Development of Small-Molecule MERS-CoV Inhibitors
Abstract
1. Introduction
2. MERS-CoV Life Cycle and Potential Targets for the Development of Small-Molecule Inhibitors Against MERS-CoV Infection
3. Current Small-Molecule Inhibitors Against MERS-CoV Infection and Their Mechanisms of Action
3.1. MERS-CoV Entry Inhibitors
3.2. MERS-CoV Replication Inhibitors
3.2.1. MERS-CoV Inhibitors Targeting Papain-Like Protease
3.2.2. MERS-CoV Inhibitors Targeting 3C-Like Protease
3.3. Other Small-Molecule Inhibitors with Defined or Undefined Mechanisms of Action
4. Strategies for Developing Small-Molecule MERS-CoV Inhibitors
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cotten, M.; Watson, S.J.; Zumla, A.I.; Makhdoom, H.Q.; Palser, A.L.; Ong, S.H.; Al Rabeeah, A.A.; Alhakeem, R.F.; Assiri, A.; Al-Tawfiq, J.A.; et al. Spread, circulation, and evolution of the middle east respiratory syndrome coronavirus. mBio 2014, 5, e01062-13. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Lau, S.K.; Woo, P.C. The emerging novel middle east respiratory syndrome coronavirus: The “knowns” and “unknowns”. J. Formos. Med. Assoc. 2013, 112, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Arabi, Y.M.; Arifi, A.A.; Balkhy, H.H.; Najm, H.; Aldawood, A.S.; Ghabashi, A.; Hawa, H.; Alothman, A.; Khaldi, A.; Al Raiy, B. Clinical course and outcomes of critically ill patients with middle east respiratory syndrome coronavirus infection. Ann. Intern. Med. 2014, 160, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Drosten, C.; Meyer, B.; Muller, M.A.; Corman, V.M.; Al-Masri, M.; Hossain, R.; Madani, H.; Sieberg, A.; Bosch, B.J.; Lattwein, E.; et al. Transmission of mers-coronavirus in household contacts. N. Engl. J. Med. 2014, 371, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Liu, Q.; Du, L.; Jiang, S. Middle east respiratory syndrome coronavirus (mers-cov): Challenges in identifying its source and controlling its spread. Microbes Infect. 2013, 15, 625–629. [Google Scholar] [CrossRef]
- Falzarano, D.; de Wit, E.; Rasmussen, A.L.; Feldmann, F.; Okumura, A.; Scott, D.P.; Brining, D.; Bushmaker, T.; Martellaro, C.; Baseler, L.; et al. Treatment with interferon-alpha2b and ribavirin improves outcome in mers-cov-infected rhesus macaques. Nat. Med. 2013, 19, 1313–1317. [Google Scholar] [CrossRef]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of clinically developed drugs for treatment of middle east respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef]
- Lu, L.; Xia, S.; Ying, T.; Jiang, S. Urgent development of effective therapeutic and prophylactic agents to control the emerging threat of middle east respiratory syndrome (mers). Emerg. Microbes Infect. 2015, 4, e37. [Google Scholar] [CrossRef]
- Gierer, S.; Bertram, S.; Kaup, F.; Wrensch, F.; Heurich, A.; Kramer-Kuhl, A.; Welsch, K.; Winkler, M.; Meyer, B.; Drosten, C.; et al. The spike protein of the emerging betacoronavirus emc uses a novel coronavirus receptor for entry, can be activated by tmprss2, and is targeted by neutralizing antibodies. J. Virol. 2013, 87, 5502–5511. [Google Scholar] [CrossRef]
- Bertram, S.; Dijkman, R.; Habjan, M.; Heurich, A.; Gierer, S.; Glowacka, I.; Welsch, K.; Winkler, M.; Schneider, H.; Hofmann-Winkler, H.; et al. Tmprss2 activates the human coronavirus 229e for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013, 87, 6150–6160. [Google Scholar] [CrossRef]
- Du, L.Y.; Yang, Y.; Zhou, Y.S.; Lu, L.; Li, F.; Jiang, S.B. Mers-cov spike protein: A key target for antivirals. Expert Opin. Ther. Target 2017, 21, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Liu, Q.; Wang, Q.; Sun, Z.W.; Su, S.; Dub, L.Y.; Ying, T.L.; Lu, L.; Jiang, S.B. Middle east respiratory syndrome coronavirus (mers-cov) entry inhibitors targeting spike protein. Virus Res. 2014, 194, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Forni, D.; Filippi, G.; Cagliani, R.; De Gioia, L.; Pozzoli, U.; Al-Daghri, N.; Clerici, M.; Sironi, M. The heptad repeat region is a major selection target in mers-cov and related coronaviruses. Sci. Rep. 2015, 5, 14480. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Lu, G.; Qi, J.; Li, Y.; Wu, Y.; Deng, Y.; Geng, H.; Li, H.; Wang, Q.; Xiao, H.; et al. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the s protein of middle east respiratory syndrome coronavirus. J. Virol. 2013, 87, 13134–13140. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-emc. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Liu, Q.; Zhu, Y.; Chan, K.H.; Qin, L.; Li, Y.; Wang, Q.; Chan, J.F.; Du, L.; Yu, F.; et al. Structure-based discovery of middle east respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014, 5, 3067. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lou, Z.; Liu, Y.; Pang, H.; Tien, P.; Gao, G.F.; Rao, Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Boil. Chem. 2004, 279, 49414–49419. [Google Scholar] [CrossRef]
- Channappanavar, R.; Lu, L.; Xia, S.; Du, L.; Meyerholz, D.K.; Perlman, S.; Jiang, S. Protective effect of intranasal regimens containing peptidic middle east respiratory syndrome coronavirus fusion inhibitor against mers-cov infection. J. Infect. Dis. 2015, 212, 1894–1903. [Google Scholar] [CrossRef]
- Tao, X.; Garron, T.; Agrawal, A.S.; Algaissi, A.; Peng, B.H.; Wakamiya, M.; Chan, T.S.; Lu, L.; Du, L.; Jiang, S.; et al. Characterization and demonstration of the value of a lethal mouse model of middle east respiratory syndrome coronavirus infection and disease. J. Virol. 2016, 90, 57–67. [Google Scholar] [CrossRef]
- Wang, C.; Xia, S.; Zhang, P.; Zhang, T.; Wang, W.; Tian, Y.; Meng, G.; Jiang, S.; Liu, K. Discovery of hydrocarbon-stapled short alpha-helical peptides as promising middle east respiratory syndrome coronavirus (mers-cov) fusion inhibitors. J. Med. Chem. 2018, 61, 2018–2026. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, J.; Zhang, K.; Chu, H.; Liu, D.; Poon, V.K.; Chan, C.C.; Leung, H.C.; Fai, N.; Lin, Y.P.; et al. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016, 6, 22008. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, L.; Xia, S.; Zhang, T.; Cao, R.; Liang, G.; Li, Y.; Meng, G.; Wang, W.; Shi, W.; et al. De novo design of alpha-helical lipopeptides targeting viral fusion proteins: A promising strategy for relatively broad-spectrum antiviral drug discovery. J. Med. Chem. 2018, 61, 8734–8745. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zou, P.; Wu, F.; Lu, L.; Jiang, S. Development of small-molecule viral inhibitors targeting various stages of the life cycle of emerging and re-emerging viruses. Front. Med. 2017, 11, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.B.; Tao, X.R.; Xia, S.; Garron, T.; Yu, F.; Du, L.Y.; Lu, L.; Tseng, C.T.K. Intranasally administered peptidic viral fusion inhibitor protected hdpp4 transgenic mice from mers-cov infection. Lancet 2015, 386, S44. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef]
- Liu, Q.; Xia, S.; Sun, Z.; Wang, Q.; Du, L.; Lu, L.; Jiang, S. Testing of middle east respiratory syndrome coronavirus replication inhibitors for the ability to block viral entry. Antimicrob. Agents Chemother. 2015, 59, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Du, L.; Ma, C.; Li, Y.; Li, L.; Poon, V.K.; Wang, L.; Yu, F.; Zheng, B.J.; Jiang, S.; et al. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus mers-cov. Virol. J. 2013, 10, 266. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Glowacka, I.; Blazejewska, P.; Soilleux, E.; Allen, P.; Danisch, S.; Steffen, I.; Choi, S.Y.; Park, Y.; Schneider, H.; et al. Tmprss2 and tmprss4 facilitate trypsin-independent spread of influenza virus in caco-2 cells. J. Virol. 2010, 84, 10016–10025. [Google Scholar] [CrossRef]
- Shirogane, Y.; Takeda, M.; Iwasaki, M.; Ishiguro, N.; Takeuchi, H.; Nakatsu, Y.; Tahara, M.; Kikuta, H.; Yanagi, Y. Efficient multiplication of human metapneumovirus in vero cells expressing the transmembrane serine protease tmprss2. J. Virol. 2008, 82, 8942–8946. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease tmprss2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef]
- Shirato, K.; Kawase, M.; Matsuyama, S. Middle east respiratory syndrome coronavirus infection mediated by the transmembrane serine protease tmprss2. J. Virol. 2013, 87, 12552–12561. [Google Scholar] [CrossRef]
- Zhou, Y.; Vedantham, P.; Lu, K.; Agudelo, J.; Carrion, R., Jr.; Nunneley, J.W.; Barnard, D.; Pohlmann, S.; McKerrow, J.H.; Renslo, A.R.; et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015, 116, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.C.; Doyle, P.S.; Hsieh, I.; McKerrow, J.H. Cysteine protease inhibitors cure an experimental trypanosoma cruzi infection. J. Exp. Med. 1998, 188, 725–734. [Google Scholar] [CrossRef] [PubMed]
- De Wilde, A.H.; Jochmans, D.; Posthuma, C.C.; Zevenhoven-Dobbe, J.C.; van Nieuwkoop, S.; Bestebroer, T.M.; van den Hoogen, B.G.; Neyts, J.; Snijder, E.J. Screening of an fda-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014, 58, 4875–4884. [Google Scholar] [CrossRef] [PubMed]
- Burkard, C.; Verheije, M.H.; Haagmans, B.L.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.J.; de Haan, C.A. Atp1a1-mediated src signaling inhibits coronavirus entry into host cells. J. Virol. 2015, 89, 4434–4448. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, Y.I.; Park, S.J.; Kim, I.K.; Choi, Y.K.; Kim, S.H. Safe, high-throughput screening of natural compounds of mers-cov entry inhibitors using a pseudovirus expressing mers-cov spike protein. Int. J. Antimicrob. Agents 2018, 52, 730–732. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Host cell entry of middle east respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.Y.; Lai, H.Y.; Chen, H.Y.; Cheng, S.C.; Cheng, K.W.; Chou, Y.W. Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.S.; Chang, G.G.; Juo, C.G.; Lee, H.J.; Yeh, S.H.; Hsu, J.T.; Chen, X. Papain-like protease 2 (plp2) from severe acute respiratory syndrome coronavirus (sars-cov): Expression, purification, characterization, and inhibition. Biochemistry 2005, 44, 10349–10359. [Google Scholar] [CrossRef]
- Lee, H.; Lei, H.; Santarsiero, B.D.; Gatuz, J.L.; Cao, S.; Rice, A.J.; Patel, K.; Szypulinski, M.Z.; Ojeda, I.; Ghosh, A.K.; et al. Inhibitor recognition specificity of mers-cov papain-like protease may differ from that of sars-cov. ACS Chem. Biol. 2015, 10, 1456–1465. [Google Scholar] [CrossRef]
- Thiel, V.; Ivanov, K.A.; Putics, A.; Hertzig, T.; Schelle, B.; Bayer, S.; Weissbrich, B.; Snijder, E.J.; Rabenau, H.; Doerr, H.W.; et al. Mechanisms and enzymes involved in sars coronavirus genome expression. J. Gen. Virol. 2003, 84, 2305–2315. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, B.H.; Jukneliene, D.; Kanjanahaluethai, A.; Bechill, J.; Severson, K.M.; Smith, C.M.; Rota, P.A.; Baker, S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004, 78, 13600–13612. [Google Scholar] [CrossRef] [PubMed]
- Mielech, A.M.; Kilianski, A.; Baez-Santos, Y.M.; Mesecar, A.D.; Baker, S.C. Mers-cov papain-like protease has deisgylating and deubiquitinating activities. Virology 2014, 450–451, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, X.; Bian, G.; Tu, J.; Xing, Y.; Wang, Y.; Chen, Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of middle east respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 2014, 95, 614–626. [Google Scholar] [CrossRef] [PubMed]
- Galkin, A.; Kulakova, L.; Lim, K.; Chen, C.Z.; Zheng, W.; Turko, I.V.; Herzberg, O. Structural basis for inactivation of giardia lamblia carbamate kinase by disulfiram. J. Boil. Chem. 2014, 289, 10502–10509. [Google Scholar] [CrossRef] [PubMed]
- Paranjpe, A.; Zhang, R.; Ali-Osman, F.; Bobustuc, G.C.; Srivenugopal, K.S. Disulfiram is a direct and potent inhibitor of human o6-methylguanine-DNA methyltransferase (mgmt) in brain tumor cells and mouse brain and markedly increases the alkylating DNA damage. Carcinogenesis 2014, 35, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Moses, D.C.; Hsieh, C.H.; Cheng, S.C.; Chen, Y.H.; Sun, C.Y.; Chou, C.Y. Disulfiram can inhibit mers and sars coronavirus papain-like proteases via different modes. Antivir. Res 2018, 150, 155–163. [Google Scholar] [CrossRef]
- Kumar, V.; Tan, K.P.; Wang, Y.M.; Lin, S.W.; Liang, P.H. Identification, synthesis and evaluation of sars-cov and mers-cov 3c-like protease inhibitors. Bioorg. Med. Chem. 2016, 24, 3035–3042. [Google Scholar] [CrossRef]
- Needle, D.; Lountos, G.T.; Waugh, D.S. Structures of the middle east respiratory syndrome coronavirus 3c-like protease reveal insights into substrate specificity. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1102–1111. [Google Scholar] [CrossRef]
- Hsu, M.F.; Kuo, C.J.; Chang, K.T.; Chang, H.C.; Chou, C.C.; Ko, T.P.; Shr, H.L.; Chang, G.G.; Wang, A.H.; Liang, P.H. Mechanism of the maturation process of sars-cov 3cl protease. J. Biol. Chem. 2005, 280, 31257–31266. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, Y.; Li, L.; Wang, K.; Chen, S.; Chen, J.; Ding, J.; Jiang, H.; Shen, X. Two adjacent mutations on the dimer interface of sars coronavirus 3c-like protease cause different conformational changes in crystal structure. Virology 2009, 388, 324–334. [Google Scholar] [CrossRef]
- Galasiti Kankanamalage, A.C.; Kim, Y.; Damalanka, V.C.; Rathnayake, A.D.; Fehr, A.R.; Mehzabeen, N.; Battaile, K.P.; Lovell, S.; Lushington, G.H.; Perlman, S.; et al. Structure-guided design of potent and permeable inhibitors of mers coronavirus 3cl protease that utilize a piperidine moiety as a novel design element. Eur. J. Med. Chem. 2018, 150, 334–346. [Google Scholar] [CrossRef]
- Kilianski, A.; Mielech, A.M.; Deng, X.; Baker, S.C. Assessing activity and inhibition of middle east respiratory syndrome coronavirus papain-like and 3c-like proteases using luciferase-based biosensors. J. Virol. 2013, 87, 11955–11962. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Shin, J.S.; Shie, J.J.; Ku, K.B.; Kim, C.; Go, Y.Y.; Huang, K.F.; Kim, M.; Liang, P.H. Identification and evaluation of potent middle east respiratory syndrome coronavirus (mers-cov) 3cl(pro) inhibitors. Antivir. Res. 2017, 141, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Liu, H.; Galasiti Kankanamalage, A.C.; Weerasekara, S.; Hua, D.H.; Groutas, W.C.; Chang, K.O.; Pedersen, N.C. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016, 12, e1005531. [Google Scholar]
- Ren, Z.; Yan, L.; Zhang, N.; Guo, Y.; Yang, C.; Lou, Z.; Rao, Z. The newly emerged sars-like coronavirus hcov-emc also has an “achilles’ heel”: Current effective inhibitor targeting a 3c-like protease. Protein Cell 2013, 4, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Schulte, F.W.; Lange-Grunweller, K.; Obermann, W.; Madhugiri, R.; Pleschka, S.; Ziebuhr, J.; Hartmann, R.K.; Grunweller, A. Broad-spectrum antiviral activity of the eif4a inhibitor silvestrol against corona- and picornaviruses. Antivir. Res. 2018, 150, 123–129. [Google Scholar] [CrossRef]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus susceptibility to the antiviral remdesivir (gs-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef]
- Cong, Y.; Hart, B.J.; Gross, R.; Zhou, H.; Frieman, M.; Bollinger, L.; Wada, J.; Hensley, L.E.; Jahrling, P.B.; Dyall, J.; et al. Mers-cov pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS ONE 2018, 13, e0194868. [Google Scholar] [CrossRef]
- Hart, B.J.; Dyall, J.; Postnikova, E.; Zhou, H.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Frieman, M.B.; Holbrook, M.R.; Jahrling, P.B.; et al. Interferon-beta and mycophenolic acid are potent inhibitors of middle east respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014, 95, 571–577. [Google Scholar] [CrossRef]
- Tao, X.; Mei, F.; Agrawal, A.; Peters, C.J.; Ksiazek, T.G.; Cheng, X.; Tseng, C.T. Blocking of exchange proteins directly activated by camp leads to reduced replication of middle east respiratory syndrome coronavirus. J. Virol. 2014, 88, 3902–3910. [Google Scholar] [CrossRef] [PubMed]
- Warren, T.K.; Wells, J.; Panchal, R.G.; Stuthman, K.S.; Garza, N.L.; Van Tongeren, S.A.; Dong, L.; Retterer, C.J.; Eaton, B.P.; Pegoraro, G.; et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue bcx4430. Nature 2014, 508, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Peters, H.L.; Jochmans, D.; de Wilde, A.H.; Posthuma, C.C.; Snijder, E.J.; Neyts, J.; Seley-Radtke, K.L. Design, synthesis and evaluation of a series of acyclic fleximer nucleoside analogues with anti-coronavirus activity. Bioorg. Med. Chem. Lett. 2015, 25, 2923–2926. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.S.; Jung, E. Saracatinib inhibits middle east respiratory syndrome-coronavirus replication in vitro. Viruses 2018, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.N.; Lai, K.K.; Dai, J.; Kok, K.H.; Chen, H.; Chan, K.H.; Yuen, K.Y.; Kao, R.Y.T. Broad-spectrum inhibition of common respiratory rna viruses by a pyrimidine synthesis inhibitor with involvement of the host antiviral response. J. Gen. Virol. 2017, 98, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Ho, C.T.; Chuo, W.H.; Li, S.; Wang, T.T.; Lin, C.C. Effective inhibition of mers-cov infection by resveratrol. BMC Infect. Dis. 2017, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef]
- Huczynski, A. Polyether ionophores-promising bioactive molecules for cancer therapy. Bioorg. Med. Chem. Lett. 2012, 22, 7002–7010. [Google Scholar] [CrossRef]
- McFadden, G. Gleevec casts a pox on poxviruses. Nat. Med. 2005, 11, 711–712. [Google Scholar] [CrossRef]
- Adedeji, A.O.; Singh, K.; Kassim, A.; Coleman, C.M.; Elliott, R.; Weiss, S.R.; Frieman, M.B.; Sarafianos, S.G. Evaluation of ssya10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and middle east respiratory syndrome coronaviruses. Antimicrob. Agents Chemother. 2014, 58, 4894–4898. [Google Scholar] [CrossRef]
- Lundin, A.; Dijkman, R.; Bergstrom, T.; Kann, N.; Adamiak, B.; Hannoun, C.; Kindler, E.; Jonsdottir, H.R.; Muth, D.; Kint, J.; et al. Targeting membrane-bound viral rna synthesis reveals potent inhibition of diverse coronaviruses including the middle east respiratory syndrome virus. PLoS Pathog. 2014, 10, e1004166. [Google Scholar] [CrossRef] [PubMed]
- Li, H.S.; Kuok, D.I.T.; Cheung, M.C.; Ng, M.M.T.; Ng, K.C.; Hui, K.P.Y.; Peiris, J.S.M.; Chan, M.C.W.; Nicholls, J.M. Effect of interferon alpha and cyclosporine treatment separately and in combination on middle east respiratory syndrome coronavirus (mers-cov) replication in a human in-vitro and ex-vivo culture model. Antivir. Res. 2018, 155, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Clasman, J.R.; Baez-Santos, Y.M.; Mettelman, R.C.; O’Brien, A.; Baker, S.C.; Mesecar, A.D. X-ray structure and enzymatic activity profile of a core papain-like protease of mers coronavirus with utility for structure-based drug design. Sci. Rep. 2017, 7, 40292. [Google Scholar] [CrossRef]
- Josset, L.; Menachery, V.D.; Gralinski, L.E.; Agnihothram, S.; Sova, P.; Carter, V.S.; Yount, B.L.; Graham, R.L.; Baric, R.S.; Katze, M.G. Cell host response to infection with novel human coronavirus emc predicts potential antivirals and important differences with sars coronavirus. mBio 2013, 4, e00165-13. [Google Scholar] [CrossRef] [PubMed]



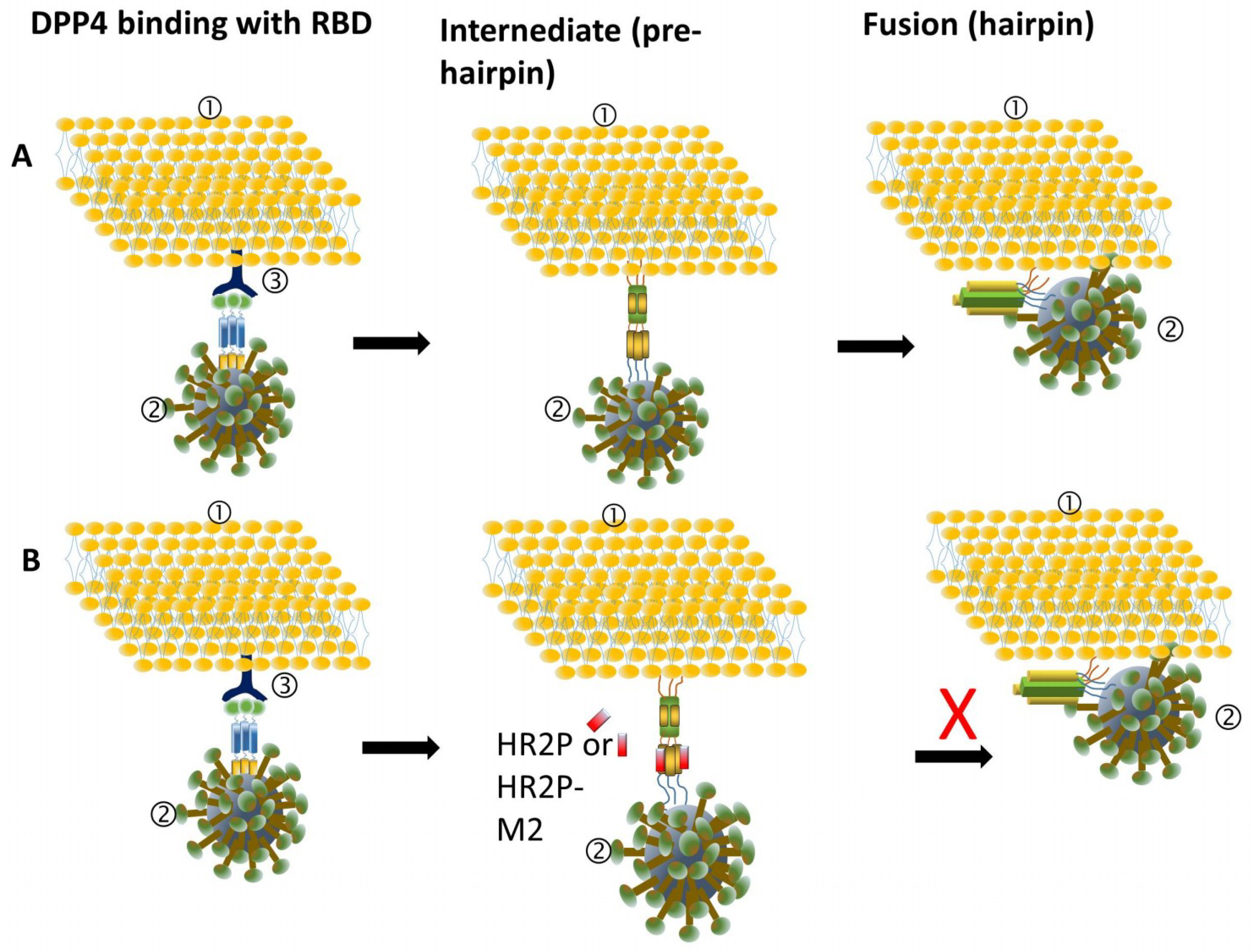






| Compound | Sequence | Testing Model | Cell Lines Tested | EC50 (μM) | CC50 (μM) | Ref. |
|---|---|---|---|---|---|---|
| Peptide inhibitors disturbing membrane fusion | ||||||
| HR2P | SLTQINTTLLDLTYEMLSLQQVVKALNESYIDLKEL | In vitro | Vero cells Huh-7 cells | 0.6 0.93 ± 0.15 b | >1000 | [16] |
| HR2P-M2 | SLTQINTTLLDLEYEMKKLEEVVKKLEESYIDLKEL | In vitro; in vivo: hDPP4 Tg mice | Calu-3 and Vero cells; Ad5-hDPP4 mice | 0.55 ± 0.04 b | - | [16,18,19] |
| P21S10 | LDLTYEM LSLQQVV K*LNE*Y | In vitro | Huh-7 cells | 0.97 ± 0.08; 0.33 ± 0.04 b | >100 | [20] |
| P21S2 | L*LTY*M LSLQQVV KALNESY | In vitro | Huh-7 cells | 3.90 ± 1.1 b | - | [20] |
| P21S4 | LDLT*EM L*LQQVV KALNESY | In vitro | Huh-7 cells | 7.14 ± 0.7 b | - | [20] |
| P21S5 | LDLTYEM *SLQ*VV KALNESY | In vitro | Huh-7 cells | 10.7 ± 2.6 b | - | [20] |
| P21S8 | LDLTYEM LSLQ*VV K*LNESY | In vitro | Huh-7 cells | 3.03 ± 0.29; 0.26 ± 0.05 b | >100 | [20] |
| P21S9 | LDLTYEM LSLQQVV *ALN*SY | In vitro | Huh-7 cells | 14.1 ± 2.3 b | - | [20] |
| P21L2 | LXLTYXM LSLQQVV KALNESY | In vitro | Huh-7 cells | 10.9 ± 1.1 b | - | [20] |
| P21L4 | LDLTXEM LXLQQVV KALNESY | In vitro | Huh-7 cells | 8.21 ± 0.9 b | - | [20] |
| P21L5 | LDLTYEM XSLQXVV KALNESY | In vitro | Huh-7 cells | 4.49 ± 0.6 b | - | [20] |
| P21L8 | LDLTYEM LSLQXVV KXLNESY | In vitro | Huh-7 cells | 20.6 ± 3.3 b | - | [20] |
| P21L9 | LDLTYEM LSLQQVV XALNXSY | In vitro | Huh-7 cells | 10.9 ± 1.0 b | - | [20] |
| P21L10 | LDLTYEM LSLQQVV KXLNEXY | In vitro | Huh-7 cells | 3.55 ± 0.2 b | - | [20] |
| P21R8 | LDLTYEM LSLQ^VV K^LNESY | In vitro | Huh-7 cells | 16.3 ± 1.1 b | - | [20] |
| P21S8Z | LDLTYEZ LSLQ*VV K*LNESY | In vitro | Huh-7 cells | 2.80 ± 0.74; 0.63 ± 0.05 b | >100 | [20] |
| P21S8F | LDLTYEM LSLQ*VV K*LNESF | In vitro | Huh-7 cells | 2.16 ± 1.1 b | - | [20] |
| P21S8ZF | LDLTYES LSLQ*VV K*LNESF | In vitro | Huh-7 cells | 3.89 ± 0.8 b | - | [20] |
| P9 a | NGAICWGPCPTAFRQIGNCGHFKVRCCKIR | In vitro | MDCK cells | 5.00 μg/mL | 380 μg/mL | [21] |
| LLS | LEELSKKLEELSKKLEELSKKLEELSKKLEELSKK-βA-K (C16) | In vitro | Huh-7 cells | 0.24 ± 0.08 b | 4.04 ± 0.4 | [22] |
| IIS | IEEISKKIEEISKKIEEISKKIEEISKKIEEISKK-βA-K (C16) | In vitro | Huh-7 cells | 0.10 ± 0.02 b | 88.8 ± 28 | [22] |
| AAS | AEEASKKAEEASKKAEEASKKAEEASKKAEEASKK-βA-K(C16) | In vitro | Huh-7 cells | 4.47 ± 1.7 b | 2.38 ± 0.9 | [22] |
| FFS | FEEFSKKFEEFSKKFEEFSKKFEEFSKKFEEFSKK-βA-K (C16) | In vitro | Huh-7 cells | 3.11 ± 0.9 b | >100 | [22] |
| YYS | YEEYSKKYEEYSKKYEEYSKKYEEYSKKYEEYSKK-βA-K(C16) | In vitro | Huh-7 cells | 6.26 ± 2.1 b | 19.8 ± 1.6 | [22] |
| IIY | IEEIYKKIEEIYKKIEEIYKKIEEIYKKIEEIYKK-βA-K (C16) | In vitro | Huh-7 cells | 0.52 ± 0.4 b | >100 | [22] |
| IIW | IEEIWKKIEEIWKKIEEIWKKIEEIWKKIEEIWKK-βA-K (C16) | In vitro | Huh-7 cells | 10.6 ± 2.4 b | >100 | [22] |
| IIH | IEEIHKKIEEIHKKIEEIHKKIEEIHKKIEEIHKK-βA-K (C16) | In vitro | Huh-7 cells | 1.68 ± 0.47 b | >100 | [22] |
| IIQ | IEEIQKKIEEIQKKIEEIQKKIEEIQKKIEEIQKK-βA-K (C16) | In vitro | Huh-7 cells | 0.13 ± 0.1; 0.11 ± 0.02 b | >100 | [22] |
| IIK | IEEIKKKIEEIKKKIEEIKKKIEEIKKKIEEIKKK-βA-K (C16) | In vitro | Huh-7 cells | 0.45 ± 0.13 b | 4.54 ± 0.6 | [22] |
| IIE | IEEIEKKIEEIEKKIEEIEKKIEEIEKKIEEIEKK-βA-K (C16) | In vitro | Huh-7 cells | 2.93 ± 0.95 b | >100 | [22] |
| Inhibitor | Testing Model | Cell Lines | EC50 (μM) | CC50 (μM) | Ref. |
|---|---|---|---|---|---|
| Inhibitors blocking the binding between virus and host cells | |||||
| ADS-J1 | In vitro | NBL-7 and Huh-7 cells | 0.6 | 26.9 | [27] |
| Inhibitors disrupting endocytosis | |||||
| Chlorpromazine | In vitro | Huh-7 cells | 23.33 ± 2.89 a; 49 ± 1.2; 9.514 | >40; 21.3 ± 1.0 | [5,6,7,26] |
| Promethazine | In vitro | Huh-7 cells | 16.67 ± 7.22 a; 11.802 | >40 | [7,26] |
| Fluphenazine | In vitro | Huh-7 cells | 15.00 ± 4.33 a; 5.868 | ~40 | [7,26] |
| K11777 | In vitro | Vero cells | 0.046 | >10 | [32] |
| Camostat | In vitro | Vero-TMPRSS2 cells | ~1 | - | [31] |
| Ouabain | In vitro | Huh-7 cells | ~0.05 | - | [35] |
| Bufalin | In vitro | Huh-7 cells | 0.01–0.015 | - | [35] |
| Dihydrotanshinone | In vitro | - | 0.5–1 μg/mL | - | [36] |
| Inhibitors interrupting MERS-CoV MERS-CoV RNA replication and translation | |||||
| Disulfiram | In vitro | - | 22.7 ± 0.5 | - | [47] |
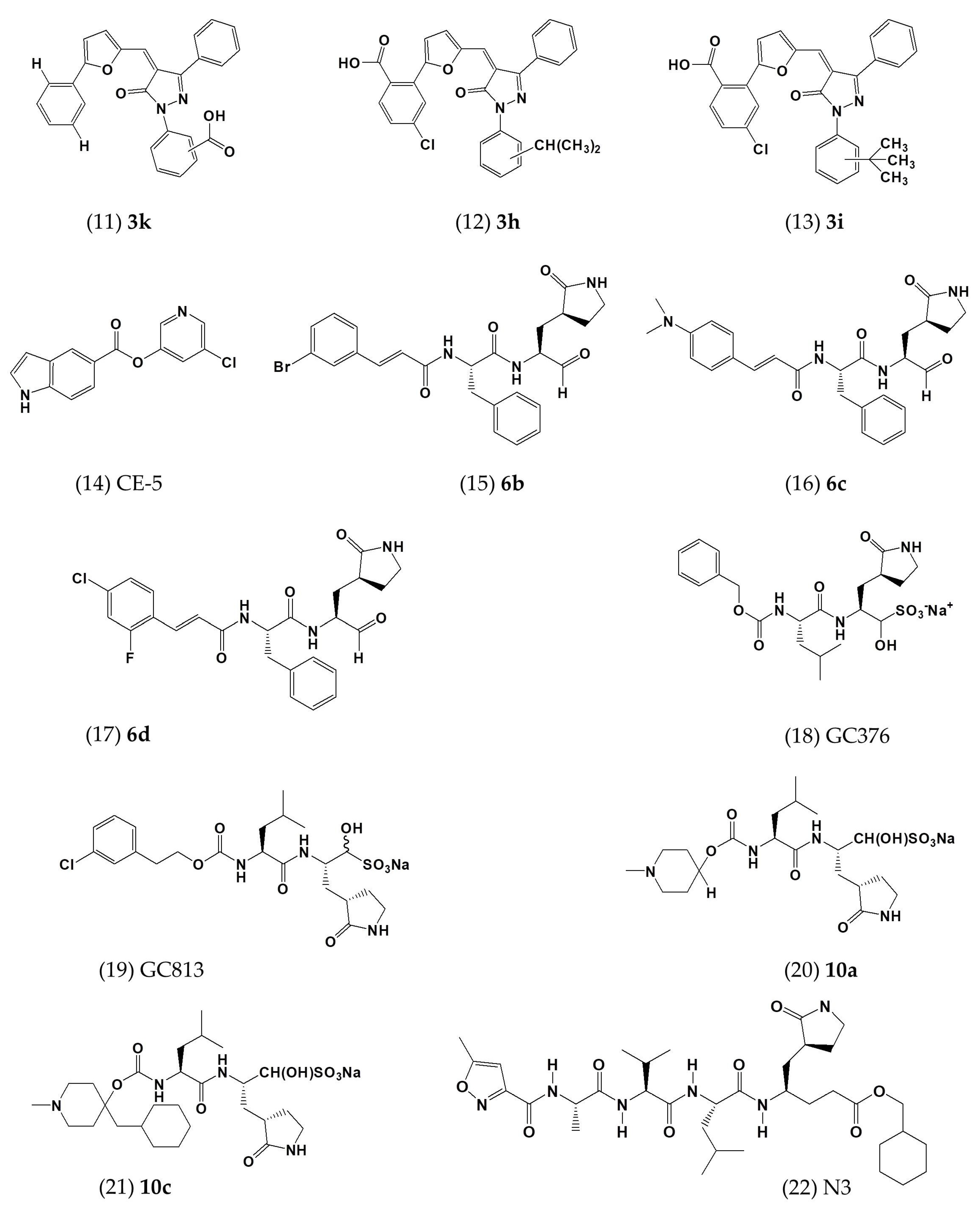
| 3k | In vitro | - | 5.8 ± 1.6 | - | [48] |
| 3h | In vitro | - | 7.3 ± 2.1 | - | [48] |
| 3i | In vitro | - | 7.4 ± 2.2 | - | [48] |
| CE-5 | In vitro | HEK293T cells | ~12.5 | - | [53] |
| 6b | In vitro | Huh-7 cells | 1.4 ± 0.0 | >100 | [54] |
| 6c | In vitro | Huh-7 cells | 1.2 ± 0.6 | >100 | [54] |
| 6d | In vitro | Huh-7 cells | 0.6 ± 0.0 | 58.6 ± 1.2 | [54] |
| GC376 | In vitro | - | 1.56 ± 0.09; 0.9 | >150 | [52,55] |
| GC813 | In vitro | - | 0.5 | - | [52] |
| 10a | In vitro | Vero81 cells | 0.5 | >100 | [52] |
| 10c | In vitro | Vero81 cells | 0.8 | >100 | [52] |
| N3 | In vitro | - | 0.28 ± 0.02 | - | [56] |
| Inhibitors with undefined mechanisms | |||||
| Silvestrol | In vitro | MRC-5 cells | 0.0013 | 0.4 | [57] |
| GS-5734 | In vitro | HAE cells | 0.074 ± 0.023 | >10 | [58] |
| GS-441524 | In vitro | HAE cells | 0.86 ± 0.78 | >100 | [58] |
| Chloroquine | In vitro | MDMs and MDDCs cells | 3.0 ± 1.1; 6.275 | 58.1 ± 1.1 | [7,59] |
| Emetine dihydrochloride hydrate | In vitro | Vero E6 cells | 0.014 | - | [7] |
| Hydroxychloroquine sulfate | In vitro | Vero E6 cells | 8.279 | - | [7] |
| Mefloquine | In vitro | Vero E6 cells | 7.416 | - | [7] |
| Amodiaquine dihydrochloride dehydrate | In vitro | Vero E6 cells | 6.212 | - | [7] |
| E-64-D | In vitro | Vero E6 cells | 1.275 | - | [7] |
| Gemcitabine hydrochloride | In vitro | Vero E6 cells | 1.216 | - | [7] |
| Tamoxifen citrate | In vitro | Vero E6 cells | 10.117 | - | [7] |
| Toremifene citrate | In vitro | Vero E6 cells | 12.915 | - | [7] |
| Terconazole | In vitro | Vero E6 cells | 12.203 | - | [7] |
| Triparanol | In vitro | Vero E6 cells | 5.283 | - | [7] |
| Anisomycin | In vitro | Vero E6 cells | 0.003 | - | [7] |
| Cycloheximide | In vitro | Vero E6 cells | 0.189 | - | [7] |
| Homoharringtonine | In vitro | Vero E6 cells | 0.0718 | - | [7] |
| Benztropine mesylate | In vitro | Vero E6 cells | 16.627 | - | [7] |
| Fluspirilene | In vitro | Vero E6 cells | 7.477 | - | [7] |
| Thiothixene | In vitro | Vero E6 cells | 9.297 | - | [7] |
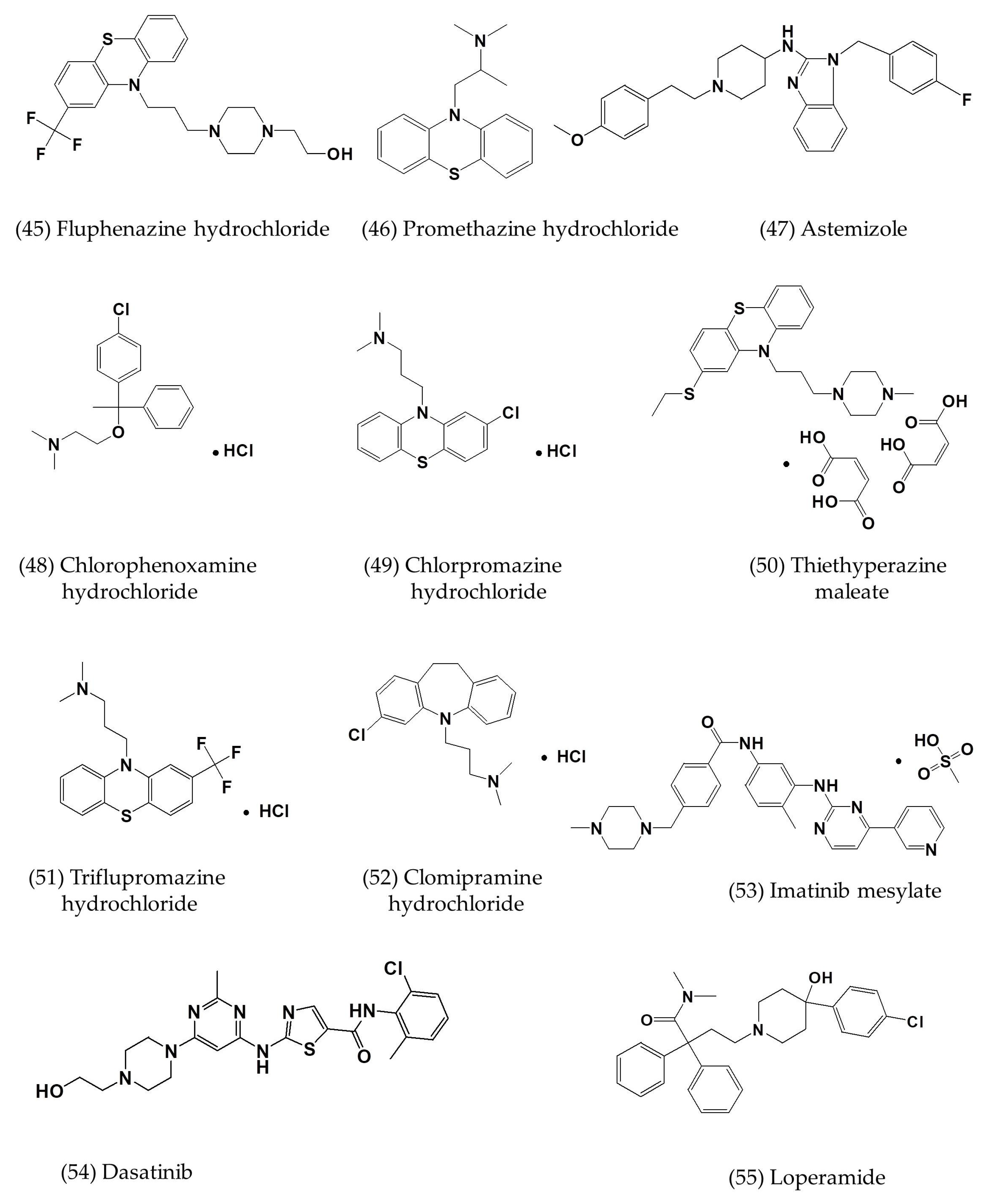
| Astemizole | In vitro | Vero E6 cells | 4.884 | - | [7] |
| Chlorphenoxamine hydrochloride | In vitro | Vero E6 cells | 12.646 | [7] | |
| Thiethylperazine maleate | In vitro | Vero E6 cells | 7.865 | - | [7] |
| Triflupromazine hydrochloride | In vitro | Vero E6 cells | 5.758 | - | [7] |
| Clomipramine hydrochloride | In vitro | Vero E6 cells | 9.332 | - | [7] |
| Imatinib mesylate | In vitro | Vero E6 cells | 17.689 | - | [7] |
| Dasatinib | In vitro | Vero E6 cells | 5.468 | - | [7] |
| Loperamide | In vitro | Vero E6 cells | 4.8 ± 1.5 | 15.5 ± 1.0 | [7] |
| Lopinavir | In vitro | Vero E6 cells | 8.0 ± 1.5 | 24.4 ± 1.0 | [7] |
| SSYA10-001 | In vitro | Vero E6 cells | ~25 | >500 | [60] |
| ESI-09 | In vitro | Calu-3 and Vero E6 cells | 5–10 | >50 | [61] |
| Mycophenolic acid | In vitro | Vero E6 cells | 2.87 | - | [60] |
| BCX4430 | In vitro | - | 68.4 | >100 | [62] |
| Fleximer analogues 2 | In vitro | Vero cells Huh-7 cells | 23 ± 0.6; 27 ± 0.0 | 71 ± 14; 149 ± 6.8 | [63] |
| Nutlin-3 | In vitro | Huh-7 cells | 6.9 ± 1.4 | 26.8 ± 1.6 | [64] |
| Amodiaquine dihydrochloride | In vitro | Huh-7 cells | 2.1 ± 0.7 | 12.3 ± 5.9 | [64] |
| Saracatinib | In vitro | Huh-7 cells | 2.9 ± 0.6 | 57 ± 5.5 | [64] |
| Sotrastaurin | In vitro | Huh-7 cells | 9.7 ± 3.3 | >50 | [64] |
| Acetophenazine maleate | In vitro | Huh-7 cells | 11.2 ± 5.0 | 23.6 ± 3.8 | [64] |
| Dosulepin hydrochloride | In vitro | Huh-7 cells | 3.4 ± 0.0 | 28.9 ± 0.0 | [64] |
| Methotrimeprazine maleate salt | In vitro | Huh-7 cells | 2.5 ± 0.0 | 24.5 ± 0.0 | [64] |
| N1-(4-pyridyl)-2-chloro-5-nitrobenzamide | In vitro | Huh-7 cells | 10.5 ± 0.3 | >50 | [64] |
| FA-613 | In vitro | Huh-7 cells | 10.2 ± 0.2 | - | [65] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, R.; Wang, L.; Zhang, N.; Deng, X.; Su, M.; Su, Y.; Hu, L.; He, C.; Ying, T.; Jiang, S.; et al. Development of Small-Molecule MERS-CoV Inhibitors. Viruses 2018, 10, 721. https://doi.org/10.3390/v10120721
Liang R, Wang L, Zhang N, Deng X, Su M, Su Y, Hu L, He C, Ying T, Jiang S, et al. Development of Small-Molecule MERS-CoV Inhibitors. Viruses. 2018; 10(12):721. https://doi.org/10.3390/v10120721
Chicago/Turabian StyleLiang, Ruiying, Lili Wang, Naru Zhang, Xiaoqian Deng, Meng Su, Yudan Su, Lanfang Hu, Chen He, Tianlei Ying, Shibo Jiang, and et al. 2018. "Development of Small-Molecule MERS-CoV Inhibitors" Viruses 10, no. 12: 721. https://doi.org/10.3390/v10120721
APA StyleLiang, R., Wang, L., Zhang, N., Deng, X., Su, M., Su, Y., Hu, L., He, C., Ying, T., Jiang, S., & Yu, F. (2018). Development of Small-Molecule MERS-CoV Inhibitors. Viruses, 10(12), 721. https://doi.org/10.3390/v10120721








