Structural Pattern Differences in Unbranched Rod-Like RNA of Hepatitis Delta Virus Affect RNA Editing
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. DNA Transfection and Posttransfection Analyses of HDV RNA and HDAg
3. Results
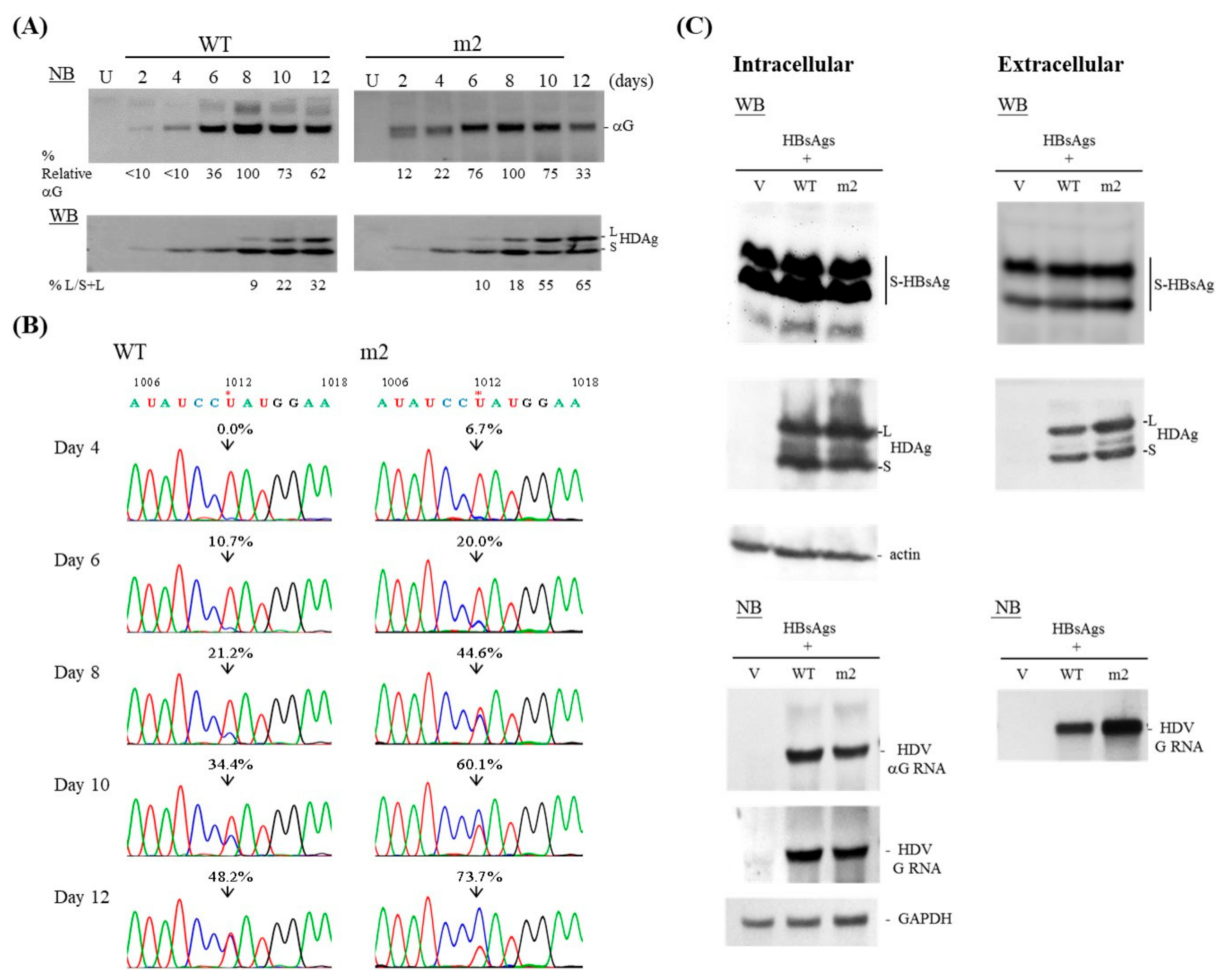
3.1. Effect of Structural Differences in the HDV Replication
3.2. Kinetics of HDV Mutant m2
3.3. Packaging Ability of the m2 Mutant
3.4. The Length, But Not the Position, of the Consecutive bp on the HDV Antigenome Affects RNA Editing
3.5. HDAg Fails to Regulate Amber/W Editing of HDV Mutants Carrying Elongated Base-Pairing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rizzetto, M.; Hoyer, B.; Canese, M.G.; Shih, J.W.; Purcell, R.H.; Gerin, J.L. delta Agent: Association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 1980, 77, 6124–6128. [Google Scholar] [CrossRef]
- Bonino, F.; Heermann, K.H.; Rizzetto, M.; Gerlich, W.H. Hepatitis delta virus: Protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 1986, 58, 945–950. [Google Scholar]
- Kos, A.; Dijkema, R.; Arnberg, A.C.; Van der Meide, P.H.; Schellekens, H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986, 323, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.S.; Choo, Q.L.; Weiner, A.J.; Ou, J.H.; Najarian, R.C.; Thayer, R.M.; Mullenbach, G.T.; Denniston, K.J.; Gerin, J.L.; Houghton, M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature 1986, 323, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Deny, P. Hepatitis delta virus genetic variability: From genotypes I, II, III to eight major clades? Curr. Top. Microbiol. Immunol. 2006, 307, 151–171. [Google Scholar] [PubMed]
- Le Gal, F.; Brichler, S.; Drugan, T.; Alloui, C.; Roulot, D.; Pawlotsky, J.M.; Deny, P.; Gordien, E. Genetic diversity and worldwide distribution of the deltavirus genus: A study of 2,152 clinical strains. Hepatology 2017, 66, 1826–1841. [Google Scholar] [CrossRef] [PubMed]
- Jayan, G.C.; Casey, J.L. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J. Virol. 2002, 76, 12399–12404. [Google Scholar] [CrossRef]
- Wong, S.K.; Lazinski, D.W. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 2002, 99, 15118–15123. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.L. Control of ADAR1 editing of hepatitis delta virus RNAs. Curr. Top. Microbiol. Immunol. 2012, 353, 123–143. [Google Scholar]
- Chang, F.L.; Chen, P.J.; Tu, S.J.; Wang, C.J.; Chen, D.S. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 1991, 88, 8490–8494. [Google Scholar] [CrossRef]
- Chao, M.; Hsieh, S.Y.; Taylor, J. Role of two forms of hepatitis delta virus antigen: Evidence for a mechanism of self-limiting genome replication. J. Virol. 1990, 64, 5066–5069. [Google Scholar] [PubMed]
- Sato, S.; Wong, S.K.; Lazinski, D.W. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 2001, 75, 8547–8555. [Google Scholar] [CrossRef]
- Defenbaugh, D.A.; Johnson, M.; Chen, R.; Zheng, Y.Y.; Casey, J.L. Hepatitis delta antigen requires a minimum length of the hepatitis delta virus unbranched rod RNA structure for binding. J. Virol. 2009, 83, 4548–4556. [Google Scholar] [CrossRef]
- Sato, S.; Cornillez-Ty, C.; Lazinski, D.W. By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J. Virol. 2004, 78, 8120–8134. [Google Scholar] [CrossRef]
- Polson, A.G.; Ley, H.L., 3rd; Bass, B.L.; Casey, J.L. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 1998, 18, 1919–1926. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.L.; Gerin, J.L. Hepatitis D virus RNA editing: Specific modification of adenosine in the antigenomic RNA. J. Virol. 1995, 69, 7593–7600. [Google Scholar] [PubMed]
- Polson, A.G.; Bass, B.L.; Casey, J.L. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 1996, 380, 454–456. [Google Scholar] [CrossRef] [PubMed]
- Jayan, G.C.; Casey, J.L. Effects of conserved RNA secondary structures on hepatitis delta virus genotype I RNA editing, replication, and virus production. J. Virol. 2005, 79, 11187–11193. [Google Scholar] [CrossRef]
- Casey, J.L. RNA editing in hepatitis delta virus genotype III requires a branched double-hairpin RNA structure. J. Virol. 2002, 76, 7385–7397. [Google Scholar] [CrossRef]
- Kuo, M.Y.; Goldberg, J.; Coates, L.; Mason, W.; Gerin, J.; Taylor, J. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: Sequence, structure, and applications. J. Virol. 1988, 62, 1855–1861. [Google Scholar]
- Chao, M.; Wang, T.C.; Lin, C.C.; Yung-Liang Wang, R.; Lin, W.B.; Lee, S.E.; Cheng, Y.Y.; Yeh, C.T.; Iang, S.B. Analyses of a whole-genome inter-clade recombination map of hepatitis delta virus suggest a host polymerase-driven and viral RNA structure-promoted template-switching mechanism for viral RNA recombination. Oncotarget 2017, 8, 60841–60859. [Google Scholar] [CrossRef] [PubMed]
- Makarova, O.; Kamberov, E.; Margolis, B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 2000, 29, 970–972. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Bellaousov, S.; Reuter, J.S.; Seetin, M.G.; Mathews, D.H. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013, 41, (Web Server issue). W471-4. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; pp. 1.1–1.170. [Google Scholar]
- Wang, T.C.; Chao, M. RNA recombination of hepatitis delta virus in natural mixed-genotype infection and transfected cultured cells. J. Virol. 2005, 79, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Gudima, S.; Wu, S.Y.; Chiang, C.M.; Moraleda, G.; Taylor, J. Origin of hepatitis delta virus mRNA. J. Virol. 2000, 74, 7204–7210. [Google Scholar] [CrossRef] [PubMed]
- Ryu, W.S.; Bayer, M.; Taylor, J. Assembly of hepatitis delta virus particles. J. Virol. 1992, 66, 2310–2315. [Google Scholar]
- Mathews, D.H.; Sabina, J.; Zuker, M.; Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999, 288, 911–940. [Google Scholar] [CrossRef]
- Beard, M.R.; MacNaughton, T.B.; Gowans, E.J. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J. Virol. 1996, 70, 4986–4995. [Google Scholar]
- Kuo, M.Y.; Sharmeen, L.; Dinter-Gottlieb, G.; Taylor, J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 1988, 62, 4439–4444. [Google Scholar]
- Wu, H.N.; Lin, Y.J.; Lin, F.P.; Makino, S.; Chang, M.F.; Lai, M.M. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc. Natl. Acad. Sci. USA 1989, 86, 1831–1835. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 2004, 10, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Griffin, B.L.; Chasovskikh, S.; Dritschilo, A.; Casey, J.L. Hepatitis delta antigen requires a flexible quasi-double-stranded RNA structure to bind and condense hepatitis delta virus RNA in a ribonucleoprotein complex. J. Virol. 2014, 88, 7402–7411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Filzmayer, C.; Ni, Y.; Sultmann, H.; Mutz, P.; Hiet, M.S.; Vondran, F.W.R.; Bartenschlager, R.; Urban, S. Hepatitis D virus replication is sensed by MDA5 and induces IFN-beta/lambda responses in hepatocytes. J. Hepatol. 2018, 69, 25–35. [Google Scholar] [CrossRef] [PubMed]



 , occurs to form high-order structured HDV RNA and to bring ADAR1 and amber/W editing site in close proximity (bottom). Individual steps are described in the text.
, occurs to form high-order structured HDV RNA and to bring ADAR1 and amber/W editing site in close proximity (bottom). Individual steps are described in the text.
 , occurs to form high-order structured HDV RNA and to bring ADAR1 and amber/W editing site in close proximity (bottom). Individual steps are described in the text.
, occurs to form high-order structured HDV RNA and to bring ADAR1 and amber/W editing site in close proximity (bottom). Individual steps are described in the text.
| Consecutive bp † | Accession no. | |||
|---|---|---|---|---|
| 9 | AM779576 | |||
| 10 | AB118849 | HQ005365 | HQ005369 | HQ005370 |
| AM779580 | KF660600 | HQ005367 | HQ005371 | |
| HQ005372 | M58629 | M28267 | ||
| 11 | AM902165 | AM902170 | JX888099 | AF425644 |
| X77627 | D01075 | AM779579 | AM902177 | |
| AM902179 | M21012 | |||
| 12 | AJ000558 | AM779574 | AM902164 | AM902167 |
| JX888101 | JX888112 | AM779594 | HM046802 | |
| M92448 | X85253 | AF098261 | ||
| 13 | U81988 | AM902166 | ||
| 14 | JX888108 | HQ005364 | AM902174 | |
| 15 | U81989 | HQ005366 | HQ005368 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, C.-W.; Juang, H.-H.; Kuo, C.-Y.; Li, H.-P.; Iang, S.-B.; Lin, S.-H.; Yeh, C.-T.; Chao, M. Structural Pattern Differences in Unbranched Rod-Like RNA of Hepatitis Delta Virus Affect RNA Editing. Viruses 2019, 11, 934. https://doi.org/10.3390/v11100934
Hsu C-W, Juang H-H, Kuo C-Y, Li H-P, Iang S-B, Lin S-H, Yeh C-T, Chao M. Structural Pattern Differences in Unbranched Rod-Like RNA of Hepatitis Delta Virus Affect RNA Editing. Viruses. 2019; 11(10):934. https://doi.org/10.3390/v11100934
Chicago/Turabian StyleHsu, Chao-Wei, Horng-Heng Juang, Chien-Yi Kuo, Hsin-Pai Li, Shan-Bei Iang, Siao-Han Lin, Chau-Ting Yeh, and Mei Chao. 2019. "Structural Pattern Differences in Unbranched Rod-Like RNA of Hepatitis Delta Virus Affect RNA Editing" Viruses 11, no. 10: 934. https://doi.org/10.3390/v11100934
APA StyleHsu, C.-W., Juang, H.-H., Kuo, C.-Y., Li, H.-P., Iang, S.-B., Lin, S.-H., Yeh, C.-T., & Chao, M. (2019). Structural Pattern Differences in Unbranched Rod-Like RNA of Hepatitis Delta Virus Affect RNA Editing. Viruses, 11(10), 934. https://doi.org/10.3390/v11100934





