The Cellular Localization of the p42 and p46 Oligoadenylate Synthetase 1 Isoforms and Their Impact on Mitochondrial Respiration
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. High Resolution Respirometry (HRR)
2.3. Simultaneous Measurement of HRR and Fluorometry
2.4. Transfection of Cells
2.5. Plasmids
2.6. Immunofluorescence Microscopy
2.7. Cell Fractionation
2.8. Immunoblot Analysis
2.9. Statistical Analysis
3. Results
3.1. Effect of IFN-β on Mitochondrial OCR in HeLa and HT1080 Cells
3.2. Effect of OAS1 p46 and p42 on Mitochondrial OCR
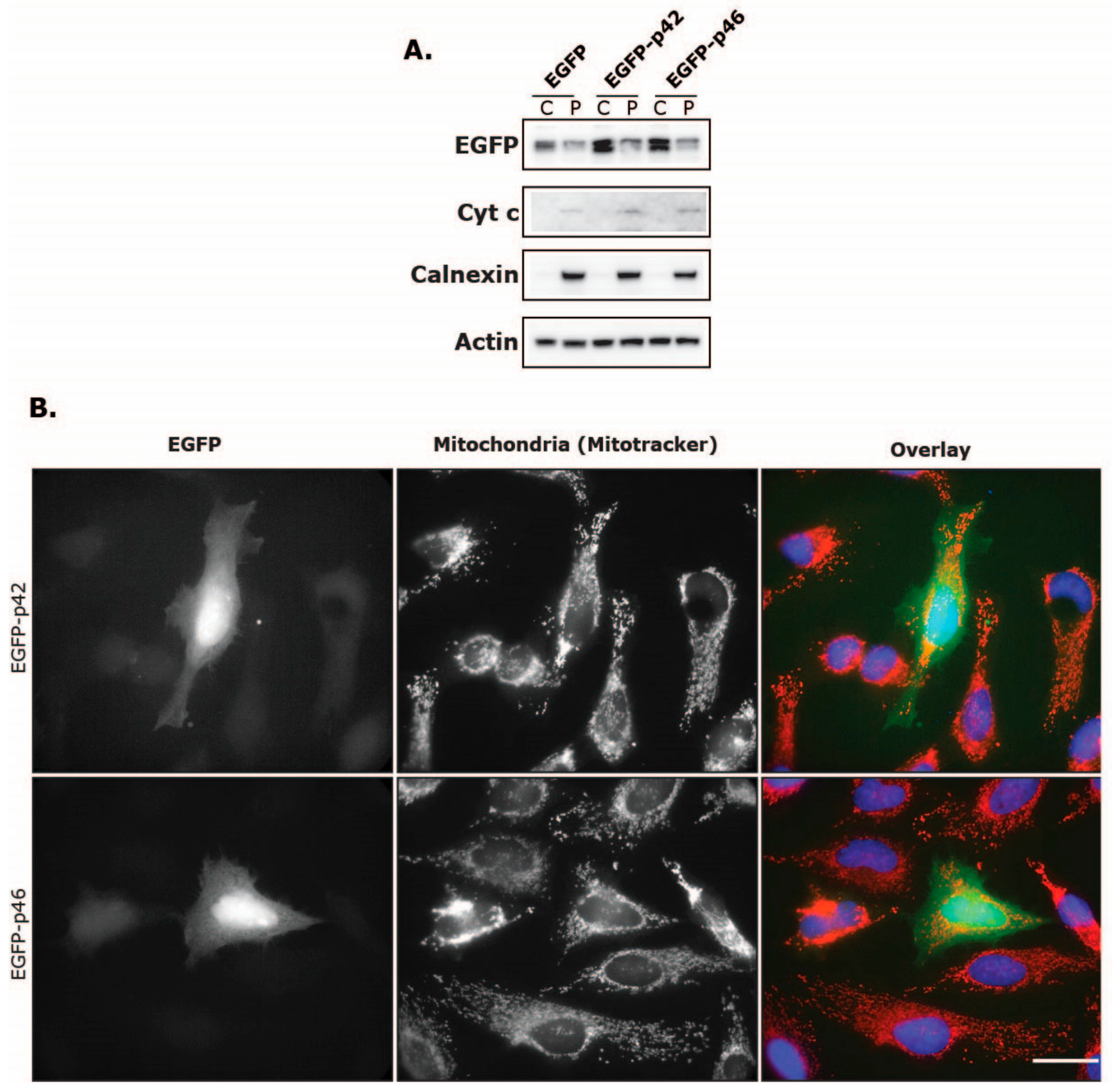
3.3. Subcellular Localization of OAS1 p46 and p42 and the Importance of the CaaX Motif
3.4. Intracellular Localization of EGFP Fused to the C-Terminus of OAS1 p46 and p42
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leisching, G.; Wiid, I.; Baker, B. The Association of OASL and Type I Interferons in the Pathogenesis and Survival of Intracellular Replicating Bacterial Species. Front. Cell. Infect. Microbiol. 2017, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; Justesen, J.; Sarkar, S.N.; Sen, G.C.; Yee, V.C. Crystal Structure of the 2′-Specific and Double-Stranded RNA-Activated Interferon-Induced Antiviral Protein 2′-5′-Oligoadenylate Synthetase. Mol. Cell 2003, 12, 1173–1185. [Google Scholar] [CrossRef]
- Donovan, J.; Dufner, M.; Korennykh, A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc. Natl. Acad. Sci. USA 2013, 110, 1652–1657. [Google Scholar] [CrossRef] [PubMed]
- Castelli, J.C.; Hassel, B.A.; Maran, A.; Paranjape, J.; Hewitt, J.A.; Li, X.-l.; Hsu, Y.-T.; Silverman, R.H.; Youle, R.J. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998, 5, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Donovan, J.; Rath, S.; Whitney, G.; Chitrakar, A.; Korennykh, A. Structure of Human RNase L Reveals the Basis for Regulated RNA Decay in the IFN Response. Science 2014, 343, 1244–1248. [Google Scholar] [CrossRef]
- Silverman, R.H. Viral Encounters with 2′,5′-Oligoadenylate Synthetase and RNase L during the Interferon Antiviral Response. J. Virol. 2007, 81, 12720–12729. [Google Scholar] [CrossRef]
- Sadler, A.J.; Williams, B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef]
- Bonnevie-Nielsen, V.; Field, L.L.; Lu, S.; Zheng, D.J.; Li, M.; Martensen, P.M.; Nielsen, T.B.; Beck-Nielsen, H.; Lau, Y.L.; Pociot, F. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am. J. Hum. Genet. 2005, 76, 623–633. [Google Scholar] [CrossRef]
- Kjær, K.H.; Pahus, J.; Hansen, M.F.; Poulsen, J.B.; Christensen, E.I.; Justesen, J.; Martensen, P.M. Mitochondrial localization of the OAS1 p46 isoform associated with a common single nucleotide polymorphism. BMC Cell Biol. 2014, 15, 33. [Google Scholar] [CrossRef]
- Lim, J.K.; Lisco, A.; McDermott, D.H.; Huynh, L.; Ward, J.M.; Johnson, B.; Johnson, H.; Pape, J.; Foster, G.A.; Krysztof, D.; et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009, 5, e1000321. [Google Scholar] [CrossRef]
- Liu, X.; Xing, H.; Gao, W.; Yu, D.; Zhao, Y.; Shi, X.; Zhang, K.; Li, P.; Yu, J.; Xu, W.; et al. A functional variant in the OAS1 gene is associated with Sjögren’s syndrome complicated with HBV infection. Sci. Rep. 2017, 7, 17571. [Google Scholar] [CrossRef] [PubMed]
- Din, N.G.B.E.; Anany, M.A.; Dawood, R.M.; Ibrahim, M.K.; El-Shenawy, R.; Abd, Y.S.E.; Awady, M.K.E. Impact of OAS1 Exon 7 rs10774671 Genetic Variation on Liver Fibrosis Progression in Egyptian HCV Genotype 4 Patients. Viral Immunol. 2015, 28, 509–516. [Google Scholar]
- El Awady, M.K.; Anany, M.A.; Esmat, G.; Zayed, N.; Tabll, A.A.; Helmy, A.; El Zayady, A.R.; Abdalla, M.S.; Sharada, H.M.; El Raziky, M.; et al. Single nucleotide polymorphism at exon 7 splice acceptor site of OAS1 gene determines response of hepatitis C virus patients to interferon therapy. J. Gastroenterol. Hepatol. 2011, 26, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, Y.; Chen, G.; Zhang, M.; Wang, M.; He, J.-Q. 2′-5′-Oligoadenylate synthetase 1 polymorphisms are associated with tuberculosis: A case-control study. BMC Pulm. Med. 2018, 18, 180. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.-C.; Kang, J.-I.; Hwang, S.B.; Ahn, B.-Y. The ribonuclease L-dependent antiviral roles of human 2’,5’-oligoadenylate synthetase family members against hepatitis C virus. FEBS Lett. 2013, 587, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Gentry, L.R.; Nishimura, A.; Cox, A.D.; Martin, T.D.; Tsygankov, D.; Nishida, M.; Elston, T.C.; Der, C.J. Divergent Roles of CAAX Motif-signaled Posttranslational Modifications in the Regulation and Subcellular Localization of Ral GTPases. J. Biol. Chem. 2015, 290, 22851–22861. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Casey, P.J. Protein prenylation: Unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 2016, 17, 110. [Google Scholar] [CrossRef]
- Le Roy, F.; Bisbal, C.; Silhol, M.; Martinand, C.; Lebleu, B.; Salehzada, T. The 2-5A/RNase L/RNase L inhibitor (RLI) [correction of (RNI)] pathway regulates mitochondrial mRNAs stability in interferon alpha-treated H9 cells. J. Biol. Chem. 2001, 276, 48473–48482. [Google Scholar] [CrossRef]
- Castellanos, E.; Lanning, N.J. Phosphorylation of OXPHOS Machinery Subunits: Functional Implications in Cell Biology and Disease. Yale J. Biol. Med. 2019, 92, 523–531. [Google Scholar]
- Dimroth, P.; Kaim, G.; Matthey, U. Crucial role of the membrane potential for ATP synthesis by F(1)F(o) ATP synthases. J. Exp. Biol. 2000, 203, 51–59. [Google Scholar]
- Wang, C.; Youle, R.J. The Role of Mitochondria in Apoptosis. Annu. Rev. Genet. 2009, 43, 95–118. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.W.G.; Green, D.R. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Ea, C.-K.; Chen, Z.J. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-κB and IRF3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Reshi, L.; Hong, J.-R. Mitochondria as a favorite organelle for invading viruses. Mol. Biol. 2017, 6, 181. [Google Scholar]
- Pesta, D.; Gnaiger, E. High-Resolution Respirometry: OXPHOS Protocols for Human Cells and Permeabilized Fibers from Small Biopsies of Human Muscle. In Mitochondrial Bioenergetics: Methods and Protocols; Palmeira, C.M., Moreno, A.J., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 25–58. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Baghirova, S.; Hughes, B.G.; Hendzel, M.J.; Schulz, R. Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX 2015, 2, 440–445. [Google Scholar] [CrossRef]
- Chebath, J.; Benech, P.; Hovanessian, A.; Galabru, J.; Revel, M. Four different forms of interferon-induced 2′,5′-oligo(A) synthetase identified by immunoblotting in human cells. J. Biol. Chem. 1987, 262, 3852–3857. [Google Scholar]
- Fritsch, S.D.; Weichhart, T. Effects of Interferons and Viruses on Metabolism. Front. Immunol. 2016, 7, 630. [Google Scholar] [CrossRef]
- Haghikia, A.; Faissner, S.; Pappas, D.; Pula, B.; Akkad, D.A.; Arning, L.; Ruhrmann, S.; Duscha, A.; Gold, R.; Baranzini, S.E.; et al. Interferon-beta affects mitochondrial activity in CD4+ lymphocytes: Implications for mechanism of action in multiple sclerosis. Mult. Scler. J. 2015, 21, 1262–1270. [Google Scholar] [CrossRef]
- Lewis, J.A.; Huq, A.; Najarro, P. Inhibition of Mitochondrial Function by Interferon. J. Biol. Chem. 1996, 271, 13184–13190. [Google Scholar] [CrossRef]
- Pitroda, S.P.; Wakim, B.T.; Sood, R.F.; Beveridge, M.G.; Beckett, M.A.; MacDermed, D.M.; Weichselbaum, R.R.; Khodarev, N.N. STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect. BMC Med. 2009, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Sanin, D.E.; Everts, B.; Chen, Q.; Qiu, J.; Buck, M.D.; Patterson, A.; Smith, A.M.; Chang, C.-H.; Liu, Z.; et al. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity 2016, 44, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Scott Reid, T.; Terry, K.L.; Casey, P.J.; Beese, L.S. Crystallographic Analysis of CaaX Prenyltransferases Complexed with Substrates Defines Rules of Protein Substrate Selectivity. J. Mol. Biol. 2004, 343, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gale, M.; Keller, B.C.; Huang, H.; Brown, M.S.; Goldstein, J.L.; Ye, J. Identification of FBL2 As a Geranylgeranylated Cellular Protein Required for Hepatitis C Virus RNA Replication. Mol. Cell 2005, 18, 425–434. [Google Scholar] [CrossRef]
- Yang, S.; Harding, A.T.; Sweeney, C.; Miao, D.; Swan, G.; Zhou, C.; Jiang, Z.; Fitzgerald, K.A.; Hammer, G.; Bergo, M.O.; et al. Control of antiviral innate immune response by protein geranylgeranylation. Sci. Adv. 2019, 5, eaav7999. [Google Scholar] [CrossRef]
- Choy, E.; Chiu, V.K.; Silletti, J.; Feoktistov, M.; Morimoto, T.; Michaelson, D.; Ivanov, I.E.; Philips, M.R. Endomembrane Trafficking of Ras: The CAAX Motif Targets Proteins to the ER and Golgi. Cell 1999, 98, 69–80. [Google Scholar] [CrossRef]
- Ramabhadran, V.; Korobova, F.; Rahme, G.J.; Higgs, H.N. Splice variant–specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell 2011, 22, 4822–4833. [Google Scholar] [CrossRef]
- Schmidt, W.K.; Tam, A.; Fujimura-Kamada, K.; Michaelis, S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 1998, 95, 11175–11180. [Google Scholar] [CrossRef]
- Ghosh, A.; Sarkar, S.N.; Guo, W.; Bandyopadhyay, S.; Sen, G.C. Enzymatic Activity of 2′–5′-Oligoadenylate Synthetase Is Impaired by Specific Mutations that Affect Oligomerization of the Protein. J. Biol. Chem. 1997, 272, 33220–33226. [Google Scholar] [CrossRef]
- Malathi, K.; Dong, B.; Gale, M.; Silverman, R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef]
- Malathi, K.; Saito, T.; Crochet, N.; Barton, D.J.; Gale, M.; Silverman, R.H. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 2010, 16, 2108–2119. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Schweizer, M.; Cossarizza, A.; Franceschi, C. Control of apoptosis by the cellular ATP level. FEBS Lett. 1996, 378, 107–110. [Google Scholar] [CrossRef]
- Fedetz, M.; Matesanz, F.; Caro-Maldonado, A.; Fernandez, O.; Tamayo, J.A.; Guerrero, M.; Delgado, C.; Lopez-Guerrero, J.A.; Alcina, A. OAS1 gene haplotype confers susceptibility to multiple sclerosis. Tissue Antigens 2006, 68, 446–449. [Google Scholar] [CrossRef] [PubMed]
- Field, L.L.; Bonnevie-Nielsen, V.; Pociot, F.; Lu, S.; Nielsen, T.B.; Beck-Nielsen, H. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes 2005, 54, 1588–1591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tessier, M.C.; Qu, H.Q.; Frechette, R.; Bacot, F.; Grabs, R.; Taback, S.P.; Lawson, M.L.; Kirsch, S.E.; Hudson, T.J.; Polychronakos, C. Type 1 diabetes and the OAS gene cluster: Association with splicing polymorphism or haplotype? J. Med. Genet. 2006, 43, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Blake, R.; Trounce, I.A. Mitochondrial dysfunction and complications associated with diabetes. Biochim. Biophys. Acta 2013, 1840, 1404–1412. [Google Scholar] [CrossRef] [PubMed]
- Su, K.; Bourdette, D.; Forte, M. Mitochondrial dysfunction and neurodegeneration in multiple sclerosis. Front. Physiol. 2013, 4, 169. [Google Scholar] [CrossRef]
- Mazeaud, C.; Freppel, W.; Chatel-Chaix, L. The Multiples Fates of the Flavivirus RNA Genome During Pathogenesis. Front. Genet. 2018, 9, 595. [Google Scholar] [CrossRef]
- Deo, S.; Patel, T.R.; Chojnowski, G.; Koul, A.; Dzananovic, E.; McEleney, K.; Bujnicki, J.M.; McKenna, S.A. Characterization of the termini of the West Nile virus genome and their interactions with the small isoform of the 2′ 5′-oligoadenylate synthetase family. J. Struct. Biol. 2015, 190, 236–249. [Google Scholar] [CrossRef]
- Courtney, S.C.; Di, H.; Stockman, B.M.; Liu, H.; Scherbik, S.V.; Brinton, M.A. Identification of Novel Host Cell Binding Partners of Oas1b, the Protein Conferring Resistance to Flavivirus-Induced Disease in Mice. J. Virol. 2012, 86, 7953–7963. [Google Scholar] [CrossRef]
- Kristiansen, H.; Scherer, C.A.; McVean, M.; Iadonato, S.P.; Vends, S.; Thavachelvam, K.; Steffensen, T.B.; Horan, K.A.; Kuri, T.; Weber, F.; et al. Extracellular 2′-5′ Oligoadenylate Synthetase Stimulates RNase L-Independent Antiviral Activity: A Novel Mechanism of Virus-Induced Innate Immunity. J. Virol. 2010, 84, 11898–11904. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, T.; Nagano-Fujii, M.; Akutsu, M.; Kadoya, H.; Ohgimoto, S.; Ishido, S.; Hotta, H. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J. Gen. Virol. 2004, 85, 959–969. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skrivergaard, S.; Jensen, M.S.; Rolander, T.B.; Nguyen, T.B.N.; Bundgaard, A.; Nejsum, L.N.; Martensen, P.M. The Cellular Localization of the p42 and p46 Oligoadenylate Synthetase 1 Isoforms and Their Impact on Mitochondrial Respiration. Viruses 2019, 11, 1122. https://doi.org/10.3390/v11121122
Skrivergaard S, Jensen MS, Rolander TB, Nguyen TBN, Bundgaard A, Nejsum LN, Martensen PM. The Cellular Localization of the p42 and p46 Oligoadenylate Synthetase 1 Isoforms and Their Impact on Mitochondrial Respiration. Viruses. 2019; 11(12):1122. https://doi.org/10.3390/v11121122
Chicago/Turabian StyleSkrivergaard, Stig, Monica Skou Jensen, Tine Breckling Rolander, Tram Bao Ngoc Nguyen, Amanda Bundgaard, Lene N. Nejsum, and Pia M. Martensen. 2019. "The Cellular Localization of the p42 and p46 Oligoadenylate Synthetase 1 Isoforms and Their Impact on Mitochondrial Respiration" Viruses 11, no. 12: 1122. https://doi.org/10.3390/v11121122
APA StyleSkrivergaard, S., Jensen, M. S., Rolander, T. B., Nguyen, T. B. N., Bundgaard, A., Nejsum, L. N., & Martensen, P. M. (2019). The Cellular Localization of the p42 and p46 Oligoadenylate Synthetase 1 Isoforms and Their Impact on Mitochondrial Respiration. Viruses, 11(12), 1122. https://doi.org/10.3390/v11121122




