marine fungi: A comprehensive update. RSC Adv. 2022, 12, 24887–24921. [Google Scholar] [CrossRef]
- Shabana, S.; Lakshmi, K.R.; Satya, A.K. An updated review of secondary metabolites from marine fungi. Mini-Rev. Med. Chem. 2021, 21, 602–642. [Google Scholar] [CrossRef]
- Jimenez, C. Marine natural products in medicinal chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2023, 40, 275–325. [Google Scholar] [CrossRef]
- Marinov, T.; Kokanova-Nedialkova, Z.; Nedialkov, P.T. Naturally occurring simple oxygenated benzophenones: Structural diversity, distribution, and biological properties. Diversity 2023, 15, 1030. [Google Scholar] [CrossRef]
- Wu, S.B.; Long, C.; Kennelly, E.J. Structural diversity and bioactivities of natural benzophenones. Nat. Prod. Rep. 2014, 31, 1158–1174. [Google Scholar] [CrossRef] [PubMed]
- Surana, K.; Chaudhary, B.; Diwaker, M.; Sharma, S. Benzophenone: A ubiquitous scaffold in medicinal chemistry. MedChemComm 2018, 9, 1803–1817. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Zhu, T.; Du, L.; Zhao, B.; Li, D.; Gu, Q. Sterigmatocystins from the deep-sea-derived fungus Aspergillus versicolor. J. Antibiot. 2011, 64, 193–196. [Google Scholar] [CrossRef]
- Wu, G.; Yu, G.; Kurtán, T.; Mándi, A.; Peng, J.; Mo, X.; Liu, M.; Li, H.; Sun, X.; Li, J.; et al. Versixanthones A–F, cytotoxic xanthone–chromanone dimers from the marine-derived fungus Aspergillus versicolor HDN1009. J. Nat. Prod. 2015, 78, 2691–2698. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.B.; Chen, W.J.; Shan, T.Z.; Sun, B.Y.; Yan, P.C.; Jiang, W. Antibacterial diphenyl ether, benzophenone and xanthone derivatives from Aspergillus flavipes. Chem. Biodivers. 2020, 17, e1900640. [Google Scholar] [CrossRef]
- Jin, Y.; Qin, S.; Gao, H.; Zhu, G.; Wang, W.; Zhu, W.; Wang, Y. An anti-HBV anthraquinone from aciduric fungus Penicillium sp. OUCMDZ-4736 under low pH stress. Extremophiles 2018, 22, 39–45. [Google Scholar] [CrossRef]
- Liu, H.X.; Tan, H.B.; Liu, Y.; Chen, Y.C.; Li, S.N.; Sun, Z.H.; Li, H.H.; Qiu, S.X.; Zhang, W.M. Three new highly-oxygenated metabolites from the endophytic fungus Cytospora rhizophorae A761. Fitoterapia 2017, 117, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Tagawa, M.; Osawa, H.; Seki, T. Agrochemical N-1477 Manufacture with Aspergillus. Japan Patent JP2001261610, 26 September 2001. [Google Scholar]
- Johnson, O.O.; Zhao, M.; Gunn, J.; Santarsiero, B.D.; Yin, Z.Q.; Ayoola, G.A.; Coker, H.A.; Che, C.T. Alpha-Glucosidase inhibitory prenylated anthranols from Harungana madagascariensis. J. Nat. Prod. 2016, 79, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Zanardi, M.M.; Sarotti, A.M. Sensitivity analysis of DP4+ with the probability distribution terms: Development of a universal and customizable method. J. Org. Chem. 2021, 86, 8544–8548. [Google Scholar] [CrossRef] [PubMed]
- Rojas, I.S.; Lotina-Hennsen, B.; Mata, R. Effect of lichen metabolites on thylakoid electron transport and photophosphorylation in isolated spinach chloroplasts. J. Nat. Prod. 2000, 63, 1396–1399. [Google Scholar] [CrossRef] [PubMed]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Intereya, K.; Kocharin, K.; National, C.F.G.E. New diphenyl ethers from the insect pathogenic fungus Cordyceps sp. BCC 1861. Chem. Pharm. Bull. 2007, 55, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, Y.; Liu, D.; Proksch, P.; Yu, S.; Lin, W. Antioxidative phenolic compounds from a marine-derived fungus Aspergillus versicolor. Tetrahedron 2016, 72, 50–57. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Xu, S.X.; Wang, X.W.; Wang, H.Y.; Wang, Z.X.; Gu, X.H.; Liu, S.Z. Two new minor compounds with inhibitory effect on K562 cells from Ventilago leiocarpa Benth. J. Herb. Pharmacother. 2001, 1, 35–41. [Google Scholar] [CrossRef]
- Tanahashi, T.; Takenaka, Y.; Nagakura, N.; Hamada, N. Dibenzofurans from the cultured lichen mycobionts of Lecanora cinereocarnea. Phytochemistry 2001, 58, 1129–1134. [Google Scholar] [CrossRef]
- Gao, L.; Xu, X.; Yang, J. Chemical constituents of the roots of Rheum officinale. Chem. Nat. Compd. 2013, 49, 603–605. [Google Scholar] [CrossRef]
- Bringmann, G.; Lang, G.; Steffens, S.; Gunther, E.; Schaumann, K. Evariquinone, isoemericellin, and stromemycin from a sponge derived strain of the fungus Emericella variecolor. Phytochemistry 2003, 63, 437–443. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumloffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Kwon, Y.J.; Seo, E.B.; Kim, S.K.; Lee, H.; Lee, J.T.; Chang, P.S.; Choi, Y.J.; Lee, S.H.; Ye, S.K. Anti-inflammatory effects of Allium cepa L. peel extracts via inhibition of JAK-STAT pathway in LPS-stimulated RAW264.7 cells. J. Ethnopharmacol. 2023, 317, 116851. [Google Scholar] [CrossRef] [PubMed]



| No. | 1 a | 2 a | 3 a | 4 a | 3-Hydroxy Microxanthone b | Cytosporaphenone A c |
|---|---|---|---|---|---|---|
| 2 | 6.81, s | 6.87, s | 7.00, s | 6.93, s | 6.90, s | 7.18, s |
| 5 | 6.87, s | 6.95, s | 6.07, s | 6.07, s | 6.90, s | 6.18, s |
| 7 | 6.61, s | 6.65, s | 6.07, s | 6.07, s | 6.64, s | 6.18, s |
| 11 | 2.39, s | 2.39, s | 2.15, s | 2.14, s | 2.40, s | 2.19, s |
| 12 | 3.91, s | 3.83, s | 3.74, s | 3.82, s | ||
| OH-8 | 12.57, br s | 12.43, br s |
| 1 a | 2 a | 3 a | 4 a | 3-Hydroxy Microxanthone b | Cytosporaphenone A c | |
|---|---|---|---|---|---|---|
| No. | δC, Type | δC, Type | δC, Type | δC, Type | δC, Type | δC, Type |
| 1 | 126.6, C | 131.4, C | 128.1, C | 126.0, C | 123.8, C | 127.1, C |
| 2 | 111.9, CH | 112.5, CH | 104.1, CH | 108.6, CH | 112.1, CH | 109.8, CH |
| 3 | 145.9, C | 156.9, C | 146.8, C | 149.5, C | 146.0, C | 144.2, C |
| 4 | 133.3, C | 134.7, C | 137.7, C | 138.6, C | 133.8, C | 137.3, C |
| 4a | 152.3, C | 150.6, C | 141.9, C | 146.6, C | 151.3, C | 142.1, C |
| 4b | 155.3, C | 155.1, C | 161.6, C | 161.7, C | 155.3, C | 162.1, C |
| 5 | 107.3, CH | 107.5, CH | 107.5, CH | 107.5, CH | 107.5, CH | 107.9, CH |
| 6 | 148.4, C | 148.6, C | 146.7, C | 146.8, C | 148.7, C | 147.1, C |
| 7 | 110.9, CH | 111.2, CH | 107.5, CH | 107.5, CH | 110.0, CH | 107.9, CH |
| 8 | 160.7, C | 160.6, C | 161.6, C | 161.7, C | 160.5, C | 162.1, C |
| 8a | 105.6, C | 105.6, C | 109.8, C | 109.6, C | 105.6, C | 109.1, C |
| 9 | 179.8, C | 179.4, C | 200.6, C | 200.2, C | 179.7, C | 200.0, C |
| 9a | 109.1, C | 109.2, C | 119.4, C | 123.7, C | 110.0, C | 118.4, C |
| 10 | 170.1, C | 169.5, C | 167.6, C | 167.1, C | 168.8, C | 166.5, C |
| 11 | 22.0, CH3 | 21.9, CH3 | 21.6, CH3 | 21.6, CH3 | 22.0, CH3 | 21.0, CH3 |
| 12 | 60.8, CH3 | 55.9, CH3 | 60.0, CH3 | 52.5, CH3 |
| 5 a | 6 a | N-1447D b | Harunganol F c | |||||
|---|---|---|---|---|---|---|---|---|
| No. | δC, Type | δH | δC, Type | δH | δC, Type | δH | δC, Type | δH |
| 1 | 151.5, C | 151.6, C | 102.2, 106.9, 109.1, 109.7, 119.9, 122.2, 138.9, 140.2, 148.8, 162.1, 165.5, 166.9, 190.0, not assigned | 166.3, C | ||||
| 2 | 133.5, C | 133.5, C | 99.5, CH | 6.36, s | ||||
| 3 | 153.7, C | 153.5, C | 168.0, C | |||||
| 4 | 107.4, CH | 6.91, s | 107.4, CH | 6.92, s | 6.97, s | 119.7, C | ||
| 4a | 130.2, C | 130.5, C | 132.9, C | |||||
| 4b | 139.2, C | 139.3, C | ||||||
| 5 | 123.5, C | 121.0, C | 121.9, C | |||||
| 6 | 149.0, C | 147.1, C | 148.2, C | |||||
| 7 | 118.6, CH | 6.83, s | 117.9, CH | 6.80, s | 6.81, s | 120.0, CH | 6.77, s | |
| 8 | 160.4, C | 159.2, C | 162.0, C | |||||
| 8a | 108.9, C | 109.4, C | 109.8, C | |||||
| 9 | 189.7, C | 189.8, C | 189.6, C | |||||
| 9a | 107.8, C | 108.1, C | 108.9, C | |||||
| 10 | 97.2, C | 97.2, C | 98.1, C | 99.4, C | ||||
| 11 | 61.7, CH | 4.52, s | 27.1, CH2 | 2.78, d, (17.4) | 63.9, CH | 4.58, s | 63.6, CH | 4.48, br d (10.0) |
| 3.04, dd, (17.4, 5.4) | ||||||||
| 12 | 87.1, CH | 4.57, s | 81.1, CH | 4.75, d, (5.4) | 87.9, CH | 4.72, s | 87.6, CH | 4.67, overlapped |
| 13 | 78.4, C | 82.1, C | 79.2, C | 78.3, C | ||||
| 14 | 29.9, CH3 | 1.38, s | 29.6, CH3 | 1.37, s | 30.2, CH3 | 1.50, s | 29.9, CH3 | 1.46, s |
| 15 | 23.1, CH3 | 1.06, s | 23.6, CH3 | 1.15, s | 23.2, CH3 | 1.17, s | 23.4, CH3 | 1.09, s |
| 16 | 18.5, CH3 | 2.41, s | 19.1, CH3 | 2.24, s | 18.9, CH3 | 2.47, s | 18.7, CH3 | 2.40, s |
| 17 | 55.9, CH3 | 3.90, s | 29.8, CH2 | 3.50, dd, (7.2, 16.7) | ||||
| 3.37 dd, (9.5, 16.7) | ||||||||
| 18 | 91.3, CH | 4.71, dd, (7.2, 9.5) | ||||||
| 19 | 71.9, C | |||||||
| 20 | 24.5, CH3 | 1.23, s | ||||||
| 21 | 25.7, CH3 | 1.35, s | ||||||
| OH-1 | 12.05, br s | |||||||
| OH-8 | 11.96, br s | 11.68, br s | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.-J.; Liang, L.-F.; Zhang, G.-S.; Li, H.-Y.; Fu, C.-Q.; Yu, Q.; Zhou, D.-M.; Su, Z.-W.; Liu, K.; Gao, C.-H.; et al. Carneusones A-F, Benzophenone Derivatives from Sponge-Derived Fungus Aspergillus carneus GXIMD00543. Mar. Drugs 2024, 22, 63. https://doi.org/10.3390/md22020063
Lu C-J, Liang L-F, Zhang G-S, Li H-Y, Fu C-Q, Yu Q, Zhou D-M, Su Z-W, Liu K, Gao C-H, et al. Carneusones A-F, Benzophenone Derivatives from Sponge-Derived Fungus Aspergillus carneus GXIMD00543. Marine Drugs. 2024; 22(2):63. https://doi.org/10.3390/md22020063
Chicago/Turabian StyleLu, Chun-Ju, Li-Fen Liang, Geng-Si Zhang, Hai-Yan Li, Chun-Qing Fu, Qin Yu, Dong-Mei Zhou, Zhi-Wei Su, Kai Liu, Cheng-Hai Gao, and et al. 2024. "Carneusones A-F, Benzophenone Derivatives from Sponge-Derived Fungus Aspergillus carneus GXIMD00543" Marine Drugs 22, no. 2: 63. https://doi.org/10.3390/md22020063
APA StyleLu, C.-J., Liang, L.-F., Zhang, G.-S., Li, H.-Y., Fu, C.-Q., Yu, Q., Zhou, D.-M., Su, Z.-W., Liu, K., Gao, C.-H., Xu, X.-Y., & Liu, Y.-H. (2024). Carneusones A-F, Benzophenone Derivatives from Sponge-Derived Fungus Aspergillus carneus GXIMD00543. Marine Drugs, 22(2), 63. https://doi.org/10.3390/md22020063