Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells and Viruses
2.3. Pigs
2.4. Experimental Design
2.5. Evaluation of Postmortem Tissues
2.6. Indirect Immunofluorescence Assay and Real-Time PCR Assay
2.7. Hemagglutination Inhibition Assay
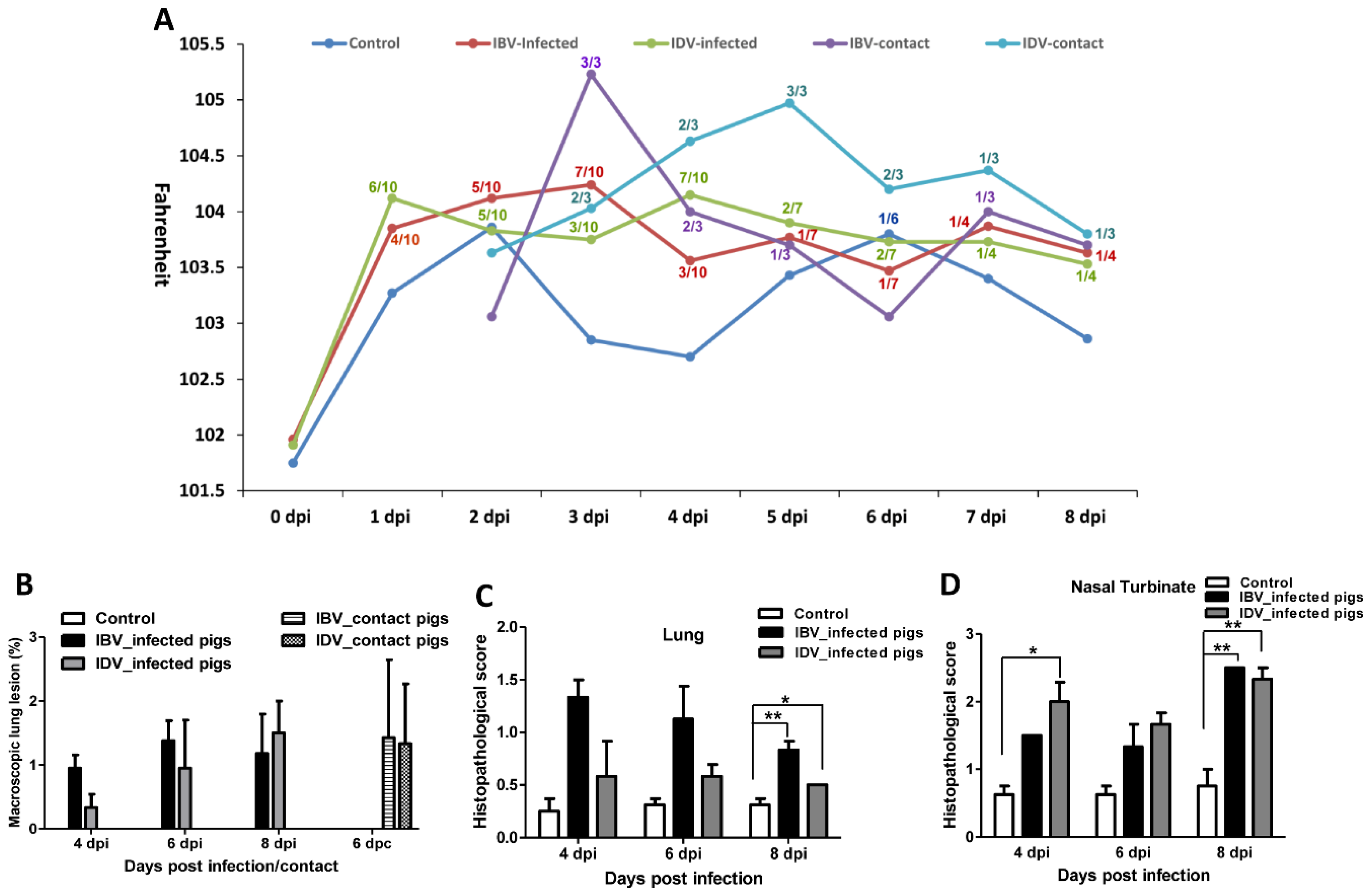
3. Results
3.1. Pathogenicity of IBV and IDV in Pigs
3.2. Transmissibility of IBV and IDV in Pigs
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kilbourne, E.D. Taxonomy and Comparative Virology of the Influenza Viruses; Springer: Boston, MA, USA, 1987; pp. 25–32. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. mBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Subbarao, K. Global epidemiology of influenza: Past and present. Annu. Rev. Med. 2000, 51, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb. Mortal. Wkly. Rep. 2010, 59, 1057–1062. [Google Scholar]
- Matsuzaki, Y.; Katsushima, N.; Nagai, Y.; Shoji, M.; Itagaki, T.; Sakamoto, M.; Kitaoka, S.; Mizuta, K.; Nishimura, H. Clinical features of influenza C virus infection in children. J. Infect. Dis. 2006, 193, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, G.C.; Lednicky, J.A. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Kanegae, Y.; Sugita, S.; Endo, A.; Ishida, M.; Senya, S.; Osako, K.; Nerome, K.; Oya, A. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: Cocirculating lineages in the same epidemic season. J. Virol. 1990, 64, 2860–2865. [Google Scholar] [PubMed]
- Rota, P.A.; Wallis, T.R.; Harmon, M.W.; Rota, J.S.; Kendal, A.P.; Nerome, K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 1990, 175, 59–68. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Sugawara, K.; Furuse, Y.; Shimotai, Y.; Hongo, S.; Oshitani, H.; Mizuta, K.; Nishimura, H. Genetic Lineage and Reassortment of Influenza C Viruses Circulating between 1947 and 2014. J. Virol. 2016, 90, 8251–8265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odagiri, T.; Ishida, H.; Li, J.Y.; Endo, M.; Kobayashi, T.; Kamiki, H.; Matsugo, H.; Takenaka-Uema, A.; Murakami, S.; Horimoto, T. Antigenic heterogeneity among phylogenetic clusters of influenza D viruses. J. Vet. Med. Sci. 2018, 80, 1241–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultz-Cherry, S.; Olsen, C.W.; Easterday, B.C. History of Swine influenza. Curr. Top. Microbiol. Immunol. 2013, 370, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Jain, S.; Finelli, L.; Shaw, M.W.; Lindstrom, S.; Garten, R.J.; Gubareva, L.V.; Xu, X.; Bridges, C.B.; Uyeki, T.M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009, 360, 2605–2615. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.J. Ecological aspects of influenza A viruses in animals and their relationship to human influenza: A review. J. R. Soc. Med. 1982, 75, 799–811. [Google Scholar] [PubMed]
- Ma, W.; Kahn, R.E.; Richt, J.A. The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J. Mol. Genet. Med. 2008, 3, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Holmes, E.C. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008, 66, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Baine, W.B.; Luby, J.P.; Martin, S.M. Severe illness with influenza B. Am. J. Med. 1980, 68, 181–189. [Google Scholar] [CrossRef]
- Dijkstra, F.; Donker, G.A.; Wilbrink, B.; Van Gageldonk-Lafeber, A.B.; Van Der Sande, M.A. Long time trends in influenza-like illness and associated determinants in The Netherlands. Epidemiol. Infect. 2009, 137, 473–479. [Google Scholar] [CrossRef]
- Chang, C.P.; New, A.E.; Taylor, J.F.; Chiang, H.S. Influenza virus isolations from dogs during a human epidemic in Taiwan. Int. J. Zoonoses 1976, 3, 61–64. [Google Scholar]
- Romvary, J.; Meszaros, J.; Barb, K. Susceptibility of birds to type-B influenza virus. Acta Microbiol. Acad. Sci. Hung. 1980, 27, 279–287. [Google Scholar]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A. Influenza B virus in seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef]
- Bodewes, R.; Morick, D.; de Mutsert, G.; Osinga, N.; Bestebroer, T.; van der Vliet, S.; Smits, S.L.; Kuiken, T.; Rimmelzwaan, G.F.; Fouchier, R.A.; et al. Recurring influenza B virus infections in seals. Emerg. Infect. Dis. 2013, 19, 511–512. [Google Scholar] [CrossRef]
- Takatsy, G.; Farkas, E.; Romvary, J. Susceptibility of the domestic pig to influenza B virus. Nature 1969, 222, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Takatsy, G.; Romvary, J.; Farkas, E. Susceptibility of the domestic swine to influenza B virus. Acta Microbiol. Acad. Sci. Hung. 1967, 14, 309–315. [Google Scholar] [PubMed]
- Ran, Z.; Shen, H.; Lang, Y.; Kolb, E.A.; Turan, N.; Zhu, L.; Ma, J.; Bawa, B.; Liu, Q.; Liu, H.; et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015, 89, 4818–4826. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.P.; Tsai, H.J. Influenza B viruses in pigs, Taiwan. Influenza Other Respir. Viruses 2019, 13, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Jin, F.G.; Wang, P.; Wang, M.; Zhu, J.M. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 1983, 64 Pt 1, 177–182. [Google Scholar] [CrossRef]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Pelletier, C.; Meyer, G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Eckard, L.; Epperson, W.B.; Long, L.P.; Smith, D.; Huston, C.; Genova, S.; Webby, R.; Wan, X.F. Influenza D virus infection in Mississippi beef cattle. Virology 2015, 486, 28–34. [Google Scholar] [CrossRef] [Green Version]
- Collin, E.A.; Sheng, Z.; Lang, Y.; Ma, W.; Hause, B.M.; Li, F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J. Virol. 2015, 89, 1036–1042. [Google Scholar] [CrossRef]
- Ferguson, L.; Olivier, A.K.; Genova, S.; Epperson, W.B.; Smith, D.R.; Schneider, L.; Barton, K.; McCuan, K.; Webby, R.J.; Wan, X.F. Pathogenesis of Influenza D Virus in Cattle. J. Virol. 2016, 90, 5636–5642. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.M.; Wang, S.C.; Peng, C.; Yu, J.M.; Zhuang, Q.Y.; Hou, G.Y.; Liu, S.; Li, J.P.; Chen, J.M. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 2014, 49, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Endoh, M.; Kobayashi, T.; Takenaka-Uema, A.; Chambers, J.K.; Uchida, K.; Nishihara, M.; Hause, B.; Horimoto, T. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg. Infect. Dis. 2016, 22, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Chiapponi, C.; Faccini, S.; De Mattia, A.; Baioni, L.; Barbieri, I.; Rosignoli, C.; Nigrelli, A.; Foni, E. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg. Infect. Dis. 2016, 22, 352–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flynn, O.; Gallagher, C.; Mooney, J.; Irvine, C.; Ducatez, M.; Hause, B.; McGrath, G.; Ryan, E. Influenza D Virus in Cattle, Ireland. Emerg. Infect. Dis. 2018, 24, 389–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, M.; Sreenivasan, C.; Sexton, G.; Nedland, H.; Singrey, A.; Fawcett, L.; Miller, G.; Lauer, D.; Voss, S.; Pollock, S.; et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015, 180, 281–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nedland, H.; Wollman, J.; Sreenivasan, C.; Quast, M.; Singrey, A.; Fawcett, L.; Christopher-Hennings, J.; Nelson, E.; Kaushik, R.S.; Wang, D.; et al. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 2018, 65, e148–e154. [Google Scholar] [CrossRef] [PubMed]
- Salem, E.; Cook, E.A.J.; Lbacha, H.A.; Oliva, J.; Awoume, F.; Aplogan, G.L.; Hymann, E.C.; Muloi, D.; Deem, S.L.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017, 23, 1556–1559. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Odagiri, T.; Melaku, S.K.; Bazartseren, B.; Ishida, H.; Takenaka-Uema, A.; Muraki, Y.; Sentsui, H.; Horimoto, T. Influenza D Virus Infection in Dromedary Camels, Ethiopia. Emerg. Infect. Dis. 2019, 25, 1224–1226. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Richt, J.A.; Lager, K.M.; Janke, B.H.; Woods, R.D.; Webster, R.G.; Webby, R.J. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 2003, 41, 3198–3205. [Google Scholar] [CrossRef]
- Ma, W.; Vincent, A.L.; Gramer, M.R.; Brockwell, C.B.; Lager, K.M.; Janke, B.H.; Gauger, P.C.; Patnayak, D.P.; Webby, R.J.; Richt, J.A. Identification of H2N3 influenza A viruses from swine in the United States. Proc. Natl. Acad. Sci. USA 2007, 104, 20949–20954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Henningson, J.; Ma, J.; Duff, M.; Lang, Y.; Li, Y.; Nagy, A.; Sunwoo, S.; Bawa, B.; Yang, J.; et al. Effects of PB1-F2 on the pathogenicity of H1N1 swine influenza virus in mice and pigs. J. Gen. Virol. 2017, 98, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, S.B.; Selvarangan, R. Evaluation of three influenza A and B real-time reverse transcription-PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J. Clin. Microbiol. 2010, 48, 3870–3875. [Google Scholar] [CrossRef] [PubMed]
- Mitra, N.; Cernicchiaro, N.; Torres, S.; Li, F.; Hause, B.M. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 2016, 97, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chung, H.K.; Chae, C. Association of porcine circovirus 2 with porcine respiratory disease complex. Vet. J. 2003, 166, 251–256. [Google Scholar] [CrossRef]
- Chae, C. Porcine respiratory disease complex: Interaction of vaccination and porcine circovirus type 2, porcine reproductive and respiratory syndrome virus, and Mycoplasma hyopneumoniae. Vet. J. 2016, 212, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, L.F.; Ramirez Nieto, G.; Alfonso, V.V.; Correa, J.J. Association of swine influenza H1N1 pandemic virus (SIV-H1N1p) with porcine respiratory disease complex in sows from commercial pig farms in Colombia. Virol. Sin. 2014, 29, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Cibulski, S.P.; Andrade, C.P.; Teixeira, T.F.; Varela, A.P.; Scheffer, C.M.; Franco, A.C.; de Almeida, L.L.; Roehe, P.M. Swine Influenza Virus and Association with the Porcine Respiratory Disease Complex in Pig Farms in Southern Brazil. Zoonoses Public Health 2016, 63, 234–240. [Google Scholar] [CrossRef]
- Rech, R.R.; Gava, D.; Silva, M.C.; Fernandes, L.T.; Haach, V.; Ciacci-Zanella, J.R.; Schaefer, R. Porcine respiratory disease complex after the introduction of H1N1/2009 influenza virus in Brazil. Zoonoses Public Health 2018, 65, e155–e161. [Google Scholar] [CrossRef]



| dpi/dpc | Nasal Swab | BALF | ||||||
|---|---|---|---|---|---|---|---|---|
| IBV | IDV | IBV | IDV | |||||
| Infected | Contact | Infected | Contact | Infected | Contact | Infected | Contact | |
| 2 dpi | ND (0/10) | ND (0/3) | 34.88 (5/10) | ND (0/3) | NA | NA | NA | NA |
| 4 dpi (2 dpc) | 34.36 (3/10) | ND (0/3) | 30.36 (8/10) | 35.89 (3/3) | 28.84 (3/3) | NA | 24.10 (3/3) | NA |
| 6 dpi (4 dpc) | ND (0/7) | ND (0/3) | 31.30 (7/7) | 35.25 (1/3) | 29.08 (2/3) | NA | 28.33 (3/3) | NA |
| 8 dpi (6 dpc) | ND (0/4) | ND (0/3) | 35.18 (2/4) | 30.02 (1/3) | ND (0/4) | ND (0/3) | 31.54 (4/4) | 34.44 (1/3) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Wang, L.; Palinski, R.; Walsh, T.; He, D.; Li, Y.; Wu, R.; Lang, Y.; Sunwoo, S.-Y.; Richt, J.A.; et al. Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs. Viruses 2019, 11, 905. https://doi.org/10.3390/v11100905
Lee J, Wang L, Palinski R, Walsh T, He D, Li Y, Wu R, Lang Y, Sunwoo S-Y, Richt JA, et al. Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs. Viruses. 2019; 11(10):905. https://doi.org/10.3390/v11100905
Chicago/Turabian StyleLee, Jinhwa, Liping Wang, Rachel Palinski, Tim Walsh, Dongchang He, Yonghai Li, Rui Wu, Yuekun Lang, Sun-Young Sunwoo, Juergen A. Richt, and et al. 2019. "Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs" Viruses 11, no. 10: 905. https://doi.org/10.3390/v11100905
APA StyleLee, J., Wang, L., Palinski, R., Walsh, T., He, D., Li, Y., Wu, R., Lang, Y., Sunwoo, S.-Y., Richt, J. A., & Ma, W. (2019). Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs. Viruses, 11(10), 905. https://doi.org/10.3390/v11100905






