Synthesis and Translation of Viral mRNA in Reovirus-Infected Cells: Progress and Remaining Questions
Abstract
:1. Introduction
2. The Reovirus Particle
3. Brief Overview of Reovirus Replication Cycle
4. Progress in Our Experimental Approaches
5. Synthesis of Viral mRNA
5.1. Transcription of Viral dsRNA Genome to mRNA by Inner Core Enzymes
5.2. Synthesis of the Cap Structure on Viral mRNA
6. Impact of Reovirus Infection on Cellular mRNA
7. Protein Synthesis in Reovirus-Infected Cells
7.1. Structure of Reovirus mRNA
7.2. Synthesis of Viral Proteins during the Viral Multiplication Cycle
7.3. Impact of Reovirus Infection on Translation of Cellular mRNAs
7.4. Interferon, PKR, Stress, and the Regulation of Protein Synthesis during Reovirus Infection
8. Final remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Kohl, C.; Kurth, A. Bat Reoviruses. In Bats and Viruses: A New Frontier of Emerging Infectious Diseases; Wang, L.-F., Cowled, C., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 203–215. ISBN 978-1-118-81872-5. [Google Scholar]
- Thimmasandra Narayanappa, A.; Sooryanarain, H.; Deventhiran, J.; Cao, D.; Ammayappan Venkatachalam, B.; Kambiranda, D.; LeRoith, T.; Heffron, C.L.; Lindstrom, N.; Hall, K.; et al. A novel pathogenic mammalian orthoreovirus from diarrheic pigs and swine blood meal in the United States. mBio 2015, 6, e00593-15. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-L.; Tan, B.; Wang, B.; Li, W.; Wang, N.; Luo, C.-M.; Wang, M.-N.; Zhang, W.; Li, B.; Peng, C.; et al. Isolation and identification of bat viruses closely related to human, porcine and mink orthoreoviruses. J. Gen. Virol. 2015, 96, 3525–3531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarty, R.; Tran, H.; Selvaggi, G.; Hagerman, A.; Thompson, B.; Coffey, M. The oncolytic virus, pelareorep, as a novel anticancer agent: A review. Investig. New Drugs 2015, 33, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chester, C.; Rajasekaran, N.; He, Z.; Kohrt, H.E. Strategic combinations: The future of oncolytic virotherapy with reovirus. Mol. Cancer Ther. 2016, 15, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Bouziat, R.; Hinterleitner, R.; Brown, J.J.; Stencel-Baerenwald, J.E.; Ikizler, M.; Mayassi, T.; Meisel, M.; Kim, S.M.; Discepolo, V.; Pruijssers, A.J.; et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 2017, 356, 44–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dermody, T.S.; Parker, J.S.L.; Sherry, B. Orthoreoviruses. In Field’s Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincot, Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1304–1346. ISBN 978-1451105636. [Google Scholar]
- Dryden, K.; Coombs, K.; Yeager, M. The structure of orthoreoviruses. In Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology; Patton, J.T., Ed.; Caister Academic Press: Norfolk, UK, 2008; pp. 3–26. ISBN 978-1-904455-21-9. [Google Scholar]
- Danthi, P.; Guglielmi, K.M.; Kirchner, E.; Mainou, B.; Stehle, T.; Dermody, T.S. From touchdown to transcription: The reovirus cell entry pathway. Curr. Top. Microbiol. Immunol. 2010, 343, 91–119. [Google Scholar] [CrossRef] [PubMed]
- Ivanovic, T.; Agosto, M.A.; Chandran, K.; Nibert, M.L. A role for molecular chaperone Hsc70 in reovirus outer capsid disassembly. J. Biol. Chem. 2007, 282, 12210–12219. [Google Scholar] [CrossRef] [PubMed]
- Farsetta, D.L.; Chandran, K.; Nibert, M.L. Transcriptional activities of reovirus RNA polymerase in recoated dores: Initiation and elongation are regulated by separate mechanisms. J. Biol. Chem. 2000, 275, 39693–39701. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.; Teicher, C.; Haefliger, S.; Shmulevitz, M. Reduction of virion-associated σ1 fibers on oncolytic reovirus variants promotes adaptation toward tumorigenic cells. J. Virol. 2015, 89, 4319–4334. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, R.M.; Lahti, L.; Boehme, K.W.; Ikizler, M.; Doyle, J.D.; Dermody, T.S.; Schiff, L.A. Genetic determinants of reovirus pathogenesis in a murine model of respiratory infection. J. Virol. 2013, 87, 9279–92889. [Google Scholar] [CrossRef] [PubMed]
- Sandekian, V.; Lemay, G. Amino acids substitutions in σ1 and μ1 outer capsid proteins of a Vero cell-adapted mammalian orthoreovirus are required for optimal virus binding and disassembly. Virus Res. 2015, 196, 20–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thete, D.; Danthi, P. Protein mismatches caused by reassortment influence functions of the reovirus capsid. J. Virol. 2018, 92, e00858-18. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.H.; Ogden, K.M.; Long, J.M.; Ebenhoch, R.; Thor, A.; Dermody, T.S.; Stehle, T. Structural and functional features of the reovirus σ1 tail. J. Virol. 2018, 92, e00336-18. [Google Scholar] [CrossRef] [PubMed]
- Larson, S.M.; Antczak, J.B.; Joklik, W.K. Reovirus exists in the form of 13 particle species that differ in their content of protein sigma σ1. Virology 1994, 201, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fang, Q. High-resolution 3D structures reveal the biological runctions of reoviruses. Virol. Sin. 2013, 28, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Antar, A.; Boehme, K.; Danthi, P.; Eby, E.; Guglielmi, K.; Holm, G.; Johnson, E.; Maginnis, M.; Naik, S.A.; et al. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 2007, 1, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Boehme, K.W.; Ikizler, M.; Kobayashi, T.; Dermody, T.S. Reverse genetics for mammalian reovirus. Methods 2011, 55, 109–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemay, G. La génétique inverse dans l’étude des réovirus: Progrès, obtacles et développements futurs. Virologie 2011, 15, 53–62. [Google Scholar] [CrossRef]
- Berard, A.R.; Cortens, J.P.; Krokhin, O.; Wilkins, J.A.; Severini, A.; Coombs, K.M. Quantification of the host response proteome after mammalian reovirus T1L infection. PLoS ONE 2012, 7, e51939. [Google Scholar] [CrossRef] [PubMed]
- Berard, A.R.; Severini, A.; Coombs, K.M. Comparative proteomic analyses of two reovirus T3D subtypes and comparison to T1L identifies multiple novel proteins in key cellular pathogenic pathways. Proteomics 2015, 15, 2113–2135. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M. HeLa cell response proteome alterations induced by mammalian reovirus T3D infection. Virol. J. 2013, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Ezzati, P.; Komher, K.; Severini, G. Comparative proteomic analyses demonstrate enhanced Interferon and STAT-1 activation in reovirus T3D-infected HeLa cells. Front. Cell. Infect. Microbiol. 2015, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Poggioli, G.J.; DeBiasi, R.L.; Bickel, R.; Jotte, R.; Spalding, A.; Johnson, G.L.; Tyler, K.L. Reovirus-induced alterations in gene expression related to cell cycle regulation. J. Virol. 2002, 76, 2585–2594. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Schmechel, S.C.; Raghavan, A.; Abelson, M.; Reilly, C.; Katze, M.G.; Kaufman, R.J.; Bohjanen, P.R.; Schiff, L.A. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 2006, 80, 2019–2033. [Google Scholar] [CrossRef] [PubMed]
- Tyler, K.L.; Leser, J.S.; Phang, T.L.; Clarke, P. Gene expression in the brain during reovirus encephalitis. J. Neurovirol. 2010, 16, 56–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, S.M.; Holm, G.H.; Pierce, J.M.; Tian, B.; Watson, M.J.; Chari, R.S.; Ballard, D.W.; Brasier, A.R.; Dermody, T.S. Identification of an NF-kappaB-dependent gene network in cells infected by mammalian reovirus. J. Virol. 2006, 80, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Pandha, H.S.; Heinemann, L.; Simpson, G.R.; Melcher, A.; Prestwich, R.; Errington, F.; Coffey, M.; Harrington, K.J.; Morgan, R. Synergistic effects of oncolytic reovirus and cisplatin chemotherapy in murine malignant melanoma. Clin. Cancer Res. 2009, 15, 6158–6166. [Google Scholar] [CrossRef] [PubMed]
- DeBiasi, R.L.; Clarke, P.; Meintzer, S.; Jotte, R.; Kleinschmidt-Demasters, B.K.; Johnson, G.L.; Tyler, K.L. Reovirus-enduced alteration in expression of apoptosis and DNA repair genes with potential roles in viral pathogenesis. J. Virol. 2003, 77, 8934–8947. [Google Scholar] [CrossRef] [PubMed]
- Boudreault, S.; Martenon-Brodeur, C.; Caron, M.; Garant, J.-M.; Tremblay, M.-P.; Armero, V.E.S.; Durand, M.; Lapointe, E.; Thibault, P.; Tremblay-Létourneau, M.; et al. Global profiling of the cellular alternative RNA splicing landscape during virus-host interactions. PLoS ONE 2016, 11, e0161914. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Serrano, E.E.; Fritch, E.J.; Scholl, E.H.; Sherry, B. A cytoplasmic RNA virus alters the function of the cell splicing protein SRSF2. J. Virol. 2017, 91, e02488-16. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, N.M.; Gillies, S.C.; Bullivant, S.; Bellamy, A.R. Electron microscopy study of reovirus reaction cores. J. Virol. 1974, 14, 315–326. [Google Scholar] [PubMed]
- Drayna, D.; Fields, B.N. Activation and characterization of the reovirus transcriptase: Genetic analysis. J. Virol. 1982, 41, 110–118. [Google Scholar] [PubMed]
- Reinisch, K.M.; Nibert, M.L.; Harrison, S.C. Structure of the reovirus core at 3.6 Å resolution. J. Gen. Virol. 2000, 404, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Farsetta, D.L.; Nibert, M.L.; Harrison, S.C. RNA synthesis in a cage—Structural studies of reovirus polymerase λ3. Cell 2002, 111, 733–745. [Google Scholar] [CrossRef]
- Zhang, X.; Walker, S.B.; Chipman, P.R.; Nibert, M.L.; Baker, T.S. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 2003, 10, 1011–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, Y.J.; Harrison, S.C. Crystal structure of reovirus Polymerase λ3. In Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology; Patton, J.T., Ed.; Caister Academic Press: Norfolk, UK, 2008; pp. 227–237. ISBN 978-1-904455-21-9. [Google Scholar]
- Te Velthuis, A.J.W. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 2014, 71, 4403–4420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkataraman, S.; Prasad, B.; Selvarajan, R. RNA dependent RNA polymerases: Insights from structure, function and evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, A.A.; Lee, J.; Powers, T.R.; Nibert, M.L. Effects of viscogens on RNA transcription inside reovirus particles. J. Biol. Chem. 2011, 286, 29521–29530. [Google Scholar] [CrossRef] [PubMed]
- Mendez, I.I.; Weiner, S.G.; She, Y.-M.; Yeager, M.; Coombs, K.M. Conformational changes accompany activation of reovirus RNA-dependent RNA transcription. J. Struct. Biol. 2008, 162, 277–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.L.; Booth, T.F.; Prasad, B.V. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat. Struct. Biol. 1999, 6, 565–568. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.V.; Rothnagel, R.; Zeng, C.; Jakana, J.; Lawton, J.A.; Chiu, W.; Estes, M.K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. J. Gen. Virol. 1996, 382, 471–473. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Diprose, J.M.; Grimes, J.M.; Malby, R.; Burroughs, J.N.; Ziéntara, S.; Stuart, D.I.; Mertens, P. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 1999, 97, 481–490. [Google Scholar] [CrossRef]
- Zhang, X.; Ding, K.; Yu, X.; Chang, W.; Sun, J.; Hong Zhou, Z. In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. J. Gen. Virol. 2015, 527, 531–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Cheng, L. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science 2015, 349, 1347–1350. [Google Scholar] [CrossRef] [PubMed]
- Periz, J.; Celma, C.; Jing, B.; Pinkney, J.N.M.; Roy, P.; Kapanidis, A.N. Rotavirus mRNAS are released by transcript-specific channels in the double-layered viral capsid. Proc. Natl. Acad. Sci. USA 2013, 110, 12042–12047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemay, G. Transcriptional and translational events during reovirus infection. Biochem. Cell Biol. 1988, 66, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Roner, M.R.; Bassett, K.; Roehr, J. Identification of the 5′ sequences required for incorporation of an engineered ssRNA into the reovirus genome. Virology 2004, 329, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Roner, M.R.; Roehr, J. The 3′ sequences required for incorporation of an engineered ssRNA into the Reovirus genome. Virol. J. 2006, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Roner, M.R.; Steele, B.G. Features of the mammalian orthoreovirus 3 Dearing l1 single-stranded RNA that direct packaging and serotype restriction. J. Gen. Virol. 2007, 88, 3401–3412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roner, M.R.; Steele, B.G. Localizing the reovirus packaging signals using an engineered m1 and s2 ssRNA. Virology 2007, 358, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, A.A.; Blattman, J.N.; Blattman, N.N.; Greenberg, P.D.; Nibert, M.L. Engineering recombinant reoviruses with tandem repeats and a tetravirus 2A-like element for exogenous polypeptide expression. Proc. Natl. Acad. Sci. USA 2013, 110, E1867–E1876. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.C.; Denison, M.R. Coronaviruses as DNA wannabes: A new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013, 9, e1003760. [Google Scholar] [CrossRef] [PubMed]
- Sanjuan, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [PubMed]
- Collier, A.M.; Lyytinen, O.L.; Guo, Y.R.; Toh, Y.; Poranen, M.M.; Tao, Y.J. Initiation of RNA polymerization and polymerase encapsidation by a small dsRNA virus. PLoS Pathog. 2016, 12, e1005523. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Mcdonald, S.M.; Tortorici, M.A.; Tao, Y.J.; Carpio, R.V.-D.; Nibert, M.L.; Patton, J.T.; Harrison, S.C. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 2008, 16, 1678–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, E.; Roy, P. Bluetongue virus VP1 polymerase activity in vitro: Template dependency, dinucleotide priming and cap dependency. PLoS ONE 2011, 6, e27702. [Google Scholar] [CrossRef] [PubMed]
- Starnes, M.C.; Joklik, W.K. Reovirus protein λ3 is a poly(C)-dependent poly(G) polymerase. Virology 1993, 193, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Cheang, M.; Coombs, K.M. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J. Virol. 1996, 70, 1223–1227. [Google Scholar] [PubMed]
- Noble, S.; Nibert, M.L. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 1997, 71, 7728–7735. [Google Scholar] [PubMed]
- Hermann, L.L.; Coombs, K.M. Inhibition of reovirus by mycophenolic acid is associated with the M1 genome segment. J. Virol. 2004, 78, 6171–6179. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M. Identification and characterization of a double-stranded RNA- reovirus temperature-sensitive mutant defective in minor core protein μ2. J. Virol. 1996, 70, 4237–4245. [Google Scholar] [PubMed]
- Ooms, L.S.; Jerome, W.G.; Dermody, T.S.; Chappell, J.D. Reovirus replication protein μ2 influences cell tropism by promoting particle assembly within viral inclusions. J. Virol. 2012, 86, 10979–10987. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.N.M.; Stanifer, M.L.; Höhn, K.; Engel, U.; Haselmann, U.; Bartenschlager, R.; Kräusslich, H.-G.; Krijnse-Locker, J.; Boulant, S. Genome packaging of reovirus is mediated by the scaffolding property of the microtubule network. Cell. Microbiol. 2017, 19, e12765. [Google Scholar] [CrossRef] [PubMed]
- Bisaillon, M.; Bergeron, J.; Lemay, G. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus λ1 protein. J. Biol. Chem. 1997, 272, 18298–18303. [Google Scholar] [CrossRef] [PubMed]
- Noble, S.; Nibert, M.L. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J. Virol. 1997, 71, 2182–2191. [Google Scholar] [PubMed]
- Kim, J.; Zhang, X.; Centonze, V.E.; Bowman, V.D.; Noble, S.; Baker, T.S.; Nibert, M.L. The hydrophilic amino-terminal arm of reovirus core shell protein λ1 is dispensable for particle assembly. J. Virol. 2002, 76, 12211–12222. [Google Scholar] [CrossRef] [PubMed]
- Dryden, K.A.; Farsetta, D.L.; Wang, G.; Keegan, J.M.; Fields, B.N.; Baker, T.S.; Nibert, M.L. Internal/structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 1998, 245, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Lanoie, D.; Lemay, G. Multiple proteins differing between laboratory stocks of mammalian orthoreoviruses affect both virus sensitivity to interferon and induction of interferon production during infection. Virus Res. 2018, 247, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Bisaillon, M.; Lemay, G. Molecular dissection of the reovirus λ1 protein nucleic acids binding site. Virus Res. 1997, 51, 231–237. [Google Scholar] [CrossRef]
- Lemay, G.; Danis, C. Reovirus λ1 protein: Affinity for double-stranded nucleic acids by a small amino-terminal region of the protein independent from the zinc finger motif. J. Gen. Virol. 1994, 75 Pt 11, 3261–3266. [Google Scholar] [CrossRef]
- Bisaillon, M.; Lemay, G. Characterization of the reovirus λ1 protein RNA 5′-triphosphatase activity. J. Biol. Chem. 1997, 272, 29954–29957. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Parker, J.S.L.; Murray, K.E.; Nibert, M.L. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. J. Biol. Chem. 2004, 279, 4394–4403. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Arnold, M.M.; Nibert, M.L. Silencing and complementation of reovirus core protein μ2: Functional correlations with μ2–microtubule association and differences between virus- and plasmid-derived μ2. Virology 2007, 364, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ooms, L.S.; Chappell, J.D.; Dermody, T.S. Identification of functional domains in reovirus replication proteins μNS and μ2. J. Virol. 2009, 83, 2892–2906. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, F.; Su, R.; Li, X.; Chen, W.; Chen, Q.; Yang, T.; Wang, J.; Liu, H.; Fang, Q.; et al. Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly. Proc. Natl. Acad. Sci. USA 2018, 115, 7344–7349. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, D.R.; Zarbl, H.; Millward, S. Reovirus guanylyltransferase is L2-gene product λ2. J. Virol. 1986, 60, 307–311. [Google Scholar] [PubMed]
- Mao, Z.; Joklik, W.K. Isolation and enzymatic characterization of protein λ2, the reovirus guanylyltransferase. Virology 1991, 185, 377–386. [Google Scholar] [CrossRef]
- Fausnaugh, J.; Shatkin, A.J. Active site localization in a viral mRNA capping enzyme. J. Biol. Chem. 1990, 265, 7669–7672. [Google Scholar] [PubMed]
- Bujnicki, J.M.; Rychlewski, L. Reassignment of specificities of two cap methyltransferase domains in the reovirus protein λ2. Genome Biol. 2001, 2. [Google Scholar] [CrossRef]
- Zhu, B.; Yang, C.; Liu, H.; Cheng, L.; Song, F.; Zeng, S.; Huang, X.; Ji, G.; Zhu, P. Identification of the active sites in the methyltransferases of a transcribing dsRNA virus. J. Mol. Biol. 2014, 426, 2167–2174. [Google Scholar] [CrossRef] [PubMed]
- Sandekian, V.; Lemay, G. A single amino acid substitution in the mRNA capping enzyme λ2 of a mammalian orthoreovirus mutant increases interferon sensitivity. Virology 2015, 483, 229–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diamond, M.S. IFIT1: A dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev. 2014, 25, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.L.; Diamond, M.S. Innate immune restriction and antagonism of viral RNA lacking 2′-O methylation. Virology 2015, 479-480, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.W.; Amarasinghe, G.K. When your cap matters: Structural insights into self vs non-self recognition of 5′ RNA by immunomodulatory host proteins. Curr. Opin. Struct. Biol. 2016, 36, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Zarbl, H.; Millward, S. The reovirus multiplication cycle. In The Reoviridae; Joklik, W.K., Ed.; Plenum Press: New York, NY, USA; London, UK, 1983; pp. 107–196. ISBN 978-1-489905-80-2. [Google Scholar]
- Shatkin, A.J.; Both, G.W. Reovirus mRNA: Transcription and translation. Cell 1976, 7, 305–313. [Google Scholar] [CrossRef]
- Werner, M.; Purta, E.; Kaminska, K.H.; Cymerman, I.A.; Campbell, D.A.; Mittra, B.; Zamudio, J.R.; Sturm, N.R.; Jaworski, J.; Bujnicki, J.M. 2′-O-ribose methylation of cap2 in human: Function and evolution in a horizontally mobile family. Nucl. Acids Res. 2011, 39, 4756–4768. [Google Scholar] [CrossRef] [PubMed]
- Byszewska, M.; Śmietański, M.; Purta, E.; Bujnicki, J.M. RNA methyltransferases involved in 5′ cap biosynthesis. RNA Biol. 2015, 11, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. mRNA capping: Biological functions and applications. Nucl. Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [PubMed]
- Langberg, S.R.; Moss, B. Purification and characterization of cap I and cap II RNA (nucleoside-2′-methyltransferase) from HeLa cells. J. Biol. Chem. 1981, 256, 10054–10060. [Google Scholar] [PubMed]
- Sen, G.C.; Shaila, S.; Lebleu, B.; Brown, G.E.; Desrosiers, R.C.; Lengyel, P. Impairment of reovirus messenger-RNA methylation in extracts of interferon-rreated Ehrlich ascites tumor cells—Further characteristics of phenomenon. J. Virol. 1977, 21, 69–83. [Google Scholar] [PubMed]
- Desrosiers, R.C.; Lengyel, P. Impairment of reovirus messenger-RNA cap methylation in interferon-treated mouse L929 cells. Biochim. Biophys. Acta 1979, 562, 471–480. [Google Scholar] [CrossRef]
- De Ferra, F.; Baglioni, C. Increase in S-adenosylhomocysteine concentration in interferon-treated HeLa cells and inhibition of methylation of vesicular stomatitis virus mRNA. J. Biol. Chem. 1983, 258, 2118–2121. [Google Scholar] [PubMed]
- Skup, D.; Millward, S. mRNA capping enzymes are masked in reovirus progeny subviral particles. J. Virol. 1980, 34, 490–496. [Google Scholar] [PubMed]
- Zarbl, H.; Skup, D.; Millward, S. Reovirus progeny subviral particles synthesize uncapped mRNA. J. Virol. 1980, 34, 497–505. [Google Scholar] [PubMed]
- Skup, D.; Zarbl, H.; Millward, S. Regulation of translation in L-cells infected with reovirus. J. Mol. Biol. 1981, 151, 35–55. [Google Scholar] [CrossRef]
- Hazelton, P.R.; Coombs, K.M. The reovirus mutant tsA279 L2 gene is associated with generation of a spikeless core particle: Implications for capsid assembly. J. Virol. 1999, 73, 2298–2308. [Google Scholar] [PubMed]
- Broering, T.J.; McCutcheon, A.M.; Centonze, V.E.; Nibert, M.L. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 2000, 74, 5516–5524. [Google Scholar] [CrossRef] [PubMed]
- Habjan, M.; Andersson, I.; Klingström, J.; Schümann, M.; Martin, A.; Zimmermann, P.; Wagner, V.; Pichlmair, A.; Schneider, U.; Mühlberger, E.; et al. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE 2008, 3, e2032. [Google Scholar] [CrossRef] [PubMed]
- Habjan, M.; Hubel, P.; Lacerda, L.; Benda, C.; Holze, C.; Eberl, C.H.; Mann, A.; Kindler, E.; Gil-Cruz, C.; Ziebuhr, J.; et al. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog. 2013, 9, e1003663. [Google Scholar] [CrossRef] [PubMed]
- Habjan, M.; Pichlmair, A. Cytoplasmic sensing of viral nucleic acids. Curr. Opin. Virol. 2015, 11, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Luecke, S.; Paludan, S.R. Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine 2016, 98, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Devarkar, S.C.; Wang, C.; Miller, M.T.; Ramanathan, A.; Jiang, F.; Khan, A.G.; Patel, S.S.; Marcotrigiano, J. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. USA 2016, 113, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Uzri, D.; Greenberg, H.B. Characterization of rotavirus RNAs that activate innate immune signaling through the RIG-I-like receptors. PLoS ONE 2013, 8, e69825. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, A.H.; Fields, B.N. Reovirus inhibition of cellular RNA and protein-synthesis—Role of the S4 gene. Virology 1982, 122, 381–391. [Google Scholar] [CrossRef]
- Zakaryan, H.; Stamminger, T. Nuclear remodeling during viral infections. Cell. Microbiol. 2011, 13, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Flather, D.; Semler, B.L. Picornaviruses and nuclear functions: Targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015, 6, 594. [Google Scholar] [CrossRef] [PubMed]
- Wulan, W.N.; Heydet, D.; Walker, E.J.; Gahan, M.E.; Ghildyal, R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. 2015, 6, 553. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.E. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology 2015, 479–480, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Rubio, R.M. Rotavirus prevents the expression of host responses by blocking the nucleo-cytoplasmic transport of polyadenylated mRNAs. J. Virol. 2013, 87, 6336–6345. [Google Scholar] [CrossRef] [PubMed]
- López, S.; Oceguera, A.; Sandoval-Jaime, C. Stress response and translation control in rotavirus infection. Viruses 2016, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Shatkin, A.J. Regulated, stable expression and nuclear presence of reovirus double-stranded RNA-binding protein σ3 in HeLa cells. J. Virol. 1996, 70, 3497–3501. [Google Scholar] [PubMed]
- Bergeron, J.; Mabrouk, T.; Garzon, S.; Lemay, G. Characterization of the thermosensitive ts453 reovirus mutant: Increased dsRNA binding of σ3 protein correlates with interferon resistance. Virology 1998, 246, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, C.C.; Bouchard, R.J.; Tyler, K.L. Novel nuclear herniations induced by nuclear localization of a viral protein. J. Virol. 2004, 78, 6360–6369. [Google Scholar] [CrossRef] [PubMed]
- Zurney, J.; Kobayashi, T.; Holm, G.H.; Dermody, T.S.; Sherry, B. Reovirus μ2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J. Virol. 2009, 83, 2178–2187. [Google Scholar] [CrossRef] [PubMed]
- Boehme, K.W.; Hammer, K.; Tollefson, W.C.; Konopka-Anstadt, J.L.; Kobayashi, T.; Dermody, T.S. Nonstructural protein σ1s mediates reovirus-induced cell cycle arrest and apoptosis. J. Virol. 2013, 87, 12967–12979. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Kuo, R.-L.; Lin, J.-Y.; Huang, P.-N.; Huang, Y.; Liu, H.; Arnold, J.J.; Chen, S.-J.; Wang, R.Y.-L.; Cameron, C.E.; et al. Cytoplasmic viral RNA-dependent RNA polymerase disrupts the intracellular splicing machinery by entering the nucleus and interfering with Prp8. PLoS Pathog. 2014, 10, e1004199. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Smith, J.A.; Abelson, M.; Vlasova-St Louis, I.; Schiff, L.A.; Bohjanen, P.R. Reovirus infection induces stabilization and up-regulation of cellular transcripts that encode regulators of TGF-β signaling. PLoS ONE 2018, 13, e0204622. [Google Scholar] [CrossRef] [PubMed]
- Haimov, O.; Sinvani, H.; Dikstein, R. Cap-dependent, scanning-free translation initiation mechanisms. Biochem. Biophys. Acta 2015, 1849, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.; Thompson, S.R. Noncanonical translation initiation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, a032672. [Google Scholar] [CrossRef] [PubMed]
- Roner, M.R.; Roner, L.A.; Joklik, W.K. Translation of reovirus RNA species m1 can initiate at either of the first two in-frame initiation codons. Proc. Natl. Acad. Sci. USA 1993, 90, 8947–8951. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Brown, E.G. Translation of the reovirus M1 gene initiates from the first AUG codon in both infected and transfected cells. Virus Res. 1996, 40, 75–89. [Google Scholar] [CrossRef]
- Swanson, M.I.; She, Y.-M.; Ens, W.; Brown, E.G.; Coombs, K.M. Mammalian reovirus core protein μ2 initiates at the first start codon and is acetylated. Rapid Commun. Mass Spectrom. 2002, 16, 2317–2324. [Google Scholar] [CrossRef] [PubMed]
- Ernst, H.; Shatkin, A.J. Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc. Natl. Acad. Sci. USA 1985, 82, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, B. L.; Samuel, C.E. Biosynthesis of reovirus-specified polypeptides—The reovirus S1 messenger-RNA encodes 2 primary translation products. Virology 1985, 143, 63–74. [Google Scholar] [CrossRef]
- Sarkar, G.; Pelletier, J.; Bassel-Duby, R.; Jayasuriya, A.; Fields, B.N.; Sonenberg, N. Identification of a new polypeptide coded by reovirus gene S1. J. Virol. 1985, 54, 720–725. [Google Scholar] [PubMed]
- Mouilleron, H.; Delcourt, V.; Roucou, X. Death of a dogma: Eukaryotic mRNAs can code for more than one protein. Nucl. Acids Res. 2016, 44, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Busch, L.K.; Rodriguez-Grille, J.; Casal, J.I.; Martinez-Costas, J.; Benavente, J. Avian and mammalian reoviruses use different molecular mechanisms to synthesize their μNS isoforms. J. Gen. Virol. 2011, 92, 2566–2574. [Google Scholar] [CrossRef] [PubMed]
- Sagar, V.; Murray, K.E. The mammalian orthoreovirus bicistronic M3 mRNA initiates translation using a 5′ end-dependent, scanning mechanism that does not require interaction of 5′-3′ untranslated regions. Virus Res. 2014, 183, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Racine, T.; Barry, C.; Roy, K.; Dawe, S.J.; Shmulevitz, M.; Duncan, R. Leaky scanning and scanning-independent ribosome migration on the tricistronic S1 mRNA of avian reovirus. J. Biol. Chem. 2007, 282, 25613–25622. [Google Scholar] [CrossRef] [PubMed]
- Racine, T.; Duncan, R. Facilitated leaky scanning and atypical ribosome shunting direct downstream translation initiation on the tricistronic S1 mRNA of avian reovirus. Nucl. Acids Res. 2010, 38, 7260–7272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roner, M.R.; Gaillard, R.K.; Joklik, W.K. Control of reovirus messenger RNA translation efficiency by the regions upstream of initiation codons. Virology 1989, 168, 292–301. [Google Scholar] [CrossRef]
- Jan, E.; Mohr, I.; Walsh, D. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Ann. Rev. Virol. 2016, 3, 283–307. [Google Scholar] [CrossRef] [PubMed]
- Merrick, W.C.; Pavitt, G.D. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb. Perspect. Biol. 2018, a033092. [Google Scholar] [CrossRef] [PubMed]
- Kahvejian, A.; Roy, G.; Sonenberg, N. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001, 66, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Lemay, G.; Millward, S. Expression of the cloned S4 gene of reovirus serotype 3 in transformed eucaryotic cells: Enrichment of the viral protein in the crude initiation factor fraction. Virus Res. 1986, 6, 133–140. [Google Scholar] [CrossRef]
- Gratia, M.; Sarot, E.; Charpilienne, A.; Baron, C.H.; Duarte, M.; Pyronnet, S.; Poncet, D. Rotavirus NSP3 is a translational surrogate of the PABP-poly(A) complex. J. Virol. 2015, 89, 8773–8782. [Google Scholar] [CrossRef] [PubMed]
- Gratia, M.; Vende, P.; Charpilienne, A.; Baron, H.C.; Laroche, C.; Sarot, E.; Pyronnet, S.; Duarte, M.; Poncet, D. Challenging the roles of NSP3 and untranslated regions in rotavirus mRNA translation. PLoS ONE 2016, 11, e0145998. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, R.; Lemay, G.; Millward, S. The viral protein σ3 participates in translation of late viral mRNA in reovirus-infected L cells. J. Virol. 1987, 61, 2472–2479. [Google Scholar] [PubMed]
- Lemay, G. Study of a Reovirus Protein Involved in Viral mRNA Translation. Ph.D. Thesis, McGill University, Montreal, QC, Canada, 1987. [Google Scholar]
- Walsh, D.; Mathews, M.B.; Mohr, I. Tinkering with translation: Protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2013, 5, a012351. [Google Scholar] [CrossRef] [PubMed]
- Stern-Ginossar, N.; Thompson, S.R.; Mathews, M.B.; Mohr, I. Translational control in virus-infected cells. Cold Spring Harb. Perspect. Biol 2018, a033001. [Google Scholar] [CrossRef] [PubMed]
- Boyce, M.; Celma, C.C.P.; Roy, P. Bluetongue virus non-structural protein 1 is a positive regulator of viral protein synthesis. Virol. J. 2012, 9, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.; Wang, Y.; Zhang, E.; Han, X.; Yu, Z.; Liu, H. VP1 and VP3 are required and sufficient for translation initiation of uncapped IBDV genomic dsRNA. J. Virol. 2017, 92, e01345-17. [Google Scholar] [CrossRef] [PubMed]
- Greco, A. Involvement of the nucleolus in replication of human viruses. Rev. Med. Virol. 2009, 19, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Hiscox, J.A.; Whitehouse, A.; Matthews, D.A. Nucleolar proteomics and viral infection. Proteomics 2010, 10, 4077–4086. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.M.; Moseley, G.W. The nucleolar interface of RNA viruses. Cell. Microbiol. 2015, 17, 1108–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiscox, J.A. RNA viruses: Hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007, 5, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, A.; Greco, A. Viruses and the nucleolus: The fatal attraction. Biochim. Biophys. Acta 2014, 1842, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.B.; Stuart, J.D.; Simon, E.J.; Boehme, K.W. Nonstructural protein σ1s is required for optimal reovirus protein expression. J. Virol. 2018, 92, e02259-17. [Google Scholar] [CrossRef] [PubMed]
- Lanoie, D.; Côté, S.; Degeorges, E.; Lemay, G. A single mutation in the mammalian orthoreovirus S1 gene is responsible for increased interferon sensitivity in a virus mutant selected in Vero cells. Virology. under revision.
- Desmet, E.A.; Anguish, L.J.; Parker, J.S.L. Virus-mediated compartmentalization of the host translational machinery. mBio 2014, 5, e01463-14. [Google Scholar] [CrossRef] [PubMed]
- Katsafanas, G.C.; Moss, B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2007, 2, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Castelló, A.; Quintas, A.; Sánchez, E.G.; Sabina, P.; Nogal, M.; Carrasco, L.; Revilla, Y. Regulation of host translational machinery by African Swine Fever Virus. PLoS Pathog. 2009, 5, e1000562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez de Castro, I.; Zamora, P.F.; Ooms, L.; Fernandez, J.J.; Lai, C.M.H.; Mainou, B.A.; Dermody, T.S.; Risco, C. Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes. mBio 2013, 5, e00931-13. [Google Scholar] [CrossRef] [PubMed]
- Schmechel, S.; Chute, M.; Skinner, P.; Anderson, R.; Schiff, L. Preferential translation of reovirus mRNA by a σ3-dependent mechanism. Virology 1997, 232, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Ray, B.K.; Brendler, T.G.; Adya, S.; Daniels-McQueen, S.; Miller, J.K.; Hershey, J.W.; Grifo, J.A.; Merrick, W.C.; Thach, R.E. Role of mRNA competition in regulating translation: Further characterization of mRNA discriminatory initiation factors. Proc. Natl. Acad. Sci. USA 1983, 80, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Detjen, B.M.; Walden, W.E.; Thach, R.E. The role of messenger-RNA competition in regulating translation: Translational specificity in reovirus-infected mouse fibroblasts. J. Biol. Chem. 1982, 257, 9855–9860. [Google Scholar] [PubMed]
- Lemieux, R.; Zarbl, H.; Millward, S. mRNA discrimination in extracts from uninfected and reovirus-infected L-cells. J. Virol. 1984, 51, 215–222. [Google Scholar] [PubMed]
- Danis, C.; Lemay, G. Protein synthesis in different cell lines infected with orthoreovirus serotype 3: Inhibition of host-cell protein synthesis correlates with accelerated viral multiplication and cell killing. Biochem. Cell Biol. 1993, 71, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Danis, C.; Mabrouk, T.; Garzon, S.; Lemay, G. Establishment of persistent reovirus infection in SC1 cells: Absence of protein synthesis inhibition and increased level of double-stranded RNA-activated protein kinase. Virus Res. 1993, 27, 253–265. [Google Scholar] [CrossRef]
- Skup, D.; Millward, S. Reovirus-induced modification of cap-dependent translation in infected L cells. Proc. Natl. Acad. Sci. USA 1980, 77, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Zarbl, H. The Structure and Translation of Late Reovirus mRNA in infected L Cells. Ph.D. Thesis, McGill University, Montreal, QC, Canada, 1983. [Google Scholar]
- Sonenberg, N.; Skup, D.; Trachsel, H.; Millward, S. In vitro translation in reovirus- and poliovirus-infected cell extracts. Effects of anti-cap binding protein monoclonal antibody. J. Biol. Chem. 1981, 256, 4138–4141. [Google Scholar] [PubMed]
- Kleijn, M.; Vrins, C.L.; Voorma, H.O.; Thomas, A.A. Phosphorylation state of the cap-binding protein eIF4E during viral infection. Virology 1996, 217, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Shatkin, A.J. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 1997, 234, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.A.; Gil, J.; Ventoso, I.; Guerra, S.; Domingo, E.; Rivas, C.; Esteban, M. Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006, 70, 1032–1060. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Berg, M. The multiple faces of protein kinase R in antiviral defense. Virulence 2014, 4, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Dalet, A.; Gatti, E.; Pierre, P. Integration of PKR-dependent translation inhibition with innate immunity is required for a coordinated anti-viral response. FEBS Lett. 2015, 589, 1539–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dzananovic, E.; McKenna, S.A.; Patel, T.R. Viral proteins targeting host protein kinase R to evade an innate immune response: A mini review. Biotechnol. Genet. Eng. Rev. 2018, 34, 33–59. [Google Scholar] [CrossRef] [PubMed]
- Hoang, H.-D.; Graber, T.E.; Alain, T. Battling for ribosomes: Translational control at the forefront of the antiviral response. J. Mol. Biol. 2018, 430, 1965–1992. [Google Scholar] [CrossRef] [PubMed]
- Imani, F.; Jacobs, B.L. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 σ3 protein. Proc. Natl. Acad. Sci. USA 1988, 85, 7887–7891. [Google Scholar] [CrossRef] [PubMed]
- Denzler, K.L.; Jacobs, B.L. Site-directed mutagenic analysis of reovirus σ3 protein binding to dsRNA. Virology 1994, 204, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, T.; Danis, C.; Lemay, G. Two basic motifs of reovirus σ3 protein are involved in double-stranded RNA binding. Biochem. Cell Biol. 1995, 73, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Beattie, E.; Denzler, K.L.; Tartaglia, J.; Perkus, M.E.; Paoletti, E.; Jacobs, B.L. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 1995, 69, 499–505. [Google Scholar] [PubMed]
- Lloyd, R.M.; Shatkin, A.J. Translational stimulation by reovirus polypeptide σ3: Substitution for VAI RNA and inhibition of phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 1992, 66, 6878–6884. [Google Scholar] [PubMed]
- Gainey, M.D.; Dillon, P.J.; Clark, K.M.; Manuse, M.J.; Parks, G.D. Paramyxovirus-induced shutoff of host and viral protein synthesis: Role of the P and V proteins in limiting PKR activation. J. Virol. 2008, 82, 828–839. [Google Scholar] [CrossRef] [PubMed]
- Spurgeon, M.E.; Ornelles, D.A. The adenovirus E1B 55-kilodalton and E4 open reading frame 6 proteins limit phosphorylation of eIF2 during the late phase of infection. J. Virol. 2009, 83, 9970–9982. [Google Scholar] [CrossRef] [PubMed]
- Danis, C.; Mabrouk, T.; Faure, M.; Lemay, G. Interferon has no protective effect during acute or persistent reovirus infection of mouse SC1 fibroblasts. Virus Res. 1997, 51, 139–149. [Google Scholar] [CrossRef]
- Strong, J.E.; Coffey, M.C.; Tang, D.; Sabinin, P.; Lee, P.W. The molecular basis of viral oncolysis: Usurpation of the Ras signaling pathway by reovirus. EMBO J. 1998, 17, 3351–3362. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Carroll, K.; Hastings, C.; Miller, C.L. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2 phosphorylation and PKR. J. Virol. 2011, 85, 8798–8810. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Schmechel, S.C.; Williams, B.R.G.; Silverman, R.H.; Schiff, L.A. Involvement of the interferon-regulated antiviral proteins PKR and RNase L in reovirus-induced shutoff of cellular translation. J. Virol. 2005, 79, 2240–2250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Samuel, C.E. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J. Virol. 2007, 81, 8192–8200. [Google Scholar] [CrossRef] [PubMed]
- Shepard, D.A.; Ehnstrom, J.G.; Skinner, P.J.; Schiff, L.A. Mutations in the zinc-binding motif of the reovirus capsid protein σ3 eliminate its ability to associate with capsid protein μ1. J. Virol. 1996, 70, 2065–2068. [Google Scholar] [PubMed]
- Schiff, L.A. Reovirus capsid proteins σ3 and μ1: Interactions that influence viral entry, assembly, and translational control. Curr. Top. Microbiol. Immunol. 1998, 233, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Schiff, L.A. Structure and functions of the orthoreovirus σ3 protein. In Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology; Patton, J.T., Ed.; Caister Academic Press: Norfolk, UK, 2008; pp. 173–188. ISBN 978-1-904455-21-9. [Google Scholar]
- Miller, J.E.; Samuel, C.E. Proteolytic cleavage of the reovirus σ3 protein results in enhanced double-stranded RNA-binding activity: Identification of a repeated basic amino acid motif within the C-terminal binding region. J. Virol. 1992, 66, 5347–5356. [Google Scholar] [PubMed]
- Wang, Q.; Bergeron, J.; Mabrouk, T.; Lemay, G. Site-directed mutagenesis of the double-stranded RNA binding domain of bacterially-expressed σ3 reovirus protein. Virus Res. 1996, 41, 141–151. [Google Scholar] [CrossRef]
- Olland, A.M.; Jané-Valbuena, J.; Schiff, L.A.; Nibert, M.L.; Harrison, S.C. Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J. 2001, 20, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Jané-Valbuena, J. Analysis of the Structural and dsRNA Binding Activities of Reovirus Protein σ3. Ph.D. Thesis, University of Wisconsin, Madison, WI, USA, 2002. [Google Scholar]
- Knowlton, J.J.; de Castro, I.F.X.N.; Ashbrook, A.W.; Gestaut, D.R.; Zamora, P.F.; Bauer, J.A.; Forrest, J.C.; Frydman, J.; Risco, C.; Dermody, T.S. The TRiC chaperonin controls reovirus replication through outer-capsid folding. Nat. Microbiol. 2018, 3, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.H.; MacDonald, M.R.; Rice, C.M. To translate, or not to translate: Viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. 2015, 25, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Goubau, D.; Schlee, M.; Deddouche, S.; Pruijssers, A.J.; Zillinger, T.; Goldeck, M.; Schuberth, C.; Van der Veen, A.G.; Fujimura, T.; Rehwinkel, J.; et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 2014, 514, 372–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shmulevitz, M.; Pan, L.-Z.; Garant, K.; Pan, D.; Lee, P.W.K. Oncogenic Ras promotes reovirus spread by suppressing IFN-β production through negative regulation of RIG-I signaling. Cancer Res. 2010, 70, 4912–4921. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P. Correlation between interferon sensitivity of reovirus isolates and ability to discriminate between normal and Ras-transformed cells. J. Gen. Virol. 2005, 86, 1489–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsoulidis, E.; Kaur, S.; Platanias, L.C. Deregulation of interferon signaling in malignant cells. Pharmaceuticals 2010, 3, 406–418. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, C.; Iankov, I.D.; Anderson, S.K.; Aderca, I.; Leontovich, A.A.; Maurer, M.J.; Oberg, A.L.; Schroeder, M.A.; Giannini, C.; Greiner, S.M.; et al. Constitutive interferon pathway activation in tumors as an efficacy determinant following oncolytic virotherapy. J. Natl. Cancer Inst. 2018, 110, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.; Johnston, R.; Shmulevitz, M. Potential for improving potency and specificity of reovirus oncolysis with next-generation reovirus variants. Viruses 2015, 7, 6251–6278. [Google Scholar] [CrossRef] [PubMed]
- Kemp, V.; Hoeben, R.; van den Wollenberg, D. Exploring reovirus plasticity for improving its use as oncolytic virus. Viruses 2016, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Norman, K.L.; Hirasawa, K.; Yang, A.-D.; Shields, M.A.; Lee, P.W.K. Reovirus oncolysis: The Ras/RalGEF/p38 pathway dictates host cell permissiveness to reovirus infection. Proc. Natl. Acad. Sci. USA 2004, 101, 11099–11104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, E.-H.; Park, E.H.; Cho, I.-R.; Srisuttee, R.; Min, H.-J.; Oh, M.-J.; Jeong, Y.-J.; Jhun, B.H.; Johnston, R.N.; Lee, S.; et al. CUG2, a novel oncogene confers reoviral replication through Ras and p38 signaling pathway. Cancer Gene Ther. 2010, 17, 307–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourhill, T.; Mori, Y.; Rancourt, D.; Shmulevitz, M.; Johnston, R. Going (reo)viral: Factors promoting successful reoviral oncolytic infection. Viruses 2018, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Hastings, C.; Miller, C.L. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 2009, 83, 11090–11101. [Google Scholar] [CrossRef] [PubMed]
- Valiente-Echeverría, F.; Melnychuk, L.; Mouland, A.J. Viral modulation of stress granules. Virus Res. 2012, 169, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Reineke, L.C.; Lloyd, R.E. Diversion of stress granules and P-bodies during viral infection. Virology 2013, 436, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-C.; Lloyd, R.E. Cytoplasmic RNA granules and viral infection. Ann. Rev. Virol. 2014, 1, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.G.; Smith, J.A.; Balachandran, S.; Perwitasari, O.; Proll, S.C.; Thomas, M.J.; Korth, M.J.; Barber, G.N.; Schiff, L.A.; Katze, M.G. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J. Virol. 2007, 81, 2221–2230. [Google Scholar] [CrossRef] [PubMed]
- Carroll, K.; Hastings, C.; Miller, C.L. Amino acids 78 and 79 of mammalian orthoreovirus protein µNS are necessary for stress granule localization, core protein λ2 interaction, and de novo virus replication. Virology 2014, 448, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, P.; Bussiere, L.; Miller, C.L. Mammalian orthoreovirus factories modulate stress granule protein localization by interaction with G3BP1. J. Virol. 2017, 91, e01298-17. [Google Scholar] [CrossRef] [PubMed]
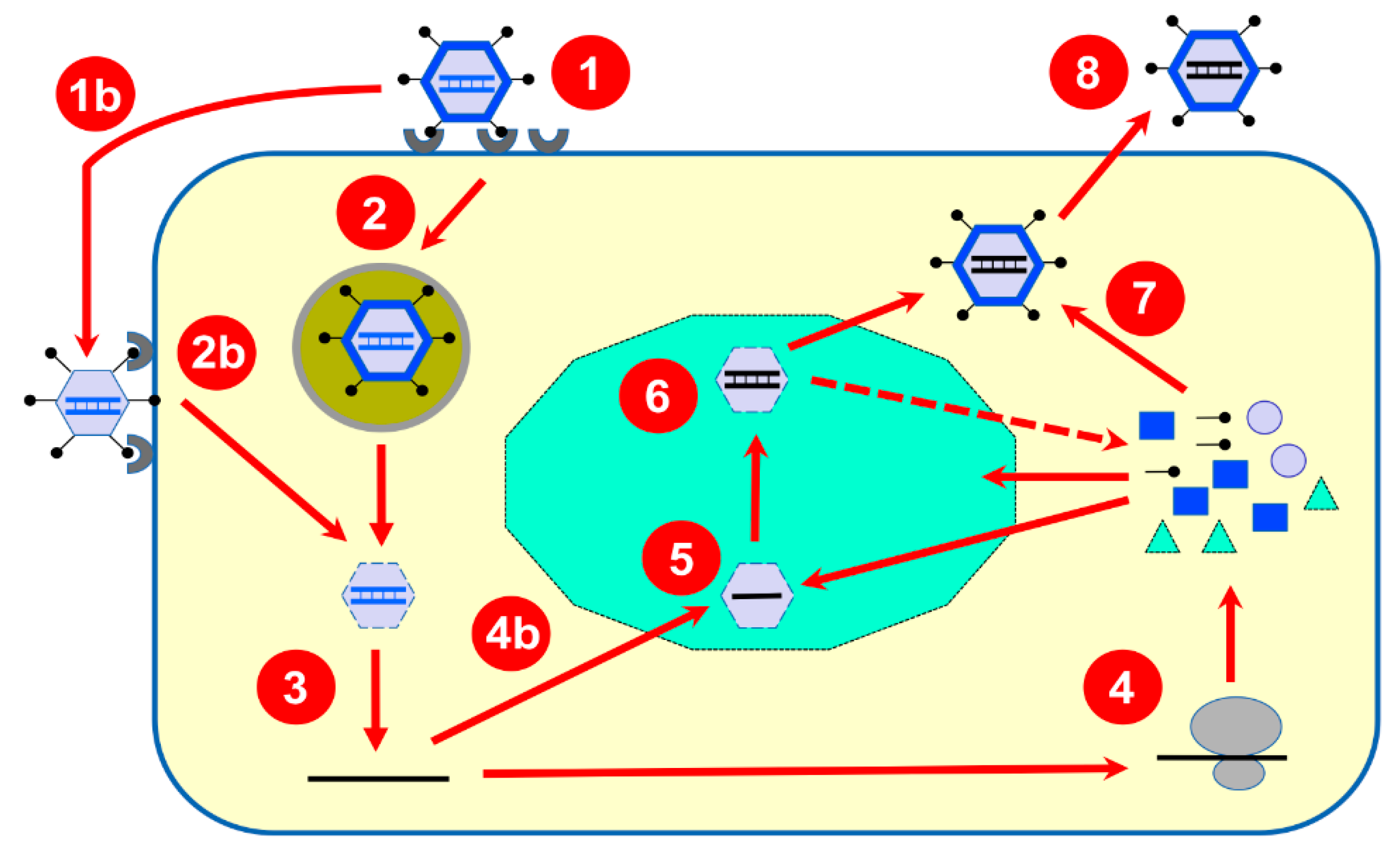



| Transcription of reovirus dsRNA genome |
| •Is there a temporal regulation in the transcription of viral genes upon infection? •Is there involvement of helicase activity of μ2 or λ1 in transcription? •Are each segment of genomic RNA transcribed by a single transcription complex? |
| Cap structure of reovirus mRNA |
| •Is μ2 or λ1 acting as the RNA triphosphatase for viral mRNA cap synthesis? •Is cap2 structure present on viral mRNA and what is its role? •Is the capping activity absent, inactive or hidden in progeny cores? |
| Impact of reovirus infection on cellular mRNA |
| •Is transcription of cellular mRNA affected by reovirus infection? •What is the exact role of modified alternative splicing during reovirus infection? |
| Synthesis of viral proteins |
| •Is a viral protein acting as a surrogate of poly(A)-binding protein? •Is a translational operator present on viral mRNA? •Is protein synthesis modified to favor translation of uncapped viral mRNA? •What is the exact involvement of σ3 in the regulation of protein synthesis? •What is the mechanism allowing σ1s to stimulate synthesis of viral proteins? |
| PKR, stress, and the regulation of protein synthesis during reovirus infection |
| •Is PKR involved in inhibition of cellular protein synthesis? •What is the exact role of stress granules formation during reovirus infection? |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemay, G. Synthesis and Translation of Viral mRNA in Reovirus-Infected Cells: Progress and Remaining Questions. Viruses 2018, 10, 671. https://doi.org/10.3390/v10120671
Lemay G. Synthesis and Translation of Viral mRNA in Reovirus-Infected Cells: Progress and Remaining Questions. Viruses. 2018; 10(12):671. https://doi.org/10.3390/v10120671
Chicago/Turabian StyleLemay, Guy. 2018. "Synthesis and Translation of Viral mRNA in Reovirus-Infected Cells: Progress and Remaining Questions" Viruses 10, no. 12: 671. https://doi.org/10.3390/v10120671
APA StyleLemay, G. (2018). Synthesis and Translation of Viral mRNA in Reovirus-Infected Cells: Progress and Remaining Questions. Viruses, 10(12), 671. https://doi.org/10.3390/v10120671





