Multiple Correspondence Analysis on Amino Acid Properties within the Variable Region of the Capsid Protein Shows Differences between Classical and Virulent Systemic Feline Calicivirus Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequences
2.2. Virus Collection
2.3. Virus Isolation
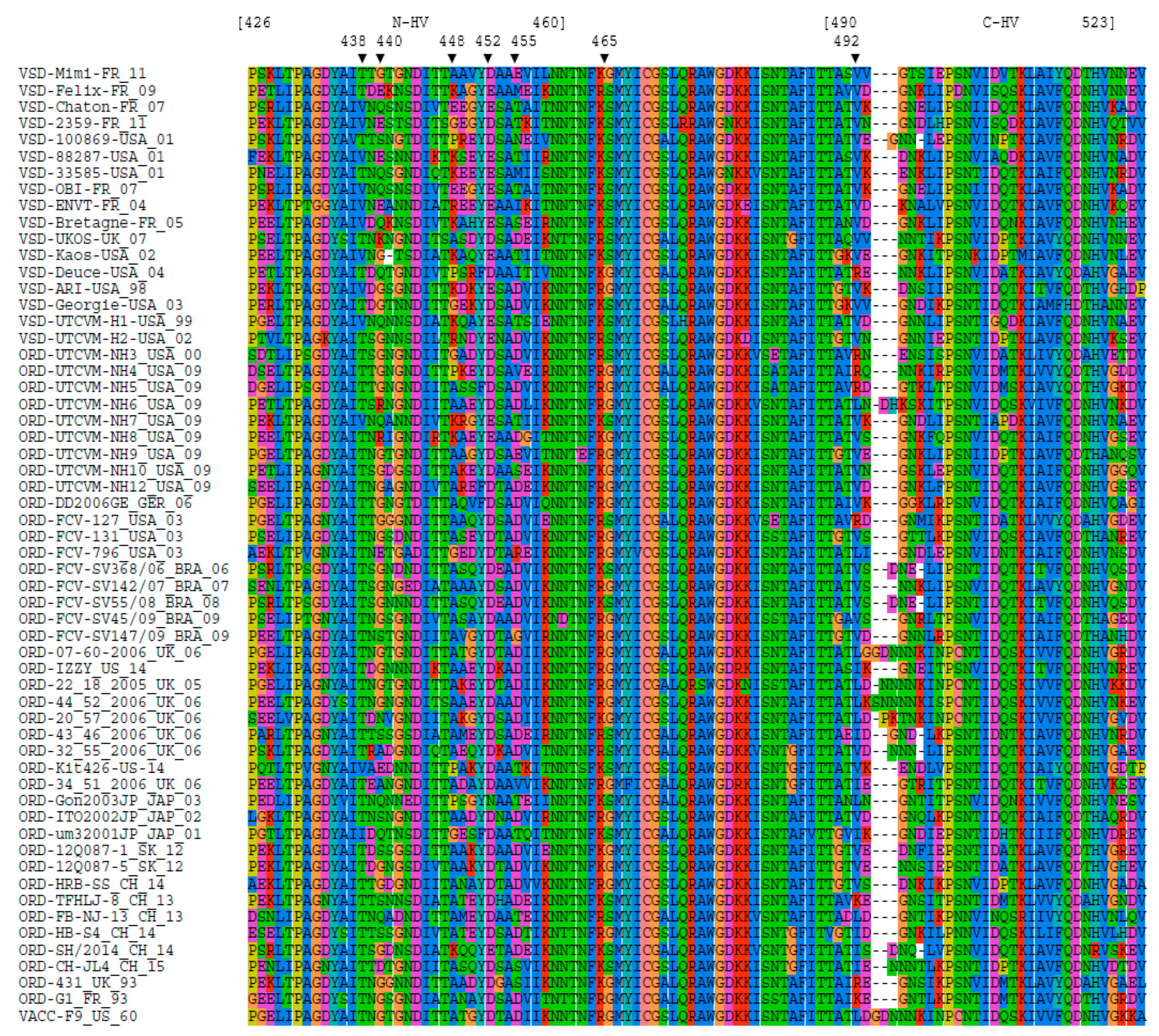
2.4. Sequencing of Region E
2.5. Phylogenic Analysis
2.6. Multiple Correspondence Analysis (MCA) of Region E
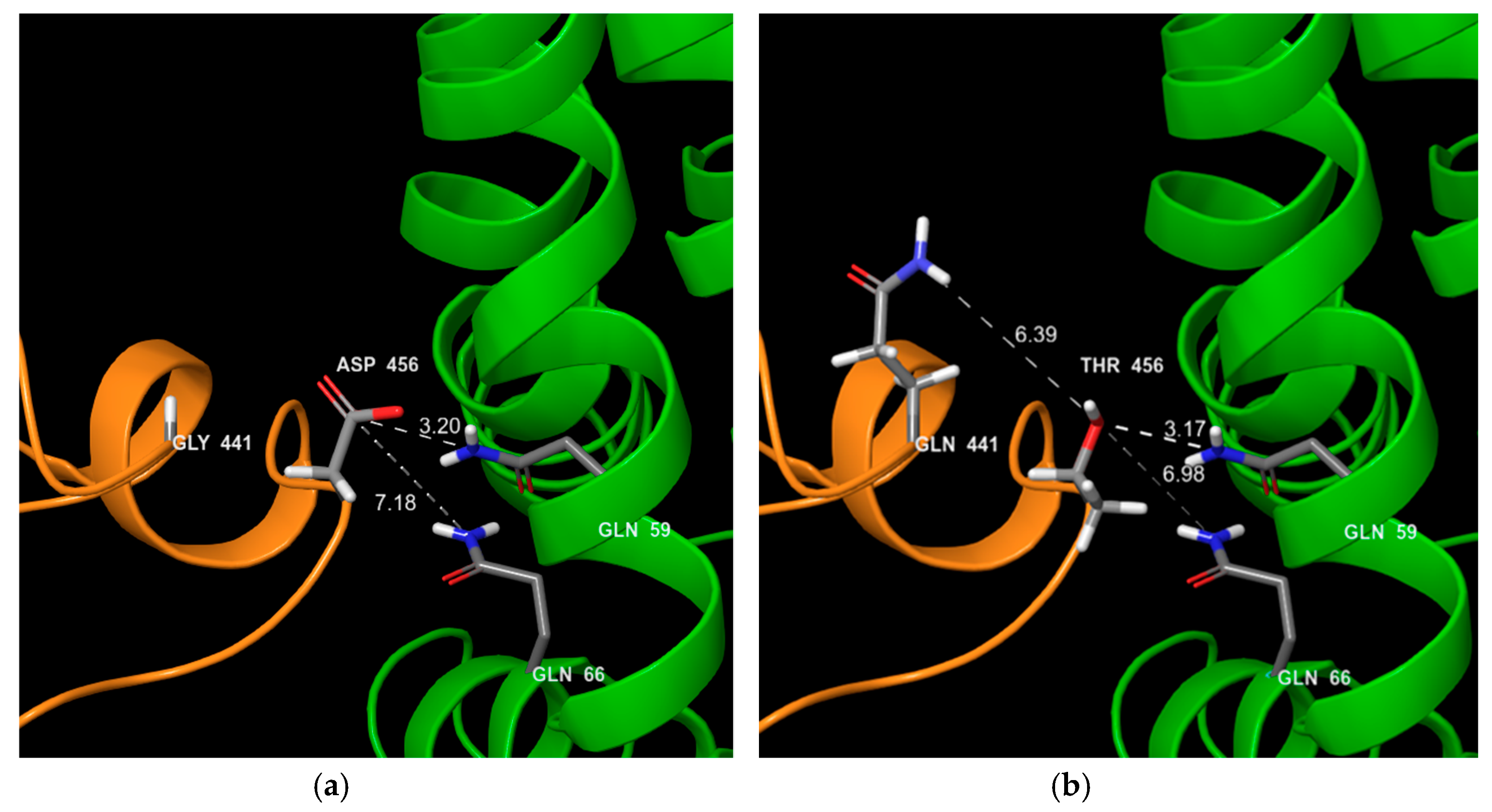
2.7. Structural Impact of Mutations
3. Results
3.1. Phylogeny
3.2. MCA Analysis on Amino Acids Properties
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bhella, D.; Goodfellow, I.G. The Cryo-Electron Microscopy Structure of Feline Calicivirus Bound to Junctional Adhesion Molecule A at 9-Angstrom Resolution Reveals Receptor-Induced Flexibility and Two Distinct Conformational Changes in the Capsid Protein VP1. J. Virol. 2011, 85, 11381–11390. [Google Scholar] [CrossRef] [PubMed]
- Sosnovtsev, S.V.; Belliot, G.; Chang, K.O.; Onwudiwe, O.; Green, K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005, 79, 4012–4024. [Google Scholar] [CrossRef] [PubMed]
- Poulet, H.; Brunet, S.; Soulier, M.; Leroy, V.; Goutebroze, S.; Chappuis, G. Comparison between acute oral/respiratory and chronic stomatitis/gingivitis isolates of feline calicivirus: Pathogenicity, antigenic profile and cross-neutralisation studies. Arch. Virol. 2000, 145, 243–261. [Google Scholar] [CrossRef] [PubMed]
- Coyne, K.P.; Christley, R.M.; Pybus, O.G.; Dawson, S.; Gaskell, R.M.; Radford, A.D. Large-scale spatial and temporal genetic diversity of feline calicivirus. J. Virol. 2012, 86, 11356–11367. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Sánchez-Vizcaíno, F.; Mcgahie, D.; Lesbros, C.; Almeras, T.; Howarth, D.; O’hara, V.; Dawson, S.; Radford, A.D. European molecular epidemiology and strain diversity of feline calicivirus. Vet. Rec. 2016, 178, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Afonso, M.M.; Pinchbeck, G.L.; Smith, S.L.; Daly, J.M.; Gaskell, R.M.; Dawson, S.; Radford, A.D. A multi-national European cross-sectional study of feline Calicivirus epidemiology, diversity and vaccine cross-reactivity. Vaccine 2017, 35, 2753–2760. [Google Scholar] [CrossRef]
- Seal, B.S.; Ridpath, J.F.; Mengeling, W.L. Analysis of feline calicivirus capsid protein genes: Identification of variable antigenic determinant regions of the protein. J. Gen. Virol. 1993, 74, 2519–2524. [Google Scholar] [CrossRef]
- Seal, B.S. Analysis of capsid protein gene variation among divergent isolates of feline calicivirus. Virus Res. 1994, 33, 39–53. [Google Scholar] [CrossRef]
- Radford, A.D.; Willoughby, K.; Dawson, S.; McCracken, C.; Gaskel, R.M. The Capsid Gene of Feline Calicivirus Contains Linear B-Cell Epitopes in both Variable and Conserved Regions. J. Virol. 1999, 73, 8496–8502. [Google Scholar]
- Tohya, Y.; Yokoyama, N.; Maeda, K.; Kawaguchi, Y.; Mikami, T. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J. Gen. Virol. 1997, 78, 303–305. [Google Scholar] [CrossRef]
- Makino, A.; Shimojima, M.; Miyazawa, T.; Kato, K.; Tohya, Y.; Akashi, H. Junctional Adhesion Molecule 1 Is a Functional Receptorfor Feline Calicivirus. J. Virol. 2006, 80, 4482–4490. [Google Scholar] [CrossRef] [PubMed]
- Ossiboff, R.J.; Parker, J.S. Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. J. Virol. 2007, 81, 13608–13621. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ledgerwood, E.D.; Hinchman, M.M.; Dick, R.; Parker, J.S.L. Conserved surface residues on the feline calicivirus capsid are essential for interaction with its receptor feline junctional adhesion molecule A (fJAM-A). J. Virol. 2018, 92, e00035-18. [Google Scholar] [CrossRef] [PubMed]
- Stuart, A.D.; Brown, T.D.K. a2,6-Linked sialic acid acts as a receptor for Feline calicivirus. J. Gen. Virol. 2007, 88, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Radford, A.D.; Coyne, K.P.; Dawson, S.; Porter, C.J.; Gaskell, R.M. Feline calicivirus. Vet. Res. 2007, 38, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Manzanilla, E.G.; Lloret, A.; León, M.; Thibault, J.C. Prevalence of feline herpesvirus-1, feline calicivirus, Chlamydophila felis and Mycoplasma felis DNA and associated risk factors in cats in Spain with upper respiratory tract disease, conjunctivitis and/or gingivostomatitis. J. Feline Med. Surg. 2017, 19, 461–469. [Google Scholar] [CrossRef]
- Reubel, G.H.; Hoffmann, D.E.; Pedersen, N.C. Acute and chronic faucitis of domestic cats. A feline calicivirus-induced disease. Feline Dentistry 1992, 22, 1347–1360. [Google Scholar]
- Waters, L.; Hopper, C.D.; Gruffydd-Jones, T.J.; Harbour, D.A. Chronic gingivitis in a colony of cats infected with feline immunodeficiency virus and feline calicivirus. Vet. Rec. 1993, 132, 340–342. [Google Scholar] [CrossRef]
- Thomas, S.; Lappin, D.F.; Spears, J.; Bennett, D.; Nile, C.; Riggio, M.P. Prevalence of feline calicivirus in cats with odontoclastic resorptive lesions and chronic gingivostomatitis. Res. Vet. Sci. 2017, 111, 124–126. [Google Scholar] [CrossRef]
- Dick, C.P.; Johnson, R.P. Sites of persistence of feline calicivirus. Res. Vet. Sci. 1989, 47, 367–373. [Google Scholar] [CrossRef]
- Wardley, R.C. Feline calicivirus carrier state. A study of the host/virus relationship. Arch. Virol. 1976, 52, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Elliott, J.B.; Glasgow, A.; Poland, A.; Keel, K. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet. Microbiol. 2000, 73, 281–300. [Google Scholar] [CrossRef]
- Schorr-Evans, E.M.; Poland, A.; Pedersen, N.C. An epizootic of highly virulent feline calicivirus disease in a hospital setting in New England. J. Feline Med. Surg. 2003, 5, 217–226. [Google Scholar] [CrossRef]
- Hurley, K.E.; Pesavento, P.A.; Pedersen, N.C.; Poland, A.M.; Wilson, E.; Foley, J.E. An outbreak of virulent systemic feline calicivirus disease. J. Am. Vet. Med. Assoc. 2004, 224, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Abd-Eldaim, M.; Potgieter, L.; Kennedy, M. Genetic analysis of feline caliciviruses associated with a hemorrhagic-like disease. J. Vet. Diagn. Invest. 2005, 17, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.; Hurley, K.; Pesavento, P.A.; Poland, A.; Pedersen, N.C. Virulent systemic feline calicivirus infection: Local cytokine modulation and contribution of viral mutants. J. Feline Med. Surg. 2006, 8, 55–61. [Google Scholar] [CrossRef]
- Coyne, K.P.; Jones, B.R.D.; Kipar, A.; Chantrey, J.; Porter, C.J.; Barber, P.J.; Dawson, S.; Gaskell, R.M.; Radford, A.D. Lethal outbreak of disease associated with feline calicivirus infection in cats. Vet. Rec. 2006, 158, 544–550. [Google Scholar] [CrossRef]
- Reynolds, B.S.; Poulet, H.; Pingret, J.L.; Jas, D.; Brunet, S.; Le Meter, C.; Etievant, M.; Boucraut-Baralon, C. A nosocomial outbreak of feline Calicivirus associated virulent systemic disease in France. J. Feline Med. Surg. 2009, 11, 633–644. [Google Scholar] [CrossRef]
- Battilani, M.; Vaccari, F.; Carelle, M.S.; Morandi, F.; Benazzi, C.; Kipar, A.; Dondi, F.; Scagliarini, A. Virulent feline calicivirus disease in a shelter in Italy: A case description. Res. Vet. Sci. 2013, 95, 283–290. [Google Scholar] [CrossRef]
- Pesavento, P.A.; MacLachlan, N.J.; Dillard-Telm, L.; Grant, C.K.; Hurley, K.F. Pathologic, immunohistochemical, and electron microscopic findings in naturally occurring virulent systemic feline calicivirus infection in cats. Vet. Pathol. 2004, 41, 257–263. [Google Scholar] [CrossRef]
- Prikhodko, V.G.; Sandoval-Jaime, C.; Abente, E.J.; Bok, K.; Parra, G.I.; Rogozin, I.B.; Ostlund, E.N.; Green, K.Y.; Sosnovtsev, S.V. Genetic characterization of feline calicivirus strains associated with varying disease manifestations during an outbreak season in Missouri (1995–1996). Virus Genes 2004, 48, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Caringella, F.; Elia, G.; Decaro, N.; Martella, V.; Lanave, G.; Varello, K.; Catella, C.; Diakoudi, G.; Carelli, G.; Loredana Colaianni, M.; et al. Feline calicivirus infection in cats with virulent systemic disease, Italy. Res. Vet. Sci. 2019, 124, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Conley, M.J.; McElwee, M.; Azmi, L.; Gabrielsen, M.; Byron, O.; Goodfellow, I.G.; Bhella, D. Calicivirus VP2 forms a portal-like assembly following receptor engagement. Nature 2019, 565, 377–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebart, L.; Morineau, A.; Warwick, K.M. Multivariate Descriptive Statistical Analysis (Correspondence Analysis and Related Techniques for Large Matrices); John Wiley & Sons: Chichester, UK, 1984. [Google Scholar]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ossiboff, R.J.; Sheh, A.; Shotton, J.; Pesavento, P.A.; Parker, J.S.L. Feline caliciviruses (FCVs) isolated from cats with virulent systemic disease possess in vitro phenotypes distinct from those of other FCV isolates. J. Gen. Virol. 2007, 88, 506–517. [Google Scholar] [CrossRef]
- Brunet, S.; Jas, D.; David, F.; Bublot, M.; Poulet, H. Feline Calicivirus: Vaccination against virulent strains. In Proceedings of the European Society for Veterinary Virology, Liverpool, UK, 20–22 June 2005; Available online: http://esvv.eu/ (accessed on 22 November 2019).
- Poulet, H.; Jas, D.; Lemeter, C.; Coupier, C.; Brunet, S. Efficacy of a bivalent inactivated non-adjuvanted feline calicivirus vaccine: Relation between in vitro cross-neutralization and heterologous protection in vivo. Vaccine 2008, 26, 3647–3654. [Google Scholar] [CrossRef]
- Declercq, J. Pustular calicivirus dermatitis on the abdomen of two cats following routine ovariectomy. Vet. Dermatol. 2005, 16, 395–400. [Google Scholar] [CrossRef]
- Willi, B.; Spiri, A.M.; Meli, M.L.; Samman, A.; Hoffmann, K.; Sydler, T.; Cattori, V.; Graf, F.; Diserens, K.A.; Padrutt, I.; et al. Molecular characterization and virus neutralization patterns of severe, non-epizootic forms of feline Calicivirus infections resembling virulent systemic disease in cats in Switzerland and in Liechtenstein. Vet. Microbiol. 2016, 182, 202–212. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Miao, Q.; Zhu, J.; Yang, Z.; Liu, G. Isolation and molecular characterization of a virulent systemic feline calicivirus isolated in China. Infect. Genet. Evol. 2018, 65, 425–429. [Google Scholar] [CrossRef]
- Rong, S.; Slade, D.; Floyd-Hawkins, K.; Wheeler, D. Characterization of a highly virulent feline calicivirus and attenuation of this virus. Virus Res. 2006, 122, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.S.; Hartmann, K.; Unterer, S.; Eichhorn, W.; Majzoub, M.; Homeier-Bachmann, T.; Truyen, U.; Ellenberger, C.; Huebner, J. Two outbreaks of virulent systemic feline calicivirus infection in cats in Germany. Berl. Munch. Tierarztl. Wochenschr. 2011, 124, 186–193. [Google Scholar] [PubMed]
- Geissler, K.; Schneider, K.; Platzer, G.; Truyen, B.; Kaaden, O.R.; Truyen, U. Genetic and antigenic heterogeneity among feline calicivirus isolates from distinct disease manifestations. Virus Res. 1997, 48, 193–206. [Google Scholar] [CrossRef]
- Bhella, D.; Gatherer, D.; Chaudry, Y.; Pink, R.; Goodfellow, I.G. Structural insights into calicivirus attachment and uncoating. J. Virol. 2008, 82, 8051–8059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbert, I.; Conley, M.; Hofmann-Lehmann, R.; Willett, B.; Hosie, M. A comparison of early entry requirements in calicivirus isolates of differing pathogenicities. In Proceedings of the International Societry for Companion Animal Infectious Diseases, Portland, OR, USA, 30 September–3 October 2018; Available online: https://iscaid.org/ (accessed on 22 November 2019).
- Poulet, H.; Brunet, S.; Leroy, V.; Chappuis, G. Immunisation with a combination of two complementary feline calicivirus strains induces a broad cross-protection against heterologous challenges. Vet. Microbiol. 2005, 106, 17–31. [Google Scholar] [CrossRef]



| Strain | Pathotype | Country | Year | Access Number | Length (aa) | Strain | Pathotype | Country | Year | Access Number | Length (aa) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mimi | VSD | France | 2011 | MN628612 | 95 | SV368 | ORD | Brazil | 2006 | AEO16222 | 95 |
| Felix | VSD | France | 2009 | MN628608 | 95 | SV142 | ORD | Brazil | 2007 | AEO16224 | 95 |
| 2359 | VSD | France | 2011 | MN628602 | 95 | SV55 | ORD | Brazil | 2008 | AEO16226 | 95 |
| 100869 | VSD | USA | 2001 | MN628605 | 95 | SV45 | ORD | Brazil | 2009 | AEO16230 | 95 |
| 88287 | VSD | USA | 2001 | MN628604 | 95 | SV147 | ORD | Brazil | 2009 | AEO16231 | 95 |
| 33585 | VSD | USA | 2001 | MN628603 | 95 | 07-60 | ORD | UK | 2006 | AFR44751 | 98 |
| OBI | VSD | France | 2007 | MN628613 | 95 | IZZY | ORD | USA | 2014 | MN628610 | 95 |
| EN | VSD | France | 2004 | MN628607 | 95 | 22-18 | ORD | UK | 2005 | AFR44753 | 97 |
| Bretagne | VSD | France | 2005 | MN628606 | 95 | 44-52 | ORD | UK | 2006 | AFR44754 | 98 |
| UKOS | VSD | UK | 2007 | ACV81957 | 95 | 20-57 | ORD | UK | 2006 | AFR44756 | 97 |
| Kaos | VSD | USA | 2002 | ABI84214 | 94 | 43-46 | ORD | UK | 2006 | AFR44762 | 95 |
| Deuce | VSD | USA | 2004 | ABI84202 | 95 | 32-55 | ORD | UK | 2006 | AFR44768 | 95 |
| ARI | VSD | USA | 1998 | ABI84212 | 95 | Kit426 | ORD | USA | 2014 | MN628611 | 95 |
| Georgie | VSD | USA | 2003 | ABI84206 | 95 | 34-51 | ORD | UK | 2006 | AFR44771 | 95 |
| H1 | VSD | USA | 1999 | AAT66087 | 95 | Gon | ORD | Japan | 2003 | AHZ59401 | 95 |
| H2 | VSD | USA | 2002 | AAT66090 | 95 | ITO | ORD | Japan | 2002 | AHZ59402 | 95 |
| NH3 | ORD | USA | 2000 | AAT66084 | 95 | um3 | ORD | Japan | 2001 | AHZ59403 | 95 |
| NH4 | ORD | USA | 2009 | AAY44304 | 95 | 12Q087-1 | ORD | South-Korea | 2012 | AIA09956 | 95 |
| NH5 | ORD | USA | 2009 | AAY44305 | 95 | 12Q087-5 | ORD | South-Korea | 2012 | AIA09959 | 95 |
| NH6 | ORD | USA | 2009 | AAY44306 | 97 | HRB-SS | ORD | China | 2014 | AII00833 | 95 |
| NH7 | ORD | USA | 2009 | AAY44307 | 95 | TFHLJ-8 | ORD | China | 2013 | AIN37114 | 95 |
| NH8 | ORD | USA | 2009 | AAY44308 | 95 | FB-NJ-13 | ORD | China | 2013 | AIS22460 | 95 |
| NH9 | ORD | USA | 2009 | AAY44309 | 95 | HB-S4 | ORD | China | 2014 | ALI87297 | 95 |
| NH10 | ORD | USA | 2009 | AAY44310 | 95 | SH-2014 | ORD | China | 2014 | ALM55428 | 95 |
| NH12 | ORD | USA | 2009 | AAY44312 | 95 | CH-JL4 | ORD | China | 2015 | ALO69832 | 96 |
| DD2006 | ORD | Germany | 2006 | ABD84433 | 95 | G1 | ORD | France | 1993 | MN628609 MN628601 | 95 |
| 127 | ORD | USA | 2004 | ABI84196 | 95 | 431 | ORD | UK | 1993 | 95 | |
| 131 | ORD | USA | 2004 | ABI84198 | 95 | F9 | Vaccine | USA | 1960 | CAA77636 | 98 |
| 796 | ORD | USA | 2003 | ABI84200 | 95 |
| Amino Acid | aa Properties | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hydrophobic | Positive | Negative | Polar | Charged | Small | Aromatic | Aliphatic | Proline | Deletion | |
| A | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| C (*) | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| D | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| E | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| F | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| G | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| H | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| K | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| I | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| L | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| M | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| P | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Q | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| R | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| S | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| T | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| V | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| W | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Y | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Variable | VSD Strain | p-Value |
|---|---|---|
| 448_POLAR | Y | <0.001 |
| 448_CHARGED | Y | <0.001 |
| 448_SMALL | N | <0.001 |
| 452_SMALL | N | <0.001 |
| 448_POSITIVE | Y | 0.005 |
| 438_POLAR | N | 0.014 |
| 438_ALIPHATIC | Y | 0.014 |
| 492_SMALL | Y | 0.014 |
| 465_HYDROPHOBIC | N | 0.025 |
| 465_POLAR | Y | 0.025 |
| 440_SMALL | N | 0.027 |
| 455_CHARGED | N | 0.034 |
| 455_NEGATIVE | N | 0.043 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunet, S.; Sigoillot-Claude, C.; Pialot, D.; Poulet, H. Multiple Correspondence Analysis on Amino Acid Properties within the Variable Region of the Capsid Protein Shows Differences between Classical and Virulent Systemic Feline Calicivirus Strains. Viruses 2019, 11, 1090. https://doi.org/10.3390/v11121090
Brunet S, Sigoillot-Claude C, Pialot D, Poulet H. Multiple Correspondence Analysis on Amino Acid Properties within the Variable Region of the Capsid Protein Shows Differences between Classical and Virulent Systemic Feline Calicivirus Strains. Viruses. 2019; 11(12):1090. https://doi.org/10.3390/v11121090
Chicago/Turabian StyleBrunet, Sylvie, Cécile Sigoillot-Claude, Daniel Pialot, and Hervé Poulet. 2019. "Multiple Correspondence Analysis on Amino Acid Properties within the Variable Region of the Capsid Protein Shows Differences between Classical and Virulent Systemic Feline Calicivirus Strains" Viruses 11, no. 12: 1090. https://doi.org/10.3390/v11121090





