The Green Valley of Drosophila melanogaster Constitutive Heterochromatin: Protein-Coding Genes Involved in Cell Division Control
Abstract
:1. Introduction
“But there’s no such thing as the unknown, only things temporarily hidden, temporarily not understood.”Captain James T. Kirk, from movie: Star Trek Beyond
2. Functions Related to Chromosome/Chromatin Organization and Gene Expression
2.1. Yeti and Nipped-A Genes Encode Two Subunits of the dTip60 Chromatin Remodeling Complex
2.1.1. Yeti
2.1.2. Nipped-A (Nip-A)
2.2. Nipped-B and Verthandi Encode Two Subunits of the Cohesin Complex
2.2.1. Nipped-B (Nip-B)
2.2.2. Verthandi (vtd)
2.3. Other Genes Involved in Chromatin Organization
2.3.1. Teashirt (tsh)
2.3.2. D4
3. Functions Related to Mitotic Apparatus/Microtubule Binding
3.1. Mitotic Genes Also Implicated in Ciliogenesis
3.1.1. Centriolar Coiled Coil Protein 110 (CP110)
3.1.2. CentrinB
3.1.3. Intraflagellar Transport 20 (IFT20)
3.1.4. Sterile Affecting Ciliogenesis (sac)
3.2. Other Genes Related to Mitotic Apparatus
3.2.1. CG10834
3.2.2. Sarcolemma Associated Protein (Slmap)
3.2.3. CG17528 (to Be Named Dmel-doublecortin)
3.2.4. Chromator (Chro)
3.2.5. Transforming Acidic Coiled-Coil Protein (tacc)
4. Functions Related to Kinase Activity and of Cell Cycle Regulation
4.1. Suppressor of Forked Gene (su(f))
4.2. Cyclin K (CycK)
4.3. Rolled (rl)
4.4. Haspin
4.5. Conundrum (conu)
4.6. Casein Kinase-II Alpha (CkIIα)
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heitz, E. Das Heterochromatin der Moose; Bornträger: Stuttgart, Germany, 1928. [Google Scholar]
- Brown, S.W. Heterochromatin. Science 1966, 151, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Elgin, S.C.; Reuter, G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 2013, 5, a017780. [Google Scholar] [CrossRef] [PubMed]
- Robert Finestra, T.; Gribnau, J. X chromosome inactivation: Silencing, topology and reactivation. Curr. Opin. Cell Biol. 2017, 46, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Nur, U. Heterochromatization and euchromatization of whole genomes in scale insects (Coccoidea: Homoptera). Development 1990, 108, 29–34. [Google Scholar] [CrossRef]
- Marsano, R.M.; Giordano, E.; Messina, G.; Dimitri, P. A New Portrait of Constitutive Heterochromatin: Lessons from Drosophila melanogaster. Trends Genet. 2019, 35, 615–631. [Google Scholar] [CrossRef]
- Hoskins, R.A.; Smith, C.D.; Carlson, J.W.; Carvalho, A.B.; Halpern, A.; Kaminker, J.S.; Kennedy, C.; Mungall, C.J.; Sullivan, B.A.; Sutton, G.G.; et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002, 3, research0085.1. [Google Scholar] [CrossRef]
- Corradini, N.; Rossi, F.; Verni, F.; Dimitri, P. FISH analysis of Drosophila melanogaster heterochromatin using BACs and P elements. Chromosoma 2003, 112, 26–37. [Google Scholar] [CrossRef]
- Smith, C.D.; Shu, S.; Mungall, C.J.; Karpen, G.H. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 2007, 316, 1586–1591. [Google Scholar] [CrossRef]
- Rossi, F.; Moschetti, R.; Caizzi, R.; Corradini, N.; Dimitri, P. Cytogenetic and molecular characterization of heterochromatin gene models in Drosophila melanogaster. Genetics 2007, 175, 595–607. [Google Scholar] [CrossRef]
- Hoskins, R.A.; Carlson, J.W.; Kennedy, C.; Acevedo, D.; Evans-Holm, M.; Frise, E.; Wan, K.H.; Park, S.; Mendez-Lago, M.; Rossi, F.; et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 2007, 316, 1625–1628. [Google Scholar] [CrossRef] [Green Version]
- Dimitri, P.; Caizzi, R.; Giordano, E.; Carmela Accardo, M.; Lattanzi, G.; Biamonti, G. Constitutive heterochromatin: A surprising variety of expressed sequences. Chromosoma 2009, 118, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Coulthard, A.B.; Alm, C.; Cealiac, I.; Sinclair, D.A.; Honda, B.M.; Rossi, F.; Dimitri, P.; Hilliker, A.J. Essential loci in centromeric heterochromatin of Drosophila melanogaster. I: The right arm of chromosome 2. Genetics 2010, 185, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015, 25, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Syrzycka, M.; Hallson, G.; Fitzpatrick, K.A.; Kim, I.; Cotsworth, S.; Hollebakken, R.E.; Simonetto, K.; Yang, L.; Luongo, S.; Beja, K.; et al. Genetic and Molecular Analysis of Essential Genes in Centromeric Heterochromatin of the Left Arm of Chromosome 3 in Drosophila melanogaster. G3 Genes Genomes Genet. 2019, 9, 1581–1595. [Google Scholar] [CrossRef]
- Gatti, M.; Pimpinelli, S. Functional elements in Drosophila melanogaster heterochromatin. Annu. Rev. Genet. 1992, 26, 239–275. [Google Scholar] [CrossRef]
- Dimitri, P. Cytogenetic analysis of the second chromosome heterochromatin of Drosophila melanogaster. Genetics 1991, 127, 553–564. [Google Scholar] [CrossRef]
- Fanti, L.; Pimpinelli, S. HP1: A functionally multifaceted protein. Curr. Opin. Genet. Dev. 2008, 18, 169–174. [Google Scholar] [CrossRef]
- Wallrath, L.L.; Rodriguez-Tirado, F.; Geyer, P.K. Shining Light on the Dark Side of the Genome. Cells 2022, 11, 330. [Google Scholar] [CrossRef]
- Saha, P.; Sowpati, D.T.; Soujanya, M.; Srivastava, I.; Mishra, R.K. Interplay of pericentromeric genome organization and chromatin landscape regulates the expression of Drosophila melanogaster heterochromatic genes. Epigenetics Chromatin 2020, 13, 41. [Google Scholar] [CrossRef]
- Lens, S.M.A.; Medema, R.H. Cytokinesis defects and cancer. Nat. Rev. Cancer 2019, 19, 32–45. [Google Scholar] [CrossRef]
- Sivakumar, S.; Gorbsky, G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Cenci, G.; Belloni, G.; Dimitri, P. 1(2)41Aa, a heterochromatic gene of Drosophila melanogaster, is required for mitotic and meiotic chromosome condensation. Genet. Res. 2003, 81, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Damia, E.; Fanti, L.; Atterrato, M.T.; Celauro, E.; Mariotti, F.R.; Accardo, M.C.; Walther, M.; Verni, F.; Picchioni, D.; et al. Yeti, an essential Drosophila melanogaster gene, encodes a protein required for chromatin organization. J. Cell Sci. 2014, 127, 2577–2588. [Google Scholar] [CrossRef] [PubMed]
- Prozzillo, Y.; Delle Monache, F.; Ferreri, D.; Cuticone, S.; Dimitri, P.; Messina, G. The True Story of Yeti, the “Abominable” Heterochromatic Gene of Drosophila melanogaster. Front. Physiol. 2019, 10, 1093. [Google Scholar] [CrossRef]
- Kusch, T.; Florens, L.; Macdonald, W.H.; Swanson, S.K.; Glaser, R.L.; Yates, J.R., 3rd; Abmayr, S.M.; Washburn, M.P.; Workman, J.L. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 2004, 306, 2084–2087. [Google Scholar] [CrossRef]
- March-Diaz, R.; Reyes, J.C. The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2009, 2, 565–577. [Google Scholar] [CrossRef]
- Messina, G.; Celauro, E.; Atterrato, M.T.; Giordano, E.; Iwashita, S.; Dimitri, P. The Bucentaur (BCNT) protein family: A long-neglected class of essential proteins required for chromatin/chromosome organization and function. Chromosoma 2015, 124, 153–162. [Google Scholar] [CrossRef]
- Messina, G.; Atterrato, M.T.; Dimitri, P. When chromatin organisation floats astray: The Srcap gene and Floating-Harbor syndrome. J. Med. Genet. 2016, 53, 793–797. [Google Scholar] [CrossRef]
- Messina, G.; Prozzillo, Y.; Delle Monache, F.; Santopietro, M.V.; Atterrato, M.T.; Dimitri, P. The ATPase SRCAP is associated with the mitotic apparatus, uncovering novel molecular aspects of Floating-Harbor syndrome. BMC Biol. 2021, 19, 184. [Google Scholar] [CrossRef]
- Billmann, M.; Horn, T.; Fischer, B.; Sandmann, T.; Huber, W.; Boutros, M. A genetic interaction map of cell cycle regulators. Mol. Biol. Cell 2016, 27, 1397–1407. [Google Scholar] [CrossRef]
- Messina, G.; Atterrato, M.T.; Fanti, L.; Giordano, E.; Dimitri, P. Expression of human Cfdp1 gene in Drosophila reveals new insights into the function of the evolutionarily conserved BCNT protein family. Sci. Rep. 2016, 6, 25511. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Prozzillo, Y.; Monache, F.D.; Santopietro, M.V.; Dimitri, P. Evolutionary conserved relocation of chromatin remodeling complexes to the mitotic apparatus. BMC Biol. 2022, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Hart, G.T.; Nepusz, T.; Yang, H.; Turinsky, A.L.; Li, Z.; Wang, P.I.; Boutz, D.R.; Fong, V.; Phanse, S.; et al. A census of human soluble protein complexes. Cell 2012, 150, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Atterrato, M.T.; Prozzillo, Y.; Piacentini, L.; Losada, A.; Dimitri, P. The human Cranio Facial Development Protein 1 (Cfdp1) gene encodes a protein required for the maintenance of higher-order chromatin organization. Sci. Rep. 2017, 7, 45022. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Bukowski-Wills, J.C.; Sanchez-Pulido, L.; Alves Fde, L.; Wood, L.; Chen, Z.A.; Platani, M.; Fischer, L.; Hudson, D.F.; Ponting, C.P.; et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 2010, 142, 810–821. [Google Scholar] [CrossRef]
- Bode-Lesniewska, B.; Fritz, C.; Exner, G.U.; Wagner, U.; Fuchs, B. EWSR1-NFATC2 and FUS-NFATC2 Gene Fusion-Associated Mesenchymal Tumors: Clinicopathologic Correlation and Literature Review. Sarcoma 2019, 2019, 9386390. [Google Scholar] [CrossRef]
- Perl, A.; Qian, Y.; Chohan, K.R.; Shirley, C.R.; Amidon, W.; Banerjee, S.; Middleton, F.A.; Conkrite, K.L.; Barcza, M.; Gonchoroff, N.; et al. Transaldolase is essential for maintenance of the mitochondrial transmembrane potential and fertility of spermatozoa. Proc. Natl. Acad. Sci. USA 2006, 103, 14813–14818. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Berkovits, B.D.; Wolgemuth, D.J. The role of the double bromodomain-containing BET genes during mammalian spermatogenesis. Curr. Top. Dev. Biol. 2013, 102, 293–326. [Google Scholar] [CrossRef] [Green Version]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667.e10. [Google Scholar] [CrossRef] [PubMed]
- Zalensky, A.O.; Siino, J.S.; Gineitis, A.A.; Zalenskaya, I.A.; Tomilin, N.V.; Yau, P.; Bradbury, E.M. Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J. Biol. Chem. 2002, 277, 43474–43480. [Google Scholar] [CrossRef] [PubMed]
- Gause, M.; Eissenberg, J.C.; Macrae, A.F.; Dorsett, M.; Misulovin, Z.; Dorsett, D. Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development. Mol. Cell. Biol. 2006, 26, 2347–2359. [Google Scholar] [CrossRef] [PubMed]
- Prozzillo, Y.; Cuticone, S.; Ferreri, D.; Fattorini, G.; Messina, G.; Dimitri, P. In Vivo Silencing of Genes Coding for dTip60 Chromatin Remodeling Complex Subunits Affects Polytene Chromosome Organization and Proper Development in Drosophila melanogaster. Int. J. Mol. Sci. 2021, 22, 4525. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.A.; Schieltz, D.; Pray-Grant, M.G.; Yates, J.R., 3rd; Workman, J.L. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 1998, 2, 863–867. [Google Scholar] [CrossRef]
- Brown, C.E.; Howe, L.; Sousa, K.; Alley, S.C.; Carrozza, M.J.; Tan, S.; Workman, J.L. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 2001, 292, 2333–2337. [Google Scholar] [CrossRef]
- Carrozza, M.J.; Utley, R.T.; Workman, J.L.; Cote, J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003, 19, 321–329. [Google Scholar] [CrossRef]
- Rollins, R.A.; Morcillo, P.; Dorsett, D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 1999, 152, 577–593. [Google Scholar] [CrossRef]
- Seitan, V.C.; Banks, P.; Laval, S.; Majid, N.A.; Dorsett, D.; Rana, A.; Smith, J.; Bateman, A.; Krpic, S.; Hostert, A.; et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006, 4, e242. [Google Scholar] [CrossRef]
- Takahashi, T.S.; Yiu, P.; Chou, M.F.; Gygi, S.; Walter, J.C. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004, 6, 991–996. [Google Scholar] [CrossRef]
- Watrin, E.; Schleiffer, A.; Tanaka, K.; Eisenhaber, F.; Nasmyth, K.; Peters, J.M. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 2006, 16, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, P.; Gruber, S.; Tanaka, K.; Haering, C.H.; Mechtler, K.; Nasmyth, K. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 2003, 13, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Ciosk, R.; Shirayama, M.; Shevchenko, A.; Tanaka, T.; Toth, A.; Shevchenko, A.; Nasmyth, K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 2000, 5, 243–254. [Google Scholar] [CrossRef]
- Gillespie, P.J.; Hirano, T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr. Biol. 2004, 14, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, T.; Nagao, K.; Kawasaki, Y.; Furuya, K.; Murakami, A.; Morishita, J.; Yuasa, T.; Sutani, T.; Kearsey, S.E.; Uhlmann, F.; et al. Characterization of fission yeast cohesin: Essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000, 14, 2757–2770. [Google Scholar] [CrossRef]
- Hirano, T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell. Biol. 2006, 7, 311–322. [Google Scholar] [CrossRef]
- Nasmyth, K.; Haering, C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005, 74, 595–648. [Google Scholar] [CrossRef]
- Huang, C.E.; Milutinovich, M.; Koshland, D. Rings, bracelet or snaps: Fashionable alternatives for Smc complexes. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2005, 360, 537–542. [Google Scholar] [CrossRef]
- Gause, M.; Webber, H.A.; Misulovin, Z.; Haller, G.; Rollins, R.A.; Eissenberg, J.C.; Bickel, S.E.; Dorsett, D. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma 2008, 117, 51–66. [Google Scholar] [CrossRef]
- Krantz, I.D.; McCallum, J.; DeScipio, C.; Kaur, M.; Gillis, L.A.; Yaeger, D.; Jukofsky, L.; Wasserman, N.; Bottani, A.; Morris, C.A.; et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 2004, 36, 631–635. [Google Scholar] [CrossRef]
- Parenti, I.; Diab, F.; Gil, S.R.; Mulugeta, E.; Casa, V.; Berutti, R.; Brouwer, R.W.W.; Dupe, V.; Eckhold, J.; Graf, E.; et al. MAU2 and NIPBL Variants Impair the Heterodimerization of the Cohesin Loader Subunits and Cause Cornelia de Lange Syndrome. Cell Rep. 2020, 31, 107647. [Google Scholar] [CrossRef]
- Rollins, R.A.; Korom, M.; Aulner, N.; Martens, A.; Dorsett, D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 2004, 24, 3100–3111. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; DeScipio, C.; McCallum, J.; Yaeger, D.; Devoto, M.; Jackson, L.G.; Spinner, N.B.; Krantz, I.D. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am. J. Med. Genet. A 2005, 138, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Luna-Pelaez, N.; March-Diaz, R.; Ceballos-Chavez, M.; Guerrero-Martinez, J.A.; Grazioli, P.; Garcia-Gutierrez, P.; Vaccari, T.; Massa, V.; Reyes, J.C.; Garcia-Dominguez, M. The Cornelia de Lange Syndrome-associated factor NIPBL interacts with BRD4 ET domain for transcription control of a common set of genes. Cell Death Dis. 2019, 10, 548. [Google Scholar] [CrossRef] [PubMed]
- Olley, G.; Ansari, M.; Bengani, H.; Grimes, G.R.; Rhodes, J.; von Kriegsheim, A.; Blatnik, A.; Stewart, F.J.; Wakeling, E.; Carroll, N.; et al. BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange-like syndrome. Nat. Genet. 2018, 50, 329–332. [Google Scholar] [CrossRef]
- Schulze, S.; Sinclair, D.A.; Silva, E.; Fitzpatrick, K.A.; Singh, M.; Lloyd, V.K.; Morin, K.A.; Kim, J.; Holm, D.G.; Kennison, J.A.; et al. Essential genes in proximal 3L heterochromatin of Drosophila melanogaster. Mol. Genet. Genom. 2001, 264, 782–789. [Google Scholar] [CrossRef]
- Fitzpatrick, K.A.; Sinclair, D.A.; Schulze, S.R.; Syrzycka, M.; Honda, B.M. A genetic and molecular profile of third chromosome centric heterochromatin in Drosophila melanogaster. Genome 2005, 48, 571–584. [Google Scholar] [CrossRef]
- Koryakov, D.E.; Zhimulev, I.F.; Dimitri, P. Cytogenetic analysis of the third chromosome heterochromatin of Drosophila melanogaster. Genetics 2002, 160, 509–517. [Google Scholar] [CrossRef]
- Hallson, G.; Syrzycka, M.; Beck, S.A.; Kennison, J.A.; Dorsett, D.; Page, S.L.; Hunter, S.M.; Keall, R.; Warren, W.D.; Brock, H.W.; et al. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc. Natl. Acad. Sci. USA 2008, 105, 12405–12410. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Kotadia, S.; Tavares, A.; Mirkovic, M.; Bowlin, K.; Eichinger, C.S.; Nasmyth, K.; Sullivan, W. Centromere-independent accumulation of cohesin at ectopic heterochromatin sites induces chromosome stretching during anaphase. PLoS Biol. 2014, 12, e1001962. [Google Scholar] [CrossRef] [Green Version]
- Valdeolmillos, A.; Rufas, J.S.; Suja, J.A.; Vass, S.; Heck, M.M.; Martinez, A.C.; Barbero, J.L. Drosophila cohesins DSA1 and Drad21 persist and colocalize along the centromeric heterochromatin during mitosis. Biol. Cell 2004, 96, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.; Althoff, F.; Oliveira, R.A.; Heidmann, S.; Schuldiner, O.; Lehner, C.F.; Dickson, B.J.; Nasmyth, K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell 2008, 14, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Urban, E.; Nagarkar-Jaiswal, S.; Lehner, C.F.; Heidmann, S.K. The cohesin subunit Rad21 is required for synaptonemal complex maintenance, but not sister chromatid cohesion, during Drosophila female meiosis. PLoS Genet. 2014, 10, e1004540. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, M.A.; Bando, M.; Nakato, R.; Watrin, E.; Itoh, T.; Minamino, M.; Saitoh, K.; Komata, M.; Katou, Y.; Clark, D.; et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 2012, 489, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, M.A.; Wilde, J.J.; Albrecht, M.; Dickinson, E.; Tennstedt, S.; Braunholz, D.; Monnich, M.; Yan, Y.; Xu, W.; Gil-Rodriguez, M.C.; et al. RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 2012, 90, 1014–1027. [Google Scholar] [CrossRef]
- Pimentel, S.A.B.J. Analysis of Teashirt Mutants Affecting Cell Proliferation in Drosophila Melanogaster; University of Lisboa: Lisboa, Portogal, 2010. [Google Scholar]
- Alexandre, E.; Graba, Y.; Fasano, L.; Gallet, A.; Perrin, L.; De Zulueta, P.; Pradel, J.; Kerridge, S.; Jacq, B. The Drosophila teashirt homeotic protein is a DNA-binding protein and modulo, a HOM-C regulated modifier of variegation, is a likely candidate for being a direct target gene. Mech. Dev. 1996, 59, 191–204. [Google Scholar] [CrossRef]
- Datta, R.R.; Lurye, J.M.; Kumar, J.P. Restriction of ectopic eye formation by Drosophila teashirt and tiptop to the developing antenna. Dev. Dyn. 2009, 238, 2202–2210. [Google Scholar] [CrossRef]
- Kajiwara, Y.; Akram, A.; Katsel, P.; Haroutunian, V.; Schmeidler, J.; Beecham, G.; Haines, J.L.; Pericak-Vance, M.A.; Buxbaum, J.D. FE65 binds Teashirt, inhibiting expression of the primate-specific caspase-4. PLoS ONE 2009, 4, e5071. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef]
- Mashtalir, N.; D’Avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; McKenzie, Z.M.; Kubiak, R.L.; St Pierre, R.; et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175, 1272–1288.e1220. [Google Scholar] [CrossRef] [Green Version]
- Barish, S.; Barakat, T.S.; Michel, B.C.; Mashtalir, N.; Phillips, J.B.; Valencia, A.M.; Ugur, B.; Wegner, J.; Scott, T.M.; Bostwick, B.; et al. BICRA, a SWI/SNF Complex Member, Is Associated with BAF-Disorder Related Phenotypes in Humans and Model Organisms. Am. J. Hum. Genet. 2020, 107, 1096–1112. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Vasileiou, G.; Vergarajauregui, S.; Endele, S.; Popp, B.; Buttner, C.; Ekici, A.B.; Gerard, M.; Bramswig, N.C.; Albrecht, B.; Clayton-Smith, J.; et al. Mutations in the BAF-Complex Subunit DPF2 Are Associated with Coffin-Siris Syndrome. Am. J. Hum. Genet. 2018, 102, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Spektor, A.; Tsang, W.Y.; Khoo, D.; Dynlacht, B.D. Cep97 and CP110 suppress a cilia assembly program. Cell 2007, 130, 678–690. [Google Scholar] [CrossRef]
- Franz, A.; Roque, H.; Saurya, S.; Dobbelaere, J.; Raff, J.W. CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J. Cell Biol. 2013, 203, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Hodges, M.E.; Scheumann, N.; Wickstead, B.; Langdale, J.A.; Gull, K. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 2010, 123, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Indjeian, V.B.; McManus, M.; Wang, L.; Dynlacht, B.D. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell 2002, 3, 339–350. [Google Scholar] [CrossRef]
- Delgehyr, N.; Rangone, H.; Fu, J.; Mao, G.; Tom, B.; Riparbelli, M.G.; Callaini, G.; Glover, D.M. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr. Biol. 2012, 22, 502–509. [Google Scholar] [CrossRef]
- Sawant, D.B.; Majumder, S.; Perkins, J.L.; Yang, C.H.; Eyers, P.A.; Fisk, H.A. Centrin 3 is an inhibitor of centrosomal Mps1 and antagonizes centrin 2 function. Mol. Biol. Cell 2015, 26, 3741–3753. [Google Scholar] [CrossRef]
- Salisbury, J.L. Centrin, centrosomes, and mitotic spindle poles. Curr. Opin. Cell Biol. 1995, 7, 39–45. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, X.; Yang, B. Calcium-induced human centrin 1 self-assembly and double-regulating the binding with peptide R18-Sfi1p. Int. J. Biol. Macromol. 2019, 128, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Schnorrer, F.; Schonbauer, C.; Langer, C.C.; Dietzl, G.; Novatchkova, M.; Schernhuber, K.; Fellner, M.; Azaryan, A.; Radolf, M.; Stark, A.; et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 2010, 464, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Avidor-Reiss, T.; Maer, A.M.; Koundakjian, E.; Polyanovsky, A.; Keil, T.; Subramaniam, S.; Zuker, C.S. Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell 2004, 117, 527–539. [Google Scholar] [CrossRef]
- Laurencon, A.; Dubruille, R.; Efimenko, E.; Grenier, G.; Bissett, R.; Cortier, E.; Rolland, V.; Swoboda, P.; Durand, B. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 2007, 8, R195. [Google Scholar] [CrossRef]
- Jonassen, J.A.; San Agustin, J.; Follit, J.A.; Pazour, G.J. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J. Cell Biol. 2008, 183, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Amador-Arjona, A.; Elliott, J.; Miller, A.; Ginbey, A.; Pazour, G.J.; Enikolopov, G.; Roberts, A.J.; Terskikh, A.V. Primary cilia regulate proliferation of amplifying progenitors in adult hippocampus: Implications for learning and memory. J. Neurosci. 2011, 31, 9933–9944. [Google Scholar] [CrossRef]
- Zhou, M.H.; Lin, Y.; Zhang, Z.G. Intraflagellar transport 20: New target for the treatment of ciliopathies. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118641. [Google Scholar] [CrossRef]
- Follit, J.A.; Tuft, R.A.; Fogarty, K.E.; Pazour, G.J. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell 2006, 17, 3781–3792. [Google Scholar] [CrossRef]
- Follit, J.A.; San Agustin, J.T.; Xu, F.; Jonassen, J.A.; Samtani, R.; Lo, C.W.; Pazour, G.J. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 2008, 4, e1000315. [Google Scholar] [CrossRef]
- Stoetzel, C.; Bar, S.; De Craene, J.O.; Scheidecker, S.; Etard, C.; Chicher, J.; Reck, J.R.; Perrault, I.; Geoffroy, V.; Chennen, K.; et al. A mutation in VPS15 (PIK3R4) causes a ciliopathy and affects IFT20 release from the cis-Golgi. Nat. Commun. 2016, 7, 13586. [Google Scholar] [CrossRef] [Green Version]
- Aoki, T.; Nishita, M.; Sonoda, J.; Ikeda, T.; Kakeji, Y.; Minami, Y. Intraflagellar transport 20 promotes collective cancer cell invasion by regulating polarized organization of Golgi-associated microtubules. Cancer Sci. 2019, 110, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Bauerly, E.; Akiyama, T.; Staber, C.; Yi, K.; Gibson, M.C. Impact of cilia-related genes on mitochondrial dynamics during Drosophila spermatogenesis. Dev. Biol. 2022, 482, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Zur Lage, P.; Newton, F.G.; Jarman, A.P. Survey of the Ciliary Motility Machinery of Drosophila Sperm and Ciliated Mechanosensory Neurons Reveals Unexpected Cell-Type Specific Variations: A Model for Motile Ciliopathies. Front. Genet. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Bowman, A.B.; Patel-King, R.S.; Benashski, S.E.; McCaffery, J.M.; Goldstein, L.S.; King, S.M. Drosophila roadblock and Chlamydomonas LC7: A conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 1999, 146, 165–180. [Google Scholar] [CrossRef]
- Terenzio, M.; Di Pizio, A.; Rishal, I.; Marvaldi, L.; Di Matteo, P.; Kawaguchi, R.; Coppola, G.; Schiavo, G.; Fisher, E.M.C.; Fainzilber, M. DYNLRB1 is essential for dynein mediated transport and neuronal survival. Neurobiol. Dis. 2020, 140, 104816. [Google Scholar] [CrossRef]
- Alliance of Genome Resources, C. Alliance of Genome Resources Portal: Unified model organism research platform. Nucleic Acids Res. 2020, 48, D650–D658. [Google Scholar] [CrossRef]
- Goudreault, M.; D’Ambrosio, L.M.; Kean, M.J.; Mullin, M.J.; Larsen, B.G.; Sanchez, A.; Chaudhry, S.; Chen, G.I.; Sicheri, F.; Nesvizhskii, A.I.; et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteom. 2009, 8, 157–171. [Google Scholar] [CrossRef]
- Byers, J.T.; Guzzo, R.M.; Salih, M.; Tuana, B.S. Hydrophobic profiles of the tail anchors in SLMAP dictate subcellular targeting. BMC Cell. Biol. 2009, 10, 48. [Google Scholar] [CrossRef]
- Frost, A.; Elgort, M.G.; Brandman, O.; Ives, C.; Collins, S.R.; Miller-Vedam, L.; Weibezahn, J.; Hein, M.Y.; Poser, I.; Mann, M.; et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell 2012, 149, 1339–1352. [Google Scholar] [CrossRef]
- Nordzieke, S.; Zobel, T.; Franzel, B.; Wolters, D.A.; Kuck, U.; Teichert, I. A fungal sarcolemmal membrane-associated protein (SLMAP) homolog plays a fundamental role in development and localizes to the nuclear envelope, endoplasmic reticulum, and mitochondria. Eukaryot. Cell 2015, 14, 345–358. [Google Scholar] [CrossRef] [Green Version]
- Guzzo, R.M.; Sevinc, S.; Salih, M.; Tuana, B.S. A novel isoform of sarcolemmal membrane-associated protein (SLMAP) is a component of the microtubule organizing centre. J. Cell Sci. 2004, 117, 2271–2281. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Takahashi, N.; Ohno, S.; Sakurada, H.; Nakamura, K.; On, Y.K.; Park, J.E.; Makiyama, T.; Horie, M.; Arimura, T.; et al. Novel SCN3B mutation associated with brugada syndrome affects intracellular trafficking and function of Nav1.5. Circ. J. 2013, 77, 959–967. [Google Scholar] [CrossRef]
- Ishikawa, T.; Sato, A.; Marcou, C.A.; Tester, D.J.; Ackerman, M.J.; Crotti, L.; Schwartz, P.J.; On, Y.K.; Park, J.E.; Nakamura, K.; et al. A novel disease gene for Brugada syndrome: Sarcolemmal membrane-associated protein gene mutations impair intracellular trafficking of hNav1.5. Circ. Arrhythmia Electrophysiol. 2012, 5, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, J.G.; Lin, P.T.; Flanagan, L.A.; Walsh, C.A. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron 1999, 23, 257–271. [Google Scholar] [CrossRef]
- Lin, P.T.; Gleeson, J.G.; Corbo, J.C.; Flanagan, L.; Walsh, C.A. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J. Neurosci. 2000, 20, 9152–9161. [Google Scholar] [CrossRef]
- Taylor, K.R.; Holzer, A.K.; Bazan, J.F.; Walsh, C.A.; Gleeson, J.G. Patient mutations in doublecortin define a repeated tubulin-binding domain. J. Biol. Chem. 2000, 275, 34442–34450. [Google Scholar] [CrossRef]
- Des Portes, V.; Pinard, J.M.; Billuart, P.; Vinet, M.C.; Koulakoff, A.; Carrie, A.; Gelot, A.; Dupuis, E.; Motte, J.; Berwald-Netter, Y.; et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell 1998, 92, 51–61. [Google Scholar] [CrossRef]
- Rath, U.; Wang, D.; Ding, Y.; Xu, Y.Z.; Qi, H.; Blacketer, M.J.; Girton, J.; Johansen, J.; Johansen, K.M. Chromator, a novel and essential chromodomain protein interacts directly with the putative spindle matrix protein skeletor. J. Cell. Biochem. 2004, 93, 1033–1047. [Google Scholar] [CrossRef]
- Rath, U.; Ding, Y.; Deng, H.; Qi, H.; Bao, X.; Zhang, W.; Girton, J.; Johansen, J.; Johansen, K.M. The chromodomain protein, Chromator, interacts with JIL-1 kinase and regulates the structure of Drosophila polytene chromosomes. J. Cell Sci. 2006, 119, 2332–2341. [Google Scholar] [CrossRef]
- Eggert, H.; Gortchakov, A.; Saumweber, H. Identification of the Drosophila interband-specific protein Z4 as a DNA-binding zinc-finger protein determining chromosomal structure. J. Cell Sci. 2004, 117, 4253–4264. [Google Scholar] [CrossRef] [Green Version]
- Pokholkova, G.V.; Demakov, S.A.; Andreenkov, O.V.; Andreenkova, N.G.; Volkova, E.I.; Belyaeva, E.S.; Zhimulev, I.F. Tethering of CHROMATOR and dCTCF proteins results in decompaction of condensed bands in the Drosophila melanogaster polytene chromosomes but does not affect their transcription and replication timing. PLoS ONE 2018, 13, e0192634. [Google Scholar] [CrossRef] [PubMed]
- Gergely, F.; Kidd, D.; Jeffers, K.; Wakefield, J.G.; Raff, J.W. D-TACC: A novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000, 19, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Rual, J.F.; Zhai, B.; Mintseris, J.; Vaidya, P.; Vaidya, N.; Beekman, C.; Wong, C.; Rhee, D.Y.; Cenaj, O.; et al. A protein complex network of Drosophila melanogaster. Cell 2011, 147, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Galletta, B.J.; Fagerstrom, C.J.; Schoborg, T.A.; McLamarrah, T.A.; Ryniawec, J.M.; Buster, D.W.; Slep, K.C.; Rogers, G.C.; Rusan, N.M. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 2016, 7, 12476. [Google Scholar] [CrossRef]
- Xu, T.; Wang, H.; Huang, X.; Li, W.; Huang, Q.; Yan, Y.; Chen, J. Gene Fusion in Malignant Glioma: An Emerging Target for Next-Generation Personalized Treatment. Transl. Oncol. 2018, 11, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Audibert, A.; Juge, F.; Simonelig, M. The suppressor of forked protein of Drosophila, a homologue of the human 77K protein required for mRNA 3’-end formation, accumulates in mitotically-active cells. Mech. Dev. 1998, 72, 53–63. [Google Scholar] [CrossRef]
- Audibert, A.; Simonelig, M. Autoregulation at the level of mRNA 3’ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc. Natl. Acad. Sci. USA 1998, 95, 14302–14307. [Google Scholar] [CrossRef]
- Sonkar, A.; Yadav, S.; Ahmed, S. Cleavage and polyadenylation factor, Rna14 is an essential protein required for the maintenance of genomic integrity in fission yeast Schizosaccharomyces pombe. Biochim. Biophys. Acta 2016, 1863, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Bartkowiak, B.; Liu, P.; Phatnani, H.P.; Fuda, N.J.; Cooper, J.J.; Price, D.H.; Adelman, K.; Lis, J.T.; Greenleaf, A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010, 24, 2303–2316. [Google Scholar] [CrossRef]
- Edwards, M.C.; Wong, C.; Elledge, S.J. Human cyclin K, a novel RNA polymerase II-associated cyclin possessing both carboxy-terminal domain kinase and Cdk-activating kinase activity. Mol. Cell. Biol. 1998, 18, 4291–4300. [Google Scholar] [CrossRef] [Green Version]
- Blazek, D.; Kohoutek, J.; Bartholomeeusen, K.; Johansen, E.; Hulinkova, P.; Luo, Z.; Cimermancic, P.; Ule, J.; Peterlin, B.M. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011, 25, 2158–2172. [Google Scholar] [CrossRef] [PubMed]
- Kohoutek, J.; Blazek, D. Cyclin K goes with Cdk12 and Cdk13. Cell Div. 2012, 7, 12. [Google Scholar] [CrossRef]
- Hilliker, A.J. Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: Deficiency mapping of EMS-induced lethal complementation groups. Genetics 1976, 83, 765–782. [Google Scholar] [CrossRef] [PubMed]
- Biggs, W.H., 3rd; Zipursky, S.L. Primary structure, expression, and signal-dependent tyrosine phosphorylation of a Drosophila homolog of extracellular signal-regulated kinase. Proc. Natl. Acad. Sci. USA 1992, 89, 6295–6299. [Google Scholar] [CrossRef]
- Nishida, Y.; Inoue, Y.H.; Tsuda, L.; Adachi-Yamada, T.; Lim, Y.M.; Hata, M.; Ha, H.Y.; Sugiyama, S. The Raf/MAP kinase cascade in cell cycle regulation and differentiation in Drosophila. Cell Struct. Funct. 1996, 21, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Saunders, R.D.; Avides, M.C.; Howard, T.; Gonzalez, C.; Glover, D.M. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 1997, 137, 881–890. [Google Scholar] [CrossRef]
- Ripoll, P.; Pimpinelli, S.; Valdivia, M.M.; Avila, J. A cell division mutant of Drosophila with a functionally abnormal spindle. Cell 1985, 41, 907–912. [Google Scholar] [CrossRef]
- Gonzalez, C.; Saunders, R.D.; Casal, J.; Molina, I.; Carmena, M.; Ripoll, P.; Glover, D.M. Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J. Cell Sci. 1990, 96 Pt 4, 605–616. [Google Scholar] [CrossRef]
- Casal, J.; Gonzalez, C.; Wandosell, F.; Avila, J.; Ripoll, P. Abnormal meiotic spindles cause a cascade of defects during spermatogenesis in asp males of Drosophila. Development 1990, 108, 251–260. [Google Scholar] [CrossRef]
- Gupta, S.; Varshney, B.; Chatterjee, S.; Ray, K. Somatic ERK activation during transit amplification is essential for maintaining the synchrony of germline divisions in Drosophila testis. Open Biol. 2018, 8, 180033. [Google Scholar] [CrossRef] [Green Version]
- Blaker-Lee, A.; Gupta, S.; McCammon, J.M.; De Rienzo, G.; Sive, H. Zebrafish homologs of genes within 16p11.2, a genomic region associated with brain disorders, are active during brain development, and include two deletion dosage sensor genes. Dis. Model. Mech. 2012, 5, 834–851. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Park, H.R.; Lee, J.H. MAPK3 at the Autism-Linked Human 16p11.2 Locus Influences Precise Synaptic Target Selection at Drosophila Larval Neuromuscular Junctions. Mol. Cells 2017, 40, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.M. Haspin-like proteins: A new family of evolutionarily conserved putative eukaryotic protein kinases. Protein Sci. 2001, 10, 1677–1684. [Google Scholar] [CrossRef] [PubMed]
- Fresan, U.; Rodriguez-Sanchez, M.A.; Reina, O.; Corces, V.G.; Espinas, M.L. Haspin kinase modulates nuclear architecture and Polycomb-dependent gene silencing. PLoS Genet. 2020, 16, e1008962. [Google Scholar] [CrossRef]
- Dai, J.; Sullivan, B.A.; Higgins, J.M. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell 2006, 11, 741–750. [Google Scholar] [CrossRef]
- Tanaka, H.; Yoshimura, Y.; Nozaki, M.; Yomogida, K.; Tsuchida, J.; Tosaka, Y.; Habu, T.; Nakanishi, T.; Okada, M.; Nojima, H.; et al. Identification and characterization of a haploid germ cell-specific nuclear protein kinase (Haspin) in spermatid nuclei and its effects on somatic cells. J. Biol. Chem. 1999, 274, 17049–17057. [Google Scholar] [CrossRef]
- Tanaka, H.; Iguchi, N.; Nakamura, Y.; Kohroki, J.; de Carvalho, C.E.; Nishimune, Y. Cloning and characterization of human haspin gene encoding haploid germ cell-specific nuclear protein kinase. Mol. Hum. Reprod. 2001, 7, 211–218. [Google Scholar] [CrossRef]
- Neisch, A.L.; Formstecher, E.; Fehon, R.G. Conundrum, an ARHGAP18 orthologue, regulates RhoA and proliferation through interactions with Moesin. Mol. Biol. Cell 2013, 24, 1420–1433. [Google Scholar] [CrossRef]
- Maeda, M.; Hasegawa, H.; Hyodo, T.; Ito, S.; Asano, E.; Yuang, H.; Funasaka, K.; Shimokata, K.; Hasegawa, Y.; Hamaguchi, M.; et al. ARHGAP18, a GTPase-activating protein for RhoA, controls cell shape, spreading, and motility. Mol. Biol. Cell 2011, 22, 3840–3852. [Google Scholar] [CrossRef]
- Li, Y.; Ji, S.; Fu, L.; Jiang, T.; Wu, D.; Meng, F. Over-expression of ARHGAP18 suppressed cell proliferation, migration, invasion, and tumor growth in gastric cancer by restraining over-activation of MAPK signaling pathways. Onco Targets Ther. 2018, 11, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Bettencourt-Dias, M.; Giet, R.; Sinka, R.; Mazumdar, A.; Lock, W.G.; Balloux, F.; Zafiropoulos, P.J.; Yamaguchi, S.; Winter, S.; Carthew, R.W.; et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature 2004, 432, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Wollman, R.; Goodwin, S.S.; Zhang, N.; Scholey, J.M.; Vale, R.D.; Stuurman, N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 2007, 316, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Ducat, D.; Kawaguchi, S.; Liu, H.; Yates, J.R., 3rd; Zheng, Y. Regulation of microtubule assembly and organization in mitosis by the AAA+ ATPase Pontin. Mol. Biol. Cell 2008, 19, 3097–3110. [Google Scholar] [CrossRef] [PubMed]
- Bonke, M.; Turunen, M.; Sokolova, M.; Vaharautio, A.; Kivioja, T.; Taipale, M.; Bjorklund, M.; Taipale, J. Transcriptional networks controlling the cell cycle. G3 Genes Genomes Genet. 2013, 3, 75–90. [Google Scholar] [CrossRef]
- Zhao, W.; Bidwai, A.P.; Glover, C.V. Interaction of casein kinase II with ribosomal protein L22 of Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2002, 298, 60–66. [Google Scholar] [CrossRef]
- Berloco, M.F.; Minervini, C.F.; Moschetti, R.; Palazzo, A.; Viggiano, L.; Marsano, R.M. Evidence of the Physical Interaction between Rpl22 and the Transposable Element Doc5, a Heterochromatic Transposon of Drosophila melanogaster. Genes 2021, 12, 1997. [Google Scholar] [CrossRef]
- Minervini, C.F.; Berloco, M.F.; Marsano, R.M.; Viggiano, L. The Ribosomal Protein RpL22 Interacts In Vitro with 5’-UTR Sequences Found in Some Drosophila melanogaster Transposons. Genes 2022, 13, 305. [Google Scholar] [CrossRef]
- Zwarts, L.; Vulsteke, V.; Buhl, E.; Hodge, J.J.L.; Callaerts, P. SlgA, encoded by the homolog of the human schizophrenia-associated gene PRODH, acts in clock neurons to regulate Drosophila aggression. Dis. Model. Mech. 2017, 10, 705–716. [Google Scholar] [CrossRef]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef]
- Chow, C.Y.; Reiter, L.T. Etiology of Human Genetic Disease on the Fly. Trends Genet. 2017, 33, 391–398. [Google Scholar] [CrossRef]
- Link, N.; Bellen, H.J. Using Drosophila to drive the diagnosis and understand the mechanisms of rare human diseases. Development 2020, 147, dev191411. [Google Scholar] [CrossRef] [PubMed]
- Caizzi, R.; Moschetti, R.; Piacentini, L.; Fanti, L.; Marsano, R.M.; Dimitri, P. Comparative Genomic Analyses Provide New Insights into the Evolutionary Dynamics of Heterochromatin in Drosophila. PLoS Genet. 2016, 12, e1006212. [Google Scholar] [CrossRef] [PubMed]
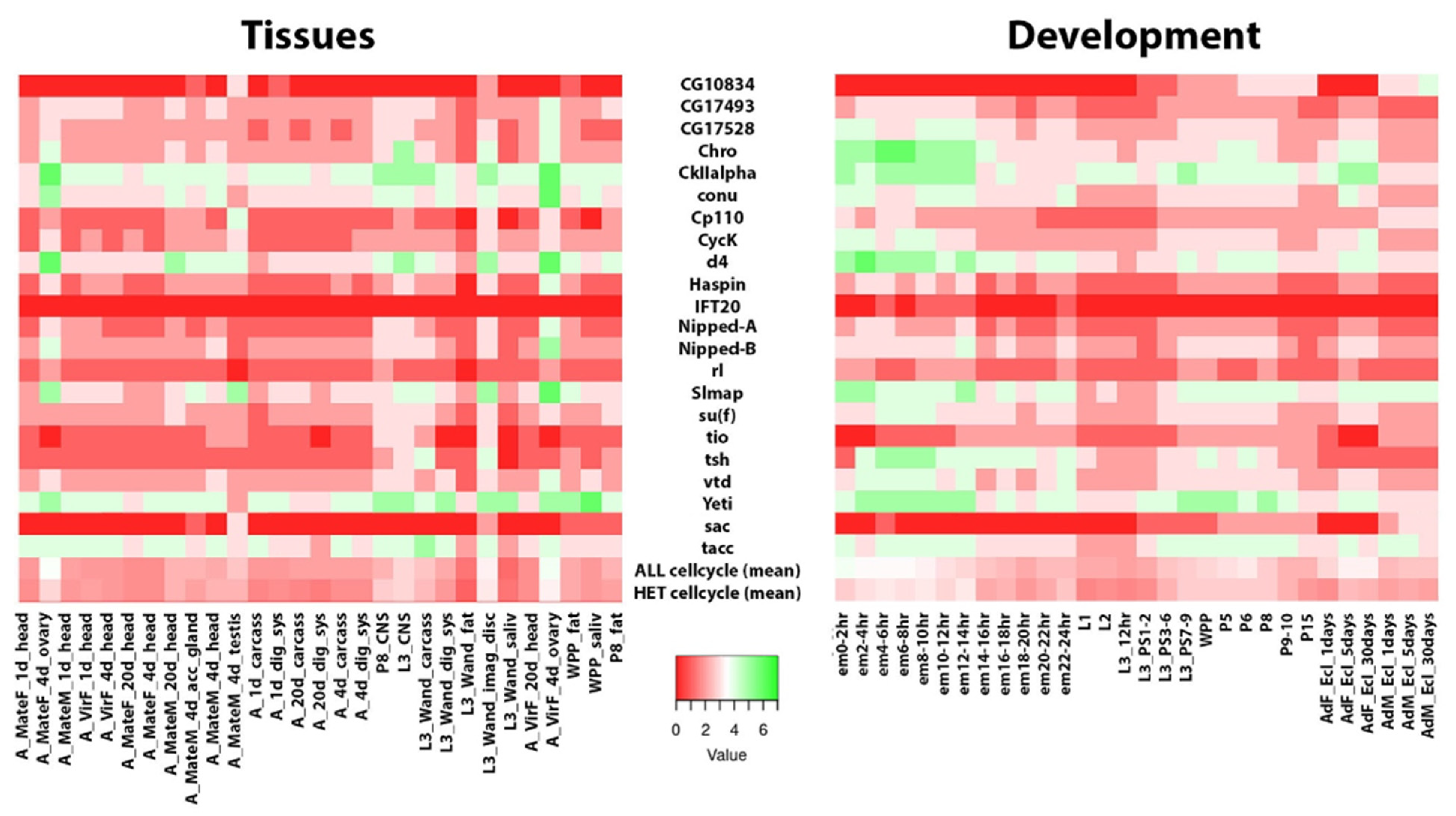

| Chrom | Name | Annotation | Polytene Map | Mitotic Map | Hsap | Ortho Map | Function |
|---|---|---|---|---|---|---|---|
| X | Cp110 | CG14617 | 20C1-20C1 | n.d. | CCP110 | 16p12.3 | centriole length regulation, ciliogenesis, cytokinesis |
| X | su(f) | CG17170 | 20E | n.d. | CSTF3 | 11p13 | mRNA binding |
| 2L | tsh | CG1374 | 40A5-40A5 | h35 [6] | TSHZ1 TSHZ2 TSHZ3 | 18q22.3 20q13.2 19q12 | chromatin organization |
| 2L | CentrinB | CG17493 | n.d. | h35 [6] | CETN1 CETN2 CETN3 | 18p11.32 Xq28 5q14.3 | ciliogenesis, centriole duplication, calcium ion binding |
| 2L | tio | CG12630 | 40D3-40D3 | h35 [6] | TSHZ1 TSHZ2 TSHZ3 | 18q22.3 20q13.2 19q12 | chromatin organization |
| 2L | CG10834 | CG10834 | 40E3-40E3 | h35 [6] | DYNLRB1 DYNLRB2 | 20q11.22 16q23.2 | dynein intermediate chain binding |
| 2L | CycK | CG15218 | 40E4-40E4 | h35 [6] | CCNK | 14q32.2 | cyclin-dependent protein serine/threonine kinase regulator |
| 2L | Slmap | CG17494 | 40F7-40F7 | h35 [6] | SLMAP | 3p14.3 | protein kinase binding |
| 2R | rl | CG12559 | 41A [8] | h41 [6] | MAPK1 | 22q11.22 | MAP kinase activity, transcription factor binding |
| 2R | Yeti | CG40218 | 41A [8] | h41 [6] | CFDP1 | 16q23.1 | chromatin remodelig, kinesin binding |
| 2R | Haspin | CG40080 | 41B-C [8] | h45 [6] | HASPIN | 17p13.2 | ATP binding, histone kinase activity, serine/threonine kinase |
| 2R | Nip-B | CG17704 | 41B3-41C1 | h46 [6] | NIPLBL | 5p13.2 | kollerin complex, sister chromatid cohesion |
| 2R | conu | CG17082 | 41C1-41C1 | h46 [6] | ARHGAP18 ARHGAP40 ARHGAP28 | 6q22.33 20q11.23 18p11.31 | GTPase activator activity |
| 2R | Dmel-doublecortin | CG17528 | 41C2-41C2 | h46 [6] | DCLK1 DCLK2 DCX | 13q13.3 4q31.23 Xq22.3 | microtubule binding, calmodulin-dependent protein kinase |
| 2R | Nip-A | CG33554 | 41C-D [8] | h46 [6] | TRRAP | 7q22.1 | chromatin remodelig, kinase activity |
| 2R | d4 | CG2682 | 41E3-41E4 | h46 [6] | DPF1 DPF3 | 11q13.1 14q24.2 | chromatin organization, zinc ion binding |
| 2R | IFT20 | CG30441 | 41E5-41E5 | n.d. | IFT20 | 17q11.2 | centrosome localization, cilium-related functions |
| 3L | Chro | CG10712 | 80B1-80B2 | eu-het junction [8] | n.d. | n.a. | cell division regulator, chromatin organization |
| 3L | CkIIα | CG17520 | 80D1-80D1 | h47 [6] | CSNK2A1 CSNK2A2 CSNK2A3 | 20p13 16q21 11p15.4 | ATP binding |
| 3L | vtd | CG17436 | 80F-80F | n.d. | RAD21 RAD21L1 | 8q24.11 20p13 | kollerin complex, sister chromatid cohesion |
| 3R | sac | CG14651 | 82B3-82B3 | h57 [6] | n.d. | n.a. | cytokinesis, ciliogenesis, microtubule motor activity |
| 3R | tacc | CG9765 | 82D2-82D2 | h57-h58 [6] | TACC1 TACC2 TACC3 | 8p11.22 10q26.13 4p16.3 | microtubule binding |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messina, G.; Prozzillo, Y.; Bizzochi, G.; Marsano, R.M.; Dimitri, P. The Green Valley of Drosophila melanogaster Constitutive Heterochromatin: Protein-Coding Genes Involved in Cell Division Control. Cells 2022, 11, 3058. https://doi.org/10.3390/cells11193058
Messina G, Prozzillo Y, Bizzochi G, Marsano RM, Dimitri P. The Green Valley of Drosophila melanogaster Constitutive Heterochromatin: Protein-Coding Genes Involved in Cell Division Control. Cells. 2022; 11(19):3058. https://doi.org/10.3390/cells11193058
Chicago/Turabian StyleMessina, Giovanni, Yuri Prozzillo, Greta Bizzochi, Renè Massimiliano Marsano, and Patrizio Dimitri. 2022. "The Green Valley of Drosophila melanogaster Constitutive Heterochromatin: Protein-Coding Genes Involved in Cell Division Control" Cells 11, no. 19: 3058. https://doi.org/10.3390/cells11193058
APA StyleMessina, G., Prozzillo, Y., Bizzochi, G., Marsano, R. M., & Dimitri, P. (2022). The Green Valley of Drosophila melanogaster Constitutive Heterochromatin: Protein-Coding Genes Involved in Cell Division Control. Cells, 11(19), 3058. https://doi.org/10.3390/cells11193058










