Exploring the Diversity and Specificity of Secondary Biosynthetic Potential in Rhodococcus
Abstract
:1. Introduction
2. Results
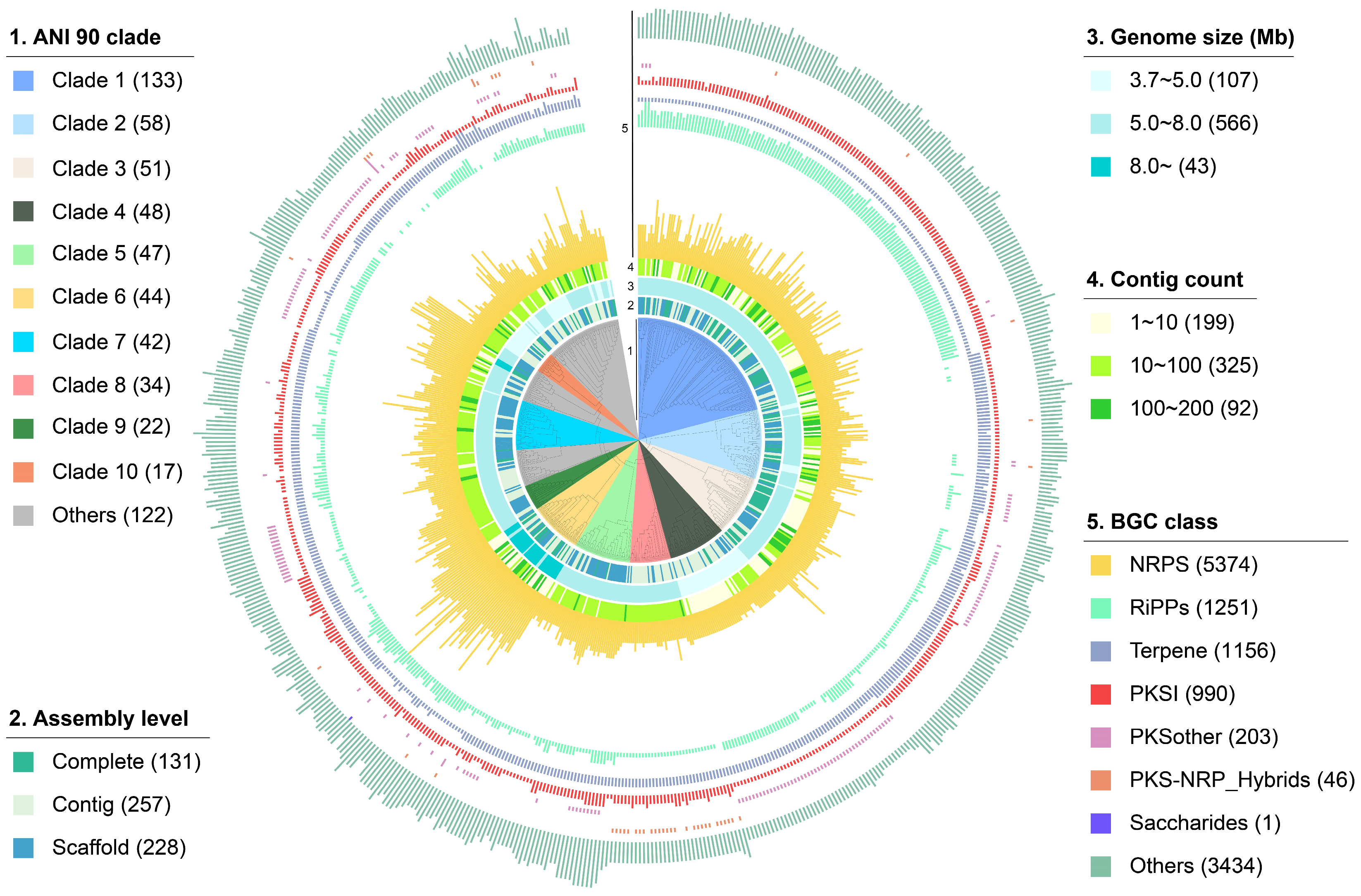
2.1. The Overall Distribution of BGCs in Rhodococcus
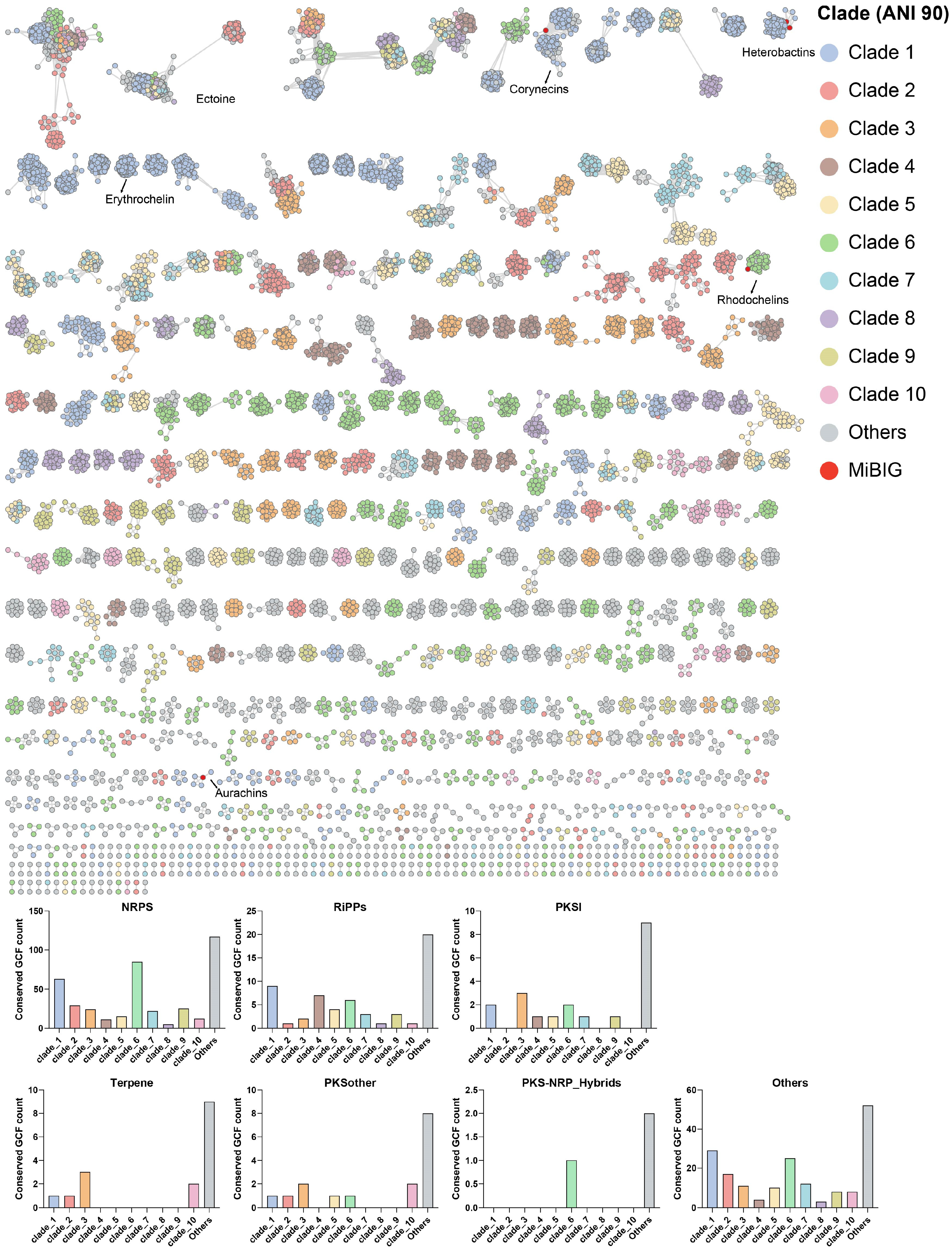
2.2. The Distribution Pattern of GCFs in Rhodococcus
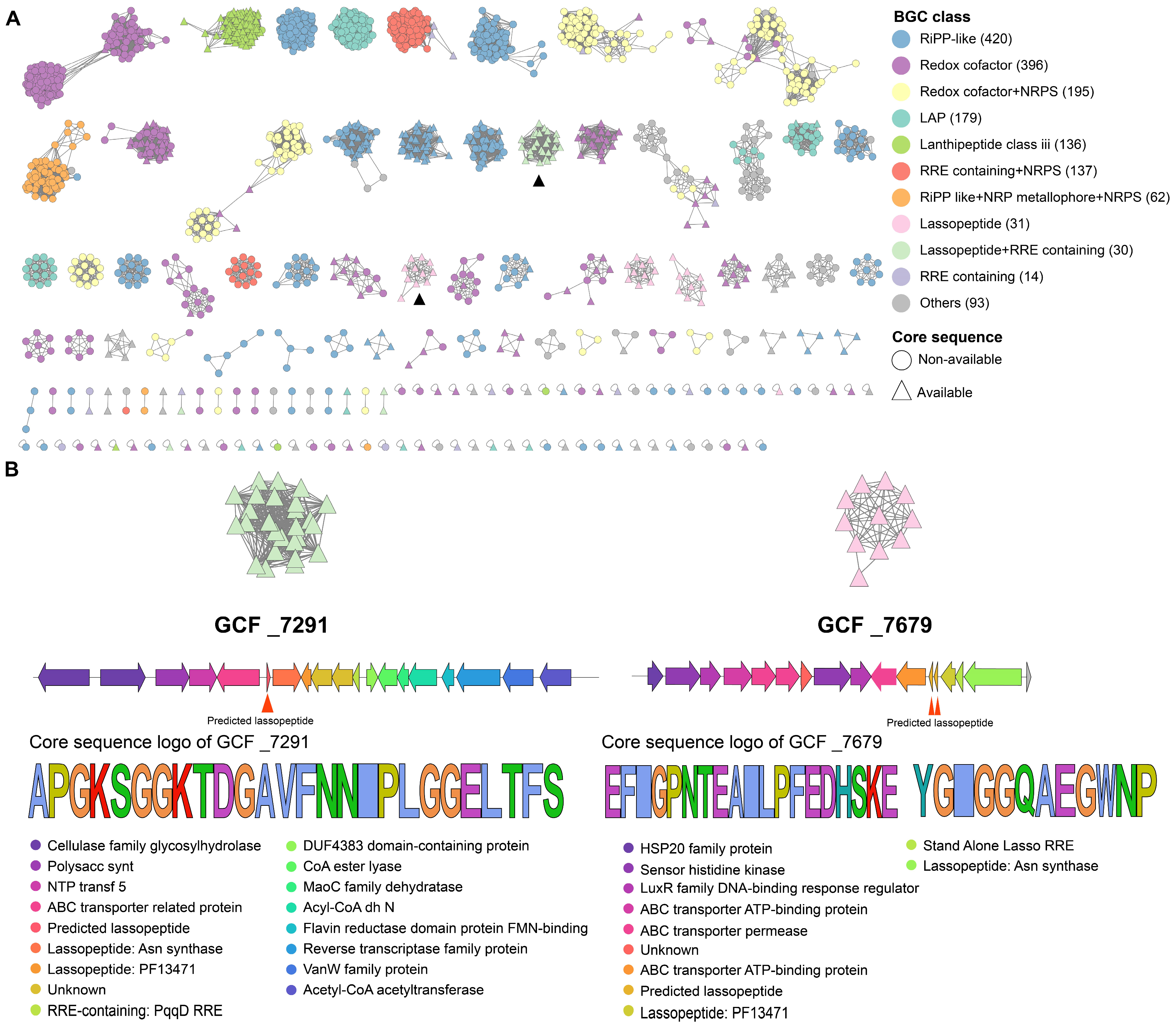
2.3. Clade Specificity of Rhodococcus in RiPP Biosynthesis
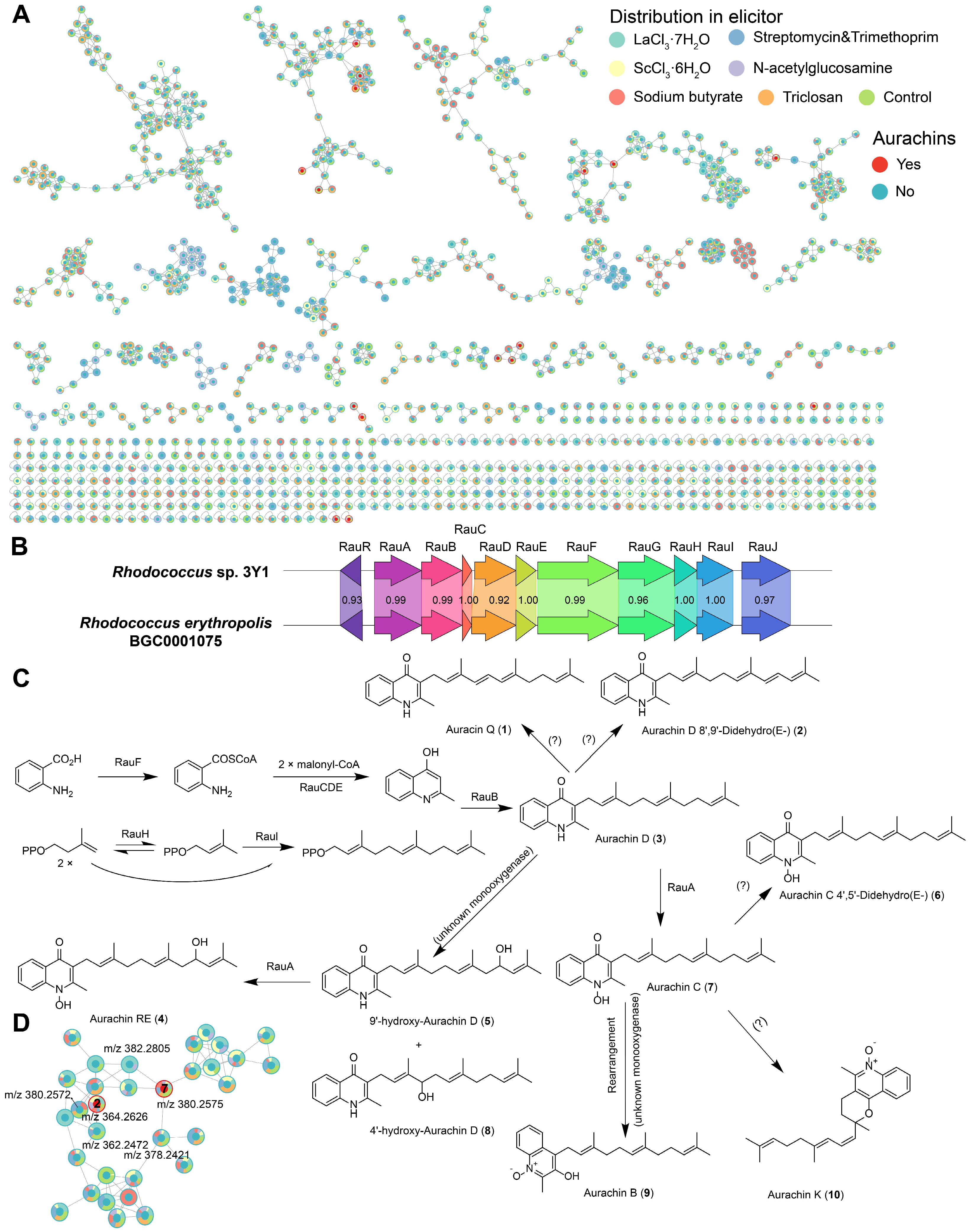
2.4. Secondary Metabolites of a Marine-Derived Rhodococcus Isolate
3. Discussion
4. Materials and Methods
4.1. Genome Collection and Phylogenomic Analysis
4.2. Diversity and Specificity of Biosynthetic Gene Clusters in Rhodococcus
4.3. Diversity and Specificity of Rhodococcus in RiPP Biosynthesis
4.4. High-Throughput Elicitor Screening (HiTES)
4.5. UPLC-QTOF-MS/MS Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Vansanten, J.A.; Poynton, E.F.; Dasha, I.; Emily, M.M.; Tylera, A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; Mccadden, C.A. The Natural Products Atlas 2.0: A Database of Microbially-Derived Natural Products. Nucleic Acids Res. 2021, 50, D1317–D1323. [Google Scholar] [CrossRef]
- Wei, B.; Luo, X.; Zhou, Z.Y.; Hu, G.A.; Li, L.; Lin, H.W.; Wang, H. Discovering the Secondary Metabolic Potential of Saccharothrix. Biotechnol. Adv. 2024, 70, 108295. [Google Scholar] [CrossRef]
- Goodfellow, M.; Alderson, G. The Actinomycete-Genus Rhodococcus: A Home for the ‘Rhodochrous’ Complex. Microbiology 1977, 100, 99–122. [Google Scholar] [CrossRef]
- Elsayed, Y.; Refaat, J.; Abdelmohsen, U.R.; Fouad, M.A. The Genus Rhodococcus as a Source of Novel Bioactive Substances: A Review. J. Pharmacogn. Phytochem. 2017, 6, 83–92. [Google Scholar]
- Chiba, H.; Agematu, H.; Kaneto, R.; Terasawa, T.; Sakai, K.; Dobashi, K.; Yoshioka, T. Rhodopeptins (Mer-N1033), Novel Cyclic Tetrapeptides with Antifungal Activity from Rhodococcus sp. I. Taxonomy, Fermentation, Isolation, Physico-Chemical Properties and Biological Activities. J. Antibiot. 1999, 52, 695–699. [Google Scholar] [CrossRef]
- Iwatsuki, M.; Uchida, R.; Takakusagi, Y.; Matsumoto, A.; Jiang, C.L.; Takahashi, Y.; Arai, M.; Kobayashi, S.; Matsumoto, M.; Inokoshi, J.; et al. Lariatins, Novel Anti-Mycobacterial Peptides with a Lasso Structure, Produced by Rhodococcus Jostii K01-B017. J. Antibiot. 2007, 60, 357–363. [Google Scholar] [CrossRef]
- Nachtigall, J.; Schneider, K.; Nicholson, G.; Goodfellow, M.; Zinecker, H.; Imhoff, J.F.; Süssmuth, R.D.; Fiedler, H.P. Two new aurachins from Rhodococcus sp. Acta 2259. J. Antibiot. Int. J. 2010, 63, 567–569. [Google Scholar] [CrossRef]
- Kurosawa, K.; Ghiviriga, I.; Sambandan, T.G.; Lessard, P.A.; Barbara, J.E.; Rha, C.; Sinskey, A.J. Rhodostreptomycins, Antibiotics Biosynthesized Following Horizontal Gene Transfer from Streptomyces Padanus to Rhodococcus fascians. J. Am. Chem. Soc. 2008, 130, 1126–1127. [Google Scholar] [CrossRef]
- Li, L.; Deng, W.; Song, J.; Ding, W.; Zhao, Q.F.; Peng, C.; Song, W.W.; Tang, G.L.; Liu, W. Characterization of the Saframycin A Gene Cluster from Streptomyces Lavendulae NRRL 11002 Revealing a Nonribosomal Peptide Synthetase System for Assembling the Unusual Tetrapeptidyl Skeleton in an Iterative Manner. J. Bacteriol. 2007, 190, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Bosello, M.; Robbel, L.; Linne, U.; Xie, X.; Marahiel, M.A. Biosynthesis of the Siderophore Rhodochelin Requires the Coordinated Expression of Three Independent Gene Clusters in Rhodococcus jostii RHA1. J. Am. Chem. Soc. 2011, 133, 4587–4595. [Google Scholar] [CrossRef] [PubMed]
- McLeod, M.P.; Warren, R.L.; Hsiao, W.W.L.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 43, 15582–15587. [Google Scholar] [CrossRef]
- Ana, C.; Lubbert, D.; Mirjan, P.; Medema, M.H. Genome-Based Exploration of the Specialized Metabolic Capacities of the Genus Rhodococcus. BMC Genomics 2017, 18, 593. [Google Scholar]
- Undabarrena, A.; Valencia, R.; Cumsille, A.; Zamora-Leiva, L.; Castro-Nallar, E.; Barona-Gómez, F.; Cámara, B. Rhodococcus Comparative Genomics Reveals a Phylogenomic-Dependent Non-Ribosomal Peptide Synthetase Distribution: Insights into Biosynthetic Gene Cluster Connection to an Orphan Metabolite. Microb. Genom. 2021, 7, 000621. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.J.; Ying, T.T.; Zhou, Z.Y.; Hu, G.A.; Yang, C.L.; Hua, Y.; Wang, H.; Wei, B. Species-Specificity of the Secondary Biosynthetic Potential in Bacillus. Front. Microbiol. 2023, 14, 1271418. [Google Scholar] [CrossRef]
- Ma, M.J.; Yu, W.C.; Sun, H.Y.; Dong, B.C.; Hu, G.A.; Zhou, Z.Y.; Hua, Y.; Basnet, B.B.; Yu, Y.L.; Wang, H.; et al. Genus-Specific Secondary Metabolome in Allokutzneria and Kibdelosporangium. Synth. Syst. Biotechnol. 2024, 9, 381–390. [Google Scholar] [CrossRef]
- Kitagawa, W.; Hata, M. Development of Efficient Genome-Reduction Tool Based on Cre/loxP System in Rhodococcus erythropolis. Microorganisms 2023, 11, 268. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, Y.; Zhang, C.; Davis, K.M.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. A Genetics-Free Method for High-Throughput Discovery of Cryptic Microbial Metabolites. Nat. Chem. Biol. 2019, 15, 161–168. [Google Scholar] [CrossRef]
- Hofmann, M.; Heine, T.; Malik, L.; Hofmann, S.; Joffroy, K.; Senges, C.H.R.; Bandow, J.E.; Tischler, D. Screening for Microbial Metal-Chelating Siderophores for the Removal of Metal Ions from Solutions. Microorganisms 2021, 9, 111. [Google Scholar] [CrossRef]
- Kitagawa, W.; Ozaki, T.; Nishioka, T.; Yasutake, Y.; Hata, M.; Nishiyama, M. Cloning and Heterologous Expression of the Aurachin RE Biosynthesis Gene Cluster Afford a New Cytochrome P450 for Quinoline N-Hydroxylation. ChemBioChem 2013, 14, 1085–1093. [Google Scholar] [CrossRef]
- Bosello, M.; Zeyadi, M.; Kraas, F.I.; Linne, U.; Marahiel, M.A. Structural Characterization of the Heterobactin Siderophores from Rhodococcus Erythropolis PR4 and Elucidation of Their Biosynthetic Machinery. J. Nat. Prod. 2013, 76, 2282–2290. [Google Scholar] [CrossRef]
- Huang, J.; Xu, Y.; Xue, Y.; Huang, Y.; Li, X.; Chen, X.; Xu, Y.; Zhang, D.; Zhang, P.; Zhao, J.; et al. Identification of Potent Antimicrobial Peptides via a Machine-Learning Pipeline That Mines the Entire Space of Peptide Sequences. Nat. Biomed. Eng. 2023, 7, 797–810. [Google Scholar] [CrossRef]
- Ma, Y.; Guo, Z.; Xia, B.; Zhang, Y.; Liu, X.; Yu, Y.; Tang, N.; Tong, X.; Wang, M.; Ye, X.; et al. Identification of Antimicrobial Peptides from the Human Gut Microbiome Using Deep Learning. Nat. Biotechnol. 2022, 40, 921–931. [Google Scholar] [CrossRef]
- Xu, J.L.; He, J.; Wang, Z.C.; Wang, K.; Li, W.J.; Tang, S.K.; Li, S.P. Rhodococcus qingshengii sp. Nov., a Carbendazim-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2007, 57, 2754. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.D.; Kim, I.S. Rhodococcus spelaei Sp. Nov., Isolated from a Cave, and Proposals That Rhodococcus biphenylivorans Is a Later Synonym of Rhodococcus pyridinivorans, Rhodococcus qingshengii and Rhodococcus baikonurensis Are Later Synonyms of Rhodococcus erythropolis, and Rhodococcus percolatus and Rhodococcus imtechensis Are Later Synonyms of Rhodococcus opacus. Int. J. Syst. Evol. Microbiol. 2021, 71, 004890. [Google Scholar]
- Pettit, R.K. Small-Molecule Elicitation of Microbial Secondary Metabolites. Microb. Biotechnol. 2011, 4, 471–478. [Google Scholar] [CrossRef]
- Kurosu, M.; Begari, E. Vitamin K2 in Electron Transport System: Are Enzymes Involved in Vitamin K2 Biosynthesis Promising Drug Targets? Molecules 2010, 15, 1531–1553. [Google Scholar] [CrossRef]
- Kitagawa, W.; Tamura, T. A Quinoline Antibiotic from Rhodococcus erythropolis JCM 6824. J. Antibiot. 2008, 61, 680–682. [Google Scholar] [CrossRef]
- Höfle, G.; Kunze, B. Biosynthesis of Aurachins A−L in Stigmatella aurantiaca: A Feeding Study. J. Nat. Prod. 2008, 71, 1843–1849. [Google Scholar] [CrossRef]
- Katsuyama, Y.; Li, X.; Müller, R.; Nay, B. Chemically Unprecedented Biocatalytic (AuaG) Retro-[2,3]-Wittig Rearrangement: A New Insight into Aurachin B Biosynthesis. ChemBioChem 2014, 15, 2349–2352. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing, Version 3.6; 2014. Available online: https://www.R-project.org (accessed on 15 May 2024).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Wei, B.; Hu, G.A.; Zhou, Z.Y.; Yu, W.C.; Du, A.Q.; Yang, C.L.; Yu, Y.L.; Chen, J.W.; Zhang, H.W.; Wu, Q.H.; et al. Global Analysis of the Biosynthetic Chemical Space of Marine Prokaryotes. Microbiome 2023, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Munoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinforma. 2014, 47, 8–13. [Google Scholar] [CrossRef]
- Merwin, N.J.; Mousa, W.K.; Dejong, C.A.; Skinnider, M.A.; Cannon, M.J.; Li, H.; Dial, K.; Gunabalasingam, M.; Johnston, C.; Magarvey, N.A. DeepRiPP Integrates Multiomics Data to Automate Discovery of Novel Ribosomally Synthesized Natural Products. Proc. Natl. Acad. Sci. USA 2020, 117, 371–380. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Adusumilli, R.; Mallick, P. Data Conversion with ProteoWizard msConvert. In Proteomics: Methods and Protocols; Comai, L., Katz, J.E., Mallick, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1550, pp. 339–368. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Duehrkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.-F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of Microbial Metabolites through Database Search of Mass Spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, G.-A.; Song, Y.; Liu, S.-Y.; Yu, W.-C.; Yu, Y.-L.; Chen, J.-W.; Wang, H.; Wei, B. Exploring the Diversity and Specificity of Secondary Biosynthetic Potential in Rhodococcus. Mar. Drugs 2024, 22, 409. https://doi.org/10.3390/md22090409
Hu G-A, Song Y, Liu S-Y, Yu W-C, Yu Y-L, Chen J-W, Wang H, Wei B. Exploring the Diversity and Specificity of Secondary Biosynthetic Potential in Rhodococcus. Marine Drugs. 2024; 22(9):409. https://doi.org/10.3390/md22090409
Chicago/Turabian StyleHu, Gang-Ao, Yue Song, Shi-Yi Liu, Wen-Chao Yu, Yan-Lei Yu, Jian-Wei Chen, Hong Wang, and Bin Wei. 2024. "Exploring the Diversity and Specificity of Secondary Biosynthetic Potential in Rhodococcus" Marine Drugs 22, no. 9: 409. https://doi.org/10.3390/md22090409






