A Novel Betabaculovirus Isolated from the Monocot Pest Mocis latipes (Lepidoptera: Noctuidae) and the Evolution of Multiple-Copy Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Sample
2.2. Viral Genomic DNA Extraction and Amplification
2.3. Genome Sequencing, Assembly, and Annotation
2.4. Phylogenetic Analyses and Genome Comparison
2.5. Gene Amplification, Shuttle Vectors, and Recombinant AcMNPV Virus Construction
2.6. Recombinant Protein Analysis by SDS-PAGE
2.7. Microscopy
3. Results and Discussion
3.1. Sample Evaluation and Genome Sequencing
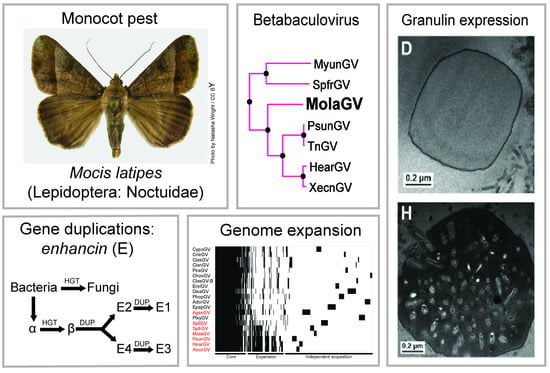
3.2. Virus Phylogeny
3.3. Gene Content
3.4. Evolution of the Betabaculovirus Multiple Copy Genes: Enhancin and a Endonuclease-Like
3.5. Genome Expansion
3.6. Genomic Analysis
3.7. Characterization of the MolaGV Granulin Gene
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Assunção-Albuquerque, M.J.T.; Peso-Aguiar, M.C.; Albuquerque, F.S. Using energy budget data to assess the most damaging life-stage of an agricultural pest Mocis latipes (Guenèe, 1982) (Lepidoptera-Noctuidae). Braz. J. Biol. 2010, 70, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Silvie, P.; Menzel, C.A.; Mello, A.; Coelho, A.G. Population dynamics of caterpillars on three cover crops before sowing cotton in Mato Grosso (Brazil). Commun. Agric. Appl. Biol. Sci. 2010, 75, 329–336. [Google Scholar] [PubMed]
- Jehle, J.A.; Blissard, G.W.; Bonning, B.C.; Cory, J.S.; Herniou, E.A.; Rohrmann, G.F.; Theilmann, D.A.; Thiem, S.M.; Vlak, J.M. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006, 151, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, D.R.; Miller, L.K.; Luckow, V.A. Baculovirus Expression Vectors: A Laboratory Manual; Oxford University Press on Demand, Freeman and Company: New York, NY, USA, 1992. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Pring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Pinto, F.A.; Mattos, M.V.; Silva, F.W.; Rocha, S.L.; Elliot, S.L. The Spread of Helicoverpa armigera (Lepidoptera: Noctuidae) and Coexistence with Helicoverpa zea in Southeastern Brazil. Insects 2017, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gil, M.; Dufayard, J.F.; Dessimoz, C.; Gascuel, O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, J.L.; Goodwin, R.H.; Tompkins, G.J.; McCawley, P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). Vitro 1977, 13, 213–217. [Google Scholar] [CrossRef]
- Ardisson-Araújo, D.M.P.; de Melo, F.L.; de Souza Andrade, M.; Sihler, W.; Báo, S.N.; Ribeiro, B.M.; de Souza, M.L. Genome sequence of Erinnyis ello granulovirus (ErelGV), a natural cassava hornworm pesticide and the first sequenced sphingid-infecting betabaculovirus. BMC Genom. 2014, 15, 856. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.A.; Lange, M.; Wang, H.; Hu, Z.; Wang, Y.; Hauschild, R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 2006, 346, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Ferrelli, M.L.; Salvador, R.; Biedma, M.E.; Berretta, M.F.; Haase, S.; Sciocco-Cap, A.; Ghiringhelli, P.D.; Romanowski, V. Genome of Epinotia aporema granulovirus (EpapGV), a polyorganotropic fast killing betabaculovirus with a novel thymidylate kinase gene. BMC Genom. 2012, 13, 548. [Google Scholar] [CrossRef] [PubMed]
- Clem, R.J. Viral IAPs, then and now. Semin. Cell Dev. Biol. 2015, 39, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Hinds, M.G.; Norton, R.S.; Vaux, D.L.; Day, C.L. Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat. Nat. Struct. Biol. 1999, 6, 648–651. [Google Scholar] [PubMed]
- Harrison, R.L.; Rowley, D.L.; Funk, C.J. The complete genome sequence of Plodia interpunctella granulovirus: Evidence for horizontal gene transfer and discovery of an unusual inhibitor-of-apoptosis gene. PLoS ONE 2016, 11, e0160389. [Google Scholar] [CrossRef] [PubMed]
- Bideshi, D.K.; Renault, S.; Stasiak, K.; Federici, B.A.; Bigot, Y. Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J. Gen. Virol. 2003, 84, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, V.; Slavicek, J.; Podgwaite, J.D.; Webb, R.; Fuester, R.; Peiffer, R.A. Deletion of v-chiA from a baculovirus reduces horizontal transmission in the field. Appl. Environ. Microbiol. 2013, 79, 4056–4064. [Google Scholar] [CrossRef] [PubMed]
- Ishimwe, E.; Hodgson, J.J.; Clem, R.J.; Passarelli, A.L. Reaching the melting point: Degradative enzymes and protease inhibitors involved in baculovirus infection and dissemination. Virology 2015, 479, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Bivian-Hernández, M.A.; López-Tlacomulco, J.; Mares-Mares, E.; Ibarra, J.E.; Del Rincón-Castro, M.C. Genomic analysis of a Trichoplusia ni Betabaculovirus (TnGV) with three different viral enhancing factors and two unique genes. Arch. Virol. 2017, 162, 3705–3715. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Corsaro, B.G.; Granados, R.R. Location and nucleotide sequence of the gene encoding the viral enhancing factor of the Trichoplusia ni granulosis virus. J. Gen. Virol. 1991, 72, 2645–2651. [Google Scholar] [CrossRef] [PubMed]
- Lepore, L.S.; Roelvink, P.R.; Granados, R.R. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J. Invertebr. Pathol. 1996, 68, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, D.S.; Slavicek, J.M. Molecular analysis of an enhancin gene in the Lymantria dispar nuclear polyhedrosis virus. J. Virol. 1997, 71, 8133–8140. [Google Scholar] [PubMed]
- Popham, H.J.; Bischoff, D.S.; Slavicek, J.M. Both Lymantria dispar Nucleopolyhedrovirus Enhancin Genes Contribute to Viral Potency. J. Virol. 2001, 75, 8639–8648. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.F.; Fang, J.C.; Wang, J.P.; Zhong, W.F.; Liu, B.S. Interaction of Xestia c-nigrum granulovirus with peritrophic matrix and Spodoptera litura nucleopolyhedrovirus in Spodoptera litura. J. Econ. Entomol. 2007, 100, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhao, L.; Ma, R.; Fang, W.; Hu, J.; Lei, C.; Sun, X. Improving baculovirus infectivity by efficiently embedding enhancing factors into occlusion bodies. Appl. Environ. Microbiol. 2017, 83, e00595-17. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhong, J.; Granados, R.R. A baculovirus enhancin alters the permeability of a mucosal midgut peritrophic matrix from lepidopteran larvae. J. Insect Physiol. 1999, 45, 159–166. [Google Scholar] [CrossRef]
- Wang, P.; Granados, R.R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. USA 1997, 94, 6977–6982. [Google Scholar] [CrossRef] [PubMed]
- Tanada, Y. A synopsis of studies on the synergistic property of an insect baculoviurs: A tribute to Edward A. Steinhaus. J. Invertebr. Pathol. 1985, 45, 125–138. [Google Scholar] [CrossRef]
- Hoover, K.; Humphries, M.A.; Gendron, A.R.; Slavicek, J.M. Impact of viral enhancin genes on potency of Lymantria dispar multiple nucleopolyhedrovirus in L. dispar following disruption of the peritrophic matrix. J. Invertebr. Pathol. 2010, 104, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Donly, C.; Li, Q.; Willis, L.G.; Keddie, B.A.; Erlandson, M.A.; Theilmann, D.A. Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata. Virology 2002, 297, 226–244. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Passarelli, A.L. The Autographa californica M nucleopolyhedrovirus ac79 gene encodes an early gene product with structural similarities to UvrC and intron-encoded endonucleases that is required for efficient budded virus production. J. Virol. 2012, 86, 5614–5625. [Google Scholar] [CrossRef] [PubMed]
- Herniou, E.A.; Olszewski, J.A.; Cory, J.S.; O’Reilly, D.R. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 2003, 48, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Rowley, D.L.; Mowery, J.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The Complete Genome Sequence of a Second Distinct Betabaculovirus from the True Armyworm, Mythimna unipuncta. PLoS ONE 2017, 12, e0170510. [Google Scholar] [CrossRef] [PubMed]
- Ardisson-Araujo, D.M.; Rohrmann, G.F.; Ribeiro, B.M.; Clem, R.J. Functional characterization of hesp018, a baculovirus-encoded serpin gene. J. Gen. Virol. 2015, 96, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.E.; Ko, R.; Maeda, S. Polyhedron-like inclusion body formation by a mutant nucleopolyhedrovirus expressing the granulin gene from a granulovirus. Virology 1998, 240, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Eason, J.E.; Hice, R.H.; Johnson, J.J.; Federici, B.A. Effects of Substituting Granulin or a Granulin-Polyhedrin Chimera for Polyhedrin on Virion Occlusion and Polyhedral Morphology in Autographa californicaMultinucleocapsid Nuclear Polyhedrosis Virus. J. Virol. 1998, 72, 6237–6243. [Google Scholar] [PubMed]
- Rohrmann, G.F. Baculovirus Molecular Biology, 3rd ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013. Available online: https://www.ncbi.nlm.nih.gov/books/NBK114593/pdf/Bookshelf_NBK114593.pdf (accessed on 13 December 2017).
- Carstens, E.B.; Williams, G.V.; Faulkner, P.; Partington, S. Analysis of polyhedra morphology mutants of Autographa californica nuclear polyhedrosis virus: Molecular and ultrastructural features. J. Gen. Virol. 1992, 73, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.W.; Carner, G.R.; Fescemyer, H.W. Polyhedrin sequence determines the tetrahedral shape of occlusion bodies in Thysanoplusia orichalcea single-nucleocapsid nucleopolyhedrovirus. J. Gen. Virol. 1998, 79, 2549–2556. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Luijckx, T.; Van Dinten, L.C.; van Oers, M.M.; Haj, J.P.; Bianchi, F.J.; van Lent, J.W.; Zuidema, D.; Vlak, J.M. Specificity of polyhedrin in the generation of baculovirus occlusion bodies. J. Gen. Virol. 1999, 80, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, B.M.; Generino, A.P.M.; Acacio, C.N.L.; Kalapothakis, E.; Báo, S.N. Characterization of a new Autographa californica multiple nucleopolyhedrovirus (AcMNPV) polyhedra mutant. Virus Res. 2009, 140, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.D.; Kim, W.J.; Kim, H.S.; Jin, B.R.; Lee, Y.H.; Kang, S.K. The morphology of the polyhedra of a host range-expanded recombinant baculovirus and its parents. Arch. Virol. 1998, 143, 1209–1214. [Google Scholar] [CrossRef] [PubMed]
- Ardisson-Araújo, D.M.P.; Rocha, J.R.; Da Costa, M.H.O.; Bocca, A.L.; Dusi, A.N.; de Oliveira Resende, R.; Ribeiro, B.M. A baculovirus-mediated strategy for full-length plant virus coat protein expression and purification. Virol. J. 2013, 10, 262. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ardisson-Araújo, D.M.P.; Da Silva, A.M.R.; Melo, F.L.; Dos Santos, E.R.; Sosa-Gómez, D.R.; Ribeiro, B.M. A Novel Betabaculovirus Isolated from the Monocot Pest Mocis latipes (Lepidoptera: Noctuidae) and the Evolution of Multiple-Copy Genes. Viruses 2018, 10, 134. https://doi.org/10.3390/v10030134
Ardisson-Araújo DMP, Da Silva AMR, Melo FL, Dos Santos ER, Sosa-Gómez DR, Ribeiro BM. A Novel Betabaculovirus Isolated from the Monocot Pest Mocis latipes (Lepidoptera: Noctuidae) and the Evolution of Multiple-Copy Genes. Viruses. 2018; 10(3):134. https://doi.org/10.3390/v10030134
Chicago/Turabian StyleArdisson-Araújo, Daniel M. P., Ana Maria Rodrigues Da Silva, Fernando L. Melo, Ethiane Rozo Dos Santos, Daniel R. Sosa-Gómez, and Bergmann M. Ribeiro. 2018. "A Novel Betabaculovirus Isolated from the Monocot Pest Mocis latipes (Lepidoptera: Noctuidae) and the Evolution of Multiple-Copy Genes" Viruses 10, no. 3: 134. https://doi.org/10.3390/v10030134
APA StyleArdisson-Araújo, D. M. P., Da Silva, A. M. R., Melo, F. L., Dos Santos, E. R., Sosa-Gómez, D. R., & Ribeiro, B. M. (2018). A Novel Betabaculovirus Isolated from the Monocot Pest Mocis latipes (Lepidoptera: Noctuidae) and the Evolution of Multiple-Copy Genes. Viruses, 10(3), 134. https://doi.org/10.3390/v10030134