The Tug-of-War between Plants and Viruses: Great Progress and Many Remaining Questions
Abstract
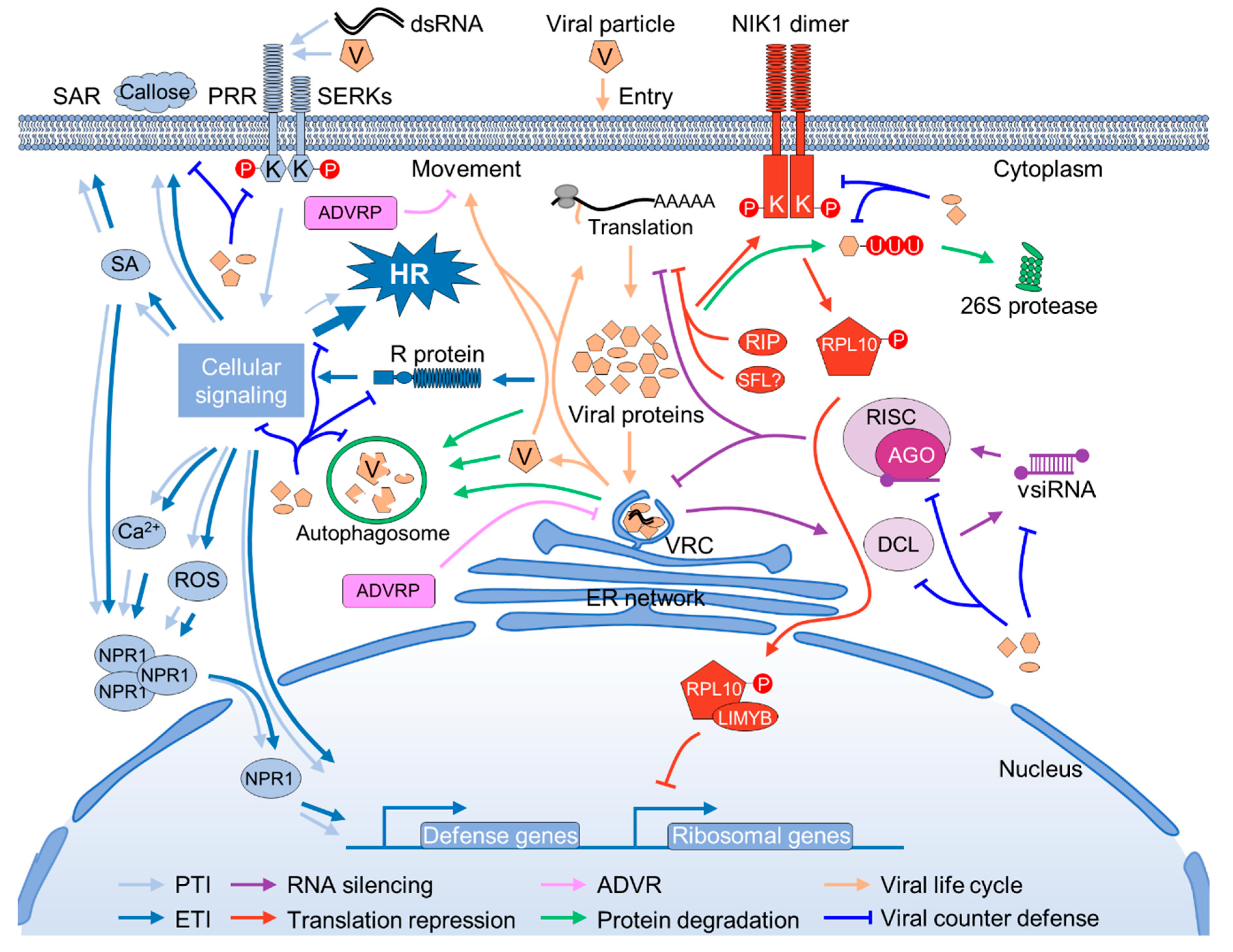
1. Introduction
2. Plant Antiviral Defenses
2.1. Innate Antiviral Immunity
2.2. RNA Silencing
2.3. Translation Repression
2.4. Atypical Dominant Viral Resistances
2.5. Ubiquitination and Autophagy-Mediated Protein Degradation
2.6. Cross-Talking between Different Antiviral Defenses
3. Strike Back from Viruses
3.1. Mutation as a Master Mechanism for Escaping Host Antiviral Mechanisms
3.2. Inhibition of Innate Immunity
3.3. Suppression and Exploitation of Host RNA Silencing
3.4. Subversion of Translation Repression
3.5. Prevention and Exploitation of Host Ubiquitination and Autophagy Pathways
4. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Pendon, J.A.; Truniger, V.; Nieto, C.; Garcia-Mas, J.; Bendahmane, A.; Aranda, M.A. Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 2004, 5, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Krishnaswamy, S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 2012, 13, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Robaglia, C.; Caranta, C. Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 2006, 11, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, K.K.; Scholthof, K.-B.G. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [PubMed]
- Soosaar, J.L.M.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Schwessinger, B.; Zipfel, C. News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008, 11, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 2009, 12, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Tang, D.; Wang, G. Receptor kinases in plant pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Kørner, C.J.; Klauser, D.; Niehl, A.; Domínguez-Ferreras, A.; Chinchilla, D.; Boller, T.; Heinlein, M.; Hann, D.R. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact. 2013, 26, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Gou, X.; He, K.; Xi, D.; Du, J.; Lin, H.; Li, J. BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to turnip crinkle virus. Eur. J. Plant Pathol. 2010, 127, 149–156. [Google Scholar] [CrossRef]
- Woo, J.Y.; Jeong, K.J.; Kim, Y.J.; Paek, K.H. CaLecRK-S.5, a pepper L-type lectin receptor kinase gene, confers broad-spectrum resistance by activating priming. J. Exp. Bot. 2016, 67, 5725–5741. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Singh, P.; Chen, M.C.; Zimmerli, L. L-Glutamine inhibits beta-aminobutyric acid-induced stress resistance and priming in Arabidopsis. J. Exp. Bot. 2010, 61, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Lapidot, M.; Culver, J.N.; Fluhr, R. An early tobacco mosaic virus-induced oxidative burst in tobacco indicates extracellular perception of the virus coat protein. Plant Physiol. 2001, 126, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Perraki, A.; Gronnier, J.; Gouguet, P.; Boudsocq, M.; Deroubaix, A.F.; Simon, V.; German-Retana, S.; Legrand, A.; Habenstein, B.; Zipfel, C.; et al. REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog. 2018, 14, e1007378. [Google Scholar] [CrossRef] [PubMed]
- Zamora, M.; Méndez-López, E.; Agirrezabala, X.; Cuesta, R.; Lavín, J.L.; Sánchez-Pina, M.A.; Aranda, M.A.; Valle, M. Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Sci. Adv. 2017, 3, eaao2182. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, B.; Calil, I.; Machado, J.P.; Santos, A.; Fontes, E. Immune receptors and co-receptors in antiviral innate immunity in plants. Front. Microbiol. 2016, 7, 2139. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Jiang, L.; Bai, B.; Zhao, W.; Chen, X.; Li, J.; Liu, Y.; Chen, Z.; Wang, B.; Wang, C.; et al. The intracellular immune receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 2017, 29, 2214–2232. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.; Krasileva, K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.; Zhu, M.; Huang, S.; Zhang, W.; Dinesh-Kumar, S.P.; Tao, X. A plant immune receptor adopts a two-step recognition mechanism to enhance viral effector perception. Mol. Plant. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Townsend, P.D.; Dixon, C.H.; Slootweg, E.J.; Sukarta, O.C.A.; Yang, A.W.H.; Hughes, T.R.; Sharples, G.J.; Palsson, L.O.; Takken, F.L.W.; Goverse, A.; et al. The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. J. Biol. Chem. 2018, 293, 3218–3233. [Google Scholar] [CrossRef] [PubMed]
- Tameling, W.I.; Nooijen, C.; Ludwig, N.; Boter, M.; Slootweg, E.; Goverse, A.; Shirasu, K.; Joosten, M.H. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 2010, 22, 4176–4194. [Google Scholar] [CrossRef] [PubMed]
- Slootweg, E.; Roosien, J.; Spiridon, L.N.; Petrescu, A.J.; Tameling, W.; Joosten, M.; Pomp, R.; van Schaik, C.; Dees, R.; Borst, J.W.; et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 2010, 22, 4195–4215. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, P.; Yoshioka, K.; Shah, J.; Dooner, H.K.; Klessig, D.F. Resistance to turnip crinkle virus in Arabidopsis is regulated by two host genes and is salicylic acid dependent but NPR1, ethylene, and jasmonate independent. Plant Cell 2000, 12, 677–690. [Google Scholar] [CrossRef] [PubMed]
- del Pozo, O.; Lam, E. Expression of the baculovirus p35 protein in tobacco affects cell death progression and compromises N gene-mediated disease resistance response to tobacco mosaic virus. Mol. Plant Microbe Interact. 2003, 16, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Okamoto, M.; Iwai, T.; Iwano, M.; Fukui, K.; Isogai, A.; Nakajima, N.; Ohashi, Y. Reduced levels of chloroplast FtsH protein in tobacco mosaic virus-infected tobacco leaves accelerate the hypersensitive reaction. Plant Cell 2000, 12, 917–932. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.M.; Duncan, G.H.; Pradel, K.S.; Carr, F.; Wood, S.; Oparka, K.J.; Cruz, S.S. Analysis of the N gene hypersensitive response induced by a fluorescently tagged tobacco mosaic virus. Plant Physiol. 2000, 123, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Takenaka, S.; Hase, S.; Kubota, M.; Ichinose, Y.; Kanayama, Y.; Nakaho, K.; Klessig, D.F.; Takahashi, H. Enhanced defense responses in Arabidopsis induced by the cell wall protein fractions from Pythium oligandrum require SGT1, RAR1, NPR1 and JAR1. Plant Cell Physiol. 2009, 50, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Amigues, B.; Peart, J.; Breuer, C.; Kadota, Y.; Casais, C.; Moore, G.; Kleanthous, C.; Ochsenbein, F.; Shirasu, K.; et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 2007, 19, 3791–3804. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Jeong, R.D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 2013, 14, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.F.; Xu, L.; Tan, W.J.; Chen, L.; Qi, H.; Xie, L.J.; Chen, M.X.; Liu, B.Y.; Yu, L.J.; Yao, N.; et al. Disruption of the Arabidopsis defense regulator genes SAG101, EDS1, and PAD4 confers enhanced freezing tolerance. Mol. Plant 2015, 8, 1536–1549. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U. Systemic acquired resistance. Plant Signal Behav. 2006, 1, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Bruce, T.J.A.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Posttranslational modifications of NPR1: A single protein playing multiple roles in plant immunity and physiology. PLoS Pathog. 2016, 12, e1005707. [Google Scholar] [CrossRef] [PubMed]
- Dong, X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004, 7, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, Y.T.; Qu, N.; Zhao, Q.; Bi, D.; Li, X. Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 2006, 48, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Schauer, S.E.; Jacobsen, S.E.; Meinke, D.W.; Ray, A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002, 7, 487–491. [Google Scholar] [CrossRef]
- Llave, C. Virus-derived small interfering RNAs at the core of plant–virus interactions. Trends Plant Sci. 2010, 15, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, V.; Szittya, G.; Burgyan, J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 2007, 81, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfacon, H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology 2014, 456–457, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Zamora, A.; Azhar, M.T.; Sacco, M.A.; Lambert, L.H.; Moffett, P. Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 2009, 58, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 2010, 22, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Kørner, C.J.; Pitzalis, N.; Peña, E.J.; Erhardt, M.; Vazquez, F.; Heinlein, M. Crosstalk between PTGS and TGS pathways in natural antiviral immunity and disease recovery. Nat. Plants 2018, 4, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Harrison, B.D.; Baulcombe, D.C. A similarity between viral defense and gene silencing in plants. Science 1997, 276, 1558–1560. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Ding, S.W. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 2001, 12, 150–154. [Google Scholar] [CrossRef]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.S.; Perry, K.L.; Moffett, P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against potato virus X in Arabidopsis. Plant Physiol. 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Maruyama, K.; Sun, L.; Kondo, H.; Tamada, T.; Suzuki, N. Differential contributions of plant Dicer-like proteins to antiviral defences against potato virus X in leaves and roots. Plant J. 2015, 81, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Moffett, P. Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. Plant Cell 2015, 27, 1742–1754. [Google Scholar] [CrossRef] [PubMed]
- Molnar, A.; Melnyk, C.W.; Bassett, A.; Hardcastle, T.J.; Dunn, R.; Baulcombe, D.C. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 2010, 328, 872–875. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.S.; Masuta, C. Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr. Opin. Plant Biol. 2014, 20, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Kanyuka, K.; Parry, M.A.; Powers, S.J.; Halford, N.G. GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in Arabidopsis. J. Exp. Bot. 2008, 59, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dickinson, J.R.; Paul, M.J.; Halford, N.G. Molecular cloning of an arabidopsis homologue of GCN2, a protein kinase involved in co-ordinated response to amino acid starvation. Planta 2003, 217, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Browning, K.S. The plant translational apparatus. Plant Mol. Biol. 1996, 32, 107–144. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, Y.; Kulasekaran, S.; Benito, P.; López, B.; Marcos, R.; Cascón, T.; Hamberg, M.; Castresana, C. Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ. 2018, 41, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, H.; Zhao, C.; Li, S.; Kong, L.; Wu, W.; Kong, W.; Liu, Y.; Wei, Y.; Zhu, J.K.; et al. The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ. 2017, 40, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Lageix, S.; Lanet, E.; Pouch-Pélissier, M.-N.; Espagnol, M.-C.; Robaglia, C.; Deragon, J.-M.; Pélissier, T. ArabidopsiseIF2α kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 2008, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Echevarria-Zomeno, S.; Yanguez, E.; Fernandez-Bautista, N.; Castro-Sanz, A.B.; Ferrando, A.; Castellano, M.M. Regulation of translation initiation under biotic and abiotic stresses. Int. J. Mol. Sci. 2013, 14, 4670–4683. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. In Advances in Virus Research; Gad, L., John, P.C., Eds.; Academic Press: New York, NY, USA, 2009; Volume 75, pp. 119–231. [Google Scholar]
- Endo, Y.; Tsurugi, K.; Lambert, J.M. The site of action of six different ribosome-inactivating proteins from plants on eukaryotic ribosomes: The RNA N-glycosidase activity of the proteins. Biochem. Biophys. Res. Commun. 1988, 150, 1032–1036. [Google Scholar] [CrossRef]
- Stirpe, F. Ribosome-inactivating proteins: From toxins to useful proteins. Toxicon 2013, 67, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Domashevskiy, A.V.; Williams, S.; Kluge, C.; Cheng, S.-Y. Plant translation initiation complex eIFiso4F directs pokeweed antiviral protein to selectively depurinate uncapped tobacco etch virus RNA. Biochemistry 2017, 56, 5980–5990. [Google Scholar] [CrossRef] [PubMed]
- Picard, D.; Kao, C.C.; Hudak, K.A. Pokeweed antiviral protein inhibits brome mosaic virus replication in plant cells. J. Biol. Chem. 2005, 280, 20069–20075. [Google Scholar] [CrossRef] [PubMed]
- Tumer, N.E.; Hwang, D.J.; Bonness, M. C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci. USA 1997, 94, 3866–3871. [Google Scholar] [CrossRef] [PubMed]
- Lodge, J.K.; Kaniewski, W.K.; Tumer, N.E. Broad-spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc. Natl. Acad. Sci. USA 1993, 90, 7089–7093. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; White, R.F.; Antoniw, J.F.; Lin, Q. Effect of pokeweed antiviral protein (PAP) on the infection of plant viruses. Plant Pathol. 1991, 40, 612–620. [Google Scholar] [CrossRef]
- Bijal, A.P.; Nilgun, E.T. Antiviral activity of ribosome inactivating proteins in medicine. Mini Rev. Med. Chem. 2004, 4, 523–543. [Google Scholar]
- Stirpe, F.; Gilabert-Oriol, R. Ribosome-inactivating proteins: An overview. In Plant Toxins; Carlini, C.R., Ligabue-Braun, R., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 153–182. [Google Scholar]
- Görschen, E.; Dunaeva, M.; Hause, B.; Reeh, I.; Wasternack, C.; Parthier, B. Expression of the ribosome-inactivating protein JIP60 from barley in transgenic tobacco leads to an abnormal phenotype and alterations on the level of translation. Planta 1997, 202, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Zorzatto, C.; Machado, J.P.B.; Lopes, K.V.G.; Nascimento, K.J.T.; Pereira, W.A.; Brustolini, O.J.B.; Reis, P.A.B.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Mariano, A.C.; Andrade, M.O.; Santos, A.A.; Carolino, S.M.; Oliveira, M.L.; Baracat-Pereira, M.C.; Brommonshenkel, S.H.; Fontes, E.P. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 2004, 318, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Fontes, E.P.B.; Santos, A.A.; Luz, D.F.; Waclawovsky, A.J.; Chory, J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004, 18, 2545–2556. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.S.; Choi, J.H.; Heinz, J.; Chetty, C.S. Domain-specific positive selection contributes to the evolution of Arabidopsis leucine-rich repeat receptor-like kinase (LRR RLK) genes. J. Mol. Evol. 2006, 63, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Santos, A.A.; Pires, S.R.; Rocha, C.S.; Saraiva, D.I.; Machado, J.P.B.; Mattos, E.C.; Fietto, L.G.; Fontes, E.P.B. Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS Pathog. 2008, 4, e1000247. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Nicole, M.C.; Meteignier, L.V.; Hong, N.; Wang, G.; Moffett, P. Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J. Exp. Bot. 2015, 66, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xuan, Y.; Han, Y.; Ding, X.; Ye, K.; Yang, F.; Gao, P.; Goff, S.P.; Gao, G. Regulation of HIV-1 Gag-Pol expression by shiftless, an inhibitor of programmed -1 ribosomal frameshifting. Cell 2019, 176, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Van Damme, E.J. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Catanzariti, A.M.; Lim, G.T.; Jones, D.A. The tomato I-3 gene: A novel gene for resistance to Fusarium wilt disease. New Phytol. 2015, 207, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S.; Gisch, N.; Schaffer, M.; Illig, T.; Westphal, L.; Knirel, Y.A.; Sanchez-Carballo, P.M.; Zahringer, U.; Huckelhoven, R.; Lee, J.; et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Mahajan, S.K.; Whitham, S.A.; Yamamoto, M.L.; Carrington, J.C. Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proc. Natl. Acad. Sci. USA 2000, 97, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Decroocq, V.; Sicard, O.; Alamillo, J.M.; Lansac, M.; Eyquard, J.P.; García, J.A.; Candresse, T.; Le Gall, O.; Revers, F. Multiple resistance traits control plum pox virus infection in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2006, 19, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.A.; Anderberg, R.J.; Chisholm, S.T.; Carrington, J.C. Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 2000, 12, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Parra, M.A.; Anderberg, R.J.; Carrington, J.C. Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 2001, 127, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Cosson, P.; Sofer, L.; Le, Q.H.; Leger, V.; Schurdi-Levraud, V.; Whitham, S.A.; Yamamoto, M.L.; Gopalan, S.; Le Gall, O.; Candresse, T.; et al. RTM3, which controls long-distance movement of potyviruses, is a member of a new plant gene family encoding a meprin and TRAF homology domain-containing protein. Plant Physiol. 2010, 154, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Cosson, P.; Schurdi-Levraud, V.; Le, Q.H.; Sicard, O.; Caballero, M.; Roux, F.; Le Gall, O.; Candresse, T.; Revers, F. The RTM resistance to potyviruses in Arabidopsis thaliana: Natural variation of the RTM genes and evidence for the implication of additional genes. PLoS ONE 2012, 7, e39169. [Google Scholar] [CrossRef] [PubMed]
- Sofer, L.; Cabanillas, D.G.; Gayral, M.; Téplier, R.; Pouzoulet, J.; Ducousso, M.; Dufin, L.; Bréhélin, C.; Ziegler-Graff, V.; Brault, V.; et al. Identification of host factors potentially involved in RTM-mediated resistance during potyvirus long distance movement. Arch. Virol. 2017, 162, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Decroocq, V.; Salvador, B.; Sicard, O.; Glasa, M.; Cosson, P.; Svanella-Dumas, L.; Revers, F.; García, J.A.; Candresse, T. The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N-terminal region of the coat protein. Mol. Plant Microbe Interact. 2009, 22, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, Y.; Maejima, K.; Komatsu, K.; Shiraishi, T.; Okano, Y.; Himeno, M.; Sugawara, K.; Neriya, Y.; Minato, N.; Miura, C.; et al. Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 2012, 24, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, K.; Shiraishi, T.; Yoshida, T.; Fujita, N.; Netsu, O.; Yamaji, Y.; Namba, S. A replicase of potato virus X acts as the resistance-breaking determinant for JAX1-mediated resistance. Mol. Plant Microbe Interact. 2013, 26, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Shiraishi, T.; Hagiwara-Komoda, Y.; Komatsu, K.; Maejima, K.; Okano, Y.; Fujimoto, Y.; Yusa, A.; Yamaji, Y.; Namba, S. The plant non-canonical antiviral resistance protein JAX1 inhibits potexviral replication by targeting the viral RNA-dependent RNA polymerase. J. Virol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.; Mishra, S.K.; Srivastava, S.; Srivastava, A. A virus inhibitory protein isolated from Cyamopsis tetragonoloba (L.) Taub. upon induction of systemic antiviral resistance shares partial amino acid sequence homology with a lectin. Plant Cell Rep. 2014, 33, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Li, H.; Zhang, W. The lectin from Musa paradisiaca binds with the capsid protein of tobacco mosaic virus and prevents viral infection. Biotechnol. Biotechnol. Equip. 2014, 28, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Masuda, K.; Naito, S.; Meshi, T.; Ishikawa, M. An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. USA 2007, 104, 13833–13838. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, H.; Gong, Y.; Tao, Y.; Jiang, L.; Zuo, W.; Yang, Q.; Ye, J.; Lai, J.; Wu, J.; et al. An atypical thioredoxin imparts early resistance to sugarcane mosaic virus in maize. Mol. Plant 2017, 10, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yan, Z.; Xia, Z.; Cheng, Y.; Jiao, Z.; Sun, B.; Zhou, T.; Fan, Z. A violaxanthin de-epoxidase interacts with a viral suppressor of RNA silencing to inhibit virus amplification. Plant Physiol. 2017, 175, 1774–1794. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Y.; He, J.; Zheng, X.; Hu, J.; Liu, Y.; Dai, H.; Zhang, Y.; Wang, B.; Wu, W.; et al. STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 2014, 5, 4768. [Google Scholar] [CrossRef] [PubMed]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y. The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA–dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [PubMed]
- Caro, M.; Verlaan, M.G.; Julian, O.; Finkers, R.; Wolters, A.A.; Hutton, S.F.; Scott, J.W.; Kormelink, R.; Visser, R.G.F.; Diez, M.J.; et al. Assessing the genetic variation of Ty-1 and Ty-3 alleles conferring resistance to tomato yellow leaf curl virus in a broad tomato germplasm. Mol. Breed. 2015, 35, 132. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Mawatari, N.; Miyashita, S.; Kishino, H.; Meshi, T.; Ishikawa, M. Coevolution and hierarchical interactions of tomato mosaic virus and the resistance gene Tm-1. PLoS Pathog. 2012, 8, e1002975. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.M.; Moreau, V.; Andre, B.; Volland, C.; Haguenauer-Tsapis, R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 1996, 271, 10946–10952. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Li, Z.; Nagy, P.D. The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 2009, 83, 11751–11764. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Cao, Y.; Sun, X.; Espinoza, C.; Nguyen, C.T.; Liang, Y.; Stacey, G. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017, 214, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Verchot, J. The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front. Plant Sci. 2014, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Alcaide-Loridan, C.; Jupin, I. Ubiquitin and plant viruses, let’s play together! Plant Physiol. 2012, 160, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Tao, X.; Xie, Y.; Fauquet, C.M.; Zhou, X. A DNAβ associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 2004, 78, 13966–13974. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, X.; Wu, J.; Briddon, R.W.; Zhou, X. βC1 encoded by tomato yellow leaf curl China betasatellite forms multimeric complexes in vitro and in vivo. Virology 2011, 409, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Hu, T.; Bao, M.; Cao, L.; Zhang, H.; Song, F.; Xie, Q.; Zhou, X. Tobacco RING E3 ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus-encoded betaC1. Mol. Plant 2016, 9, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Camborde, L.; Planchais, S.; Tournier, V.; Jakubiec, A.; Drugeon, G.; Lacassagne, E.; Pflieger, S.; Chenon, M.; Jupin, I. The ubiquitin-proteasome system regulates the accumulation of turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell 2010, 22, 3142–3152. [Google Scholar] [CrossRef] [PubMed]
- Jakubiec, A.; Drugeon, G.; Camborde, L.; Jupin, I. Proteolytic processing of turnip yellow mosaic virus replication proteins and functional impact on infectivity. J. Virol. 2007, 81, 11402–11412. [Google Scholar] [CrossRef] [PubMed]
- Reichel, C.; Beachy, R.N. Degradation of tobacco mosaicvirus movement protein by the 26S proteasome. J. Virol. 2000, 74, 3330–3337. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Amari, K.; Gereige, D.; Brandner, K.; Mély, Y.; Heinlein, M. Control of tobacco mosaic virus movement protein fate by CELL-DIVISION-CYCLE protein 48. Plant Physiol. 2012, 160, 2093–2108. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bassham, D.C. New insight into the mechanism and function of autophagy in plant cells. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, UK, 2015; Volume 320, pp. 1–40. [Google Scholar]
- Liu, Y.; Schiff, M.; Czymmek, K.; Tallóczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. Role of plant autophagy in stress response. Protein Cell 2011, 2, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Consonni, C.; Panstruga, R.; Ohsumi, Y.; Shirasu, K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009, 21, 2914–2927. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, C.; Li, Y.; Wu, G.; Hou, X.; Zhou, X.; Wang, A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 2018, 9, 1268. [Google Scholar] [CrossRef] [PubMed]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017, 6, e23897. [Google Scholar] [CrossRef] [PubMed]
- Hafrén, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [PubMed]
- Hafrén, A.; Üstün, S.; Hochmuth, A.; Svenning, S.; Johansen, T.; Hofius, D. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 2018, 176, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Hafrén, A.; Lõhmus, A.; Mäkinen, K. Formation of potato virus A-induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog. 2015, 11, e1005314. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Jeong, D.H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed]
- Boccara, M.; Sarazin, A.; Thiébeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Li, F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018, 14, e1006756. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004, 305, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Caplan, J.L.; Zhu, X.; Mamillapalli, P.; Marathe, R.; Anandalakshmi, R.; Dinesh-Kumar, S.P. Induced ER chaperones regulate a receptor-like kinase to mediate antiviral innate immune response in plants. Cell Host Microbe 2009, 6, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Weaver, N.D.; Kesarwani, M.; Dong, X. Induction of protein secretory pathway is required for systemic acquired resistance. Science 2005, 308, 1036–1040. [Google Scholar] [CrossRef] [PubMed]
- Križnik, M.; Petek, M.; Dobnik, D.; Ramšak, Ž.; Baebler, Š.; Pollmann, S.; Kreuze, J.F.; Žel, J.; Gruden, K. Salicylic acid perturbs sRNA-gibberellin regulatory network in immune response of potato to potato virus Y infection. Front. Plant Sci. 2017, 8, 2192. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.S.; Masuta, C.; Yamada, S.; Shimura, H.; Kashihara, Y.; Wada, T.S.; Meguro, A.; Goto, K.; Tadamura, K.; Sueda, K.; et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA 2012, 109, 10113–10118. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.J.; Tadamura, K.; Murakami, T.; Inaba, J.-I.; Kim, B.M.; Sato, M.; Atsumi, G.; Kuchitsu, K.; Masuta, C.; Nakahara, K.S. rgs-CaM detects and counteracts viral RNA silencing suppressors in plant immune priming. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Anandalakshmi, R.; Marathe, R.; Ge, X.; Herr, J.M., Jr.; Mau, C.; Mallory, A.; Pruss, G.; Bowman, L.; Vance, V.B. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 2000, 290, 142–144. [Google Scholar] [CrossRef] [PubMed]
- Steinhauer, D.A.; Holland, J.J. Rapid evolution of RNA viruses. Annu. Rev. Microbiol. 1987, 41, 409–431. [Google Scholar] [CrossRef] [PubMed]
- Pita, J.S.; Roossinck, M.J. Virus populations, mutation rates and frequencies. In Plant Virus Evolution; Roossinck, M.J., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 109–121. [Google Scholar]
- Ge, L.; Zhang, J.; Zhou, X.; Li, H. Genetic structure and population variability of tomato yellow leaf curl China virus. J. Virol. 2007, 81, 5902–5907. [Google Scholar] [CrossRef] [PubMed]
- Grigoras, I.; Timchenko, T.; Grande-Pérez, A.; Katul, L.; Vetten, H.-J.; Gronenborn, B. High variability and rapid evolution of a nanovirus. J. Virol. 2010, 84, 9105–9117. [Google Scholar] [CrossRef] [PubMed]
- Pinel-Galzi, A.; Dubreuil-Tranchant, C.; Hebrard, E.; Mariac, C.; Ghesquiere, A.; Albar, L. Mutations in rice yellow mottle virus polyprotein P2a involved in RYMV2 gene resistance breakdown. Front. Plant Sci. 2016, 7, 1779. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Leal, R.; Bryan, B.K.; Smith, J.T.; Rush, C.M. Breakdown of host resistance by independent evolutionary lineages of beet necrotic yellow vein virus involves a parallel c/u mutation in its p25 gene. Phytopathology 2010, 100, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Naito, S.; Meshi, T.; Ishikawa, M. An inhibitory interaction between viral and cellular proteins underlies the resistance of tomato to nonadapted tobamoviruses. Proc. Natl. Acad. Sci. USA 2009, 106, 8778–8783. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; Candresse, T. Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol. Plant Pathol. 2016, 18, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Wei, M.; Li, G.; Lei, R.; Qiu, Y.; Wang, C.; Li, Z.-H.; Zhu, S. The cucumber mosaic virus movement protein suppresses PAMP-triggered immune responses in Arabidopsis and tobacco. Biochem. Biophys. Res. Commun. 2018, 498, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, A.S.; Golyaev, V.; Turco, S.; Gubaeva, E.G.; Rajeswaran, R.; Schepetilnikov, M.V.; Srour, O.; Ryabova, L.A.; Boller, T.; Pooggin, M.M. Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 2016, 211, 1020–1034. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Xiong, R.; Li, Y.; Li, F.; Zhou, X.; Wang, A. Sumoylation of turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 2017, 29, 508–525. [Google Scholar] [CrossRef] [PubMed]
- van den Burg, H.A.; Kini, R.K.; Schuurink, R.C.; Takken, F.L.W. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 2010, 22, 1998–2016. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Withers, J.; Mohan, R.; Marqués, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 2015, 18, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Cao, X.; Wang, X.; Jiang, J.; Wan, J.; Laliberté, J.F.; Zhang, Y. Three-dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front. Plant Sci. 2018, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Harak, C.; Lohmann, V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 2015, 479–480, 418–433. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Nagy, P.D. Expanding use of multi-origin subcellular membranes by positive-strand RNA viruses during replication. Curr. Opin. Virol. 2014, 9, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Laliberté, J.F.; Zheng, H. Viral manipulation of plant host membranes. Annu. Rev. Virol. 2014, 1, 237–259. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.; McCartney, A.; Gidda, S.; Mullen, R. Localization of the carnation Italian ringspot virus replication protein p36 to the mitochondrial outer membrane is mediated by an internal targeting signal and the TOM complex. BMC Cell Biol. 2008, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Barajas, D.; Jiang, Y.; Nagy, P.D. A unique role for the host ESCRT proteins in replication of tomato bushy stunt virus. PLoS Pathog. 2009, 5, e1000705. [Google Scholar] [CrossRef] [PubMed]
- Restrepo-Hartwig, M.A.; Ahlquist, P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 1996, 70, 8908–8916. [Google Scholar] [PubMed]
- Cotton, S.; Grangeon, R.; Thivierge, K.; Mathieu, I.; Ide, C.; Wei, T.; Wang, A.; Laliberté, J.F. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 2009, 83, 10460–10471. [Google Scholar] [CrossRef] [PubMed]
- Más, P.; Beachy, R.N. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 1999, 147, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Howell, S.H. The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front. Plant Sci. 2017, 8, 344. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Dickman, M.B.; Whitham, S.A.; Payton, M.; Verchot, J. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 2011, 156, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.M.; Chen, S.; Payton, M.; Dickman, M.B.; Verchot, J. TGBp3 triggers the unfolded protein response and SKP1-dependent programmed cell death. Mol. Plant Pathol. 2013, 14, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, H.; Brandizzi, F.; Verchot, J.; Wang, A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 2015, 11, e1005164. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yang, D.; Xie, L.; Sun, L.; Zhang, S.; Zhu, Q.; Li, J.; Wang, X.; Chen, J. Rice black-streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana. Virology 2013, 447, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Shine, M.B.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Pendon, J.A.; Li, F.; Li, W.X.; Ding, S.W. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 2007, 19, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Hong, J.-S.; Tabayashi, N.; Ito, A.; Masuta, C.; Matsumura, T. Development of cucumber mosaic virus as a vector modifiable for different host species to produce therapeutic proteins. Planta 2007, 225, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, X.; Ding, S.W. Viral suppressors of RNA-based viral immunity: Host targets. Cell Host Microbe 2010, 8, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Burgyán, J.; Havelda, Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011, 16, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Valli, A.A.; Gallo, A.; Rodamilans, B.; López-Moya, J.J.; García, J.A. The HcPro from the Potyviridae family: An enviable multitasking helper component that every virus would like to have. Mol. Plant Pathol. 2018, 19, 744–763. [Google Scholar] [CrossRef] [PubMed]
- Várallyay, É.; Válóczi, A.; Ágyi, Á.; Burgyán, J.; Havelda, Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010, 29, 3507–3519. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, L.; Yu, K.; Wang, A. Pathogenesis of soybean mosaic virus in soybean carrying Rsv1 gene is associated with miRNA and siRNA pathways, and breakdown of AGO1 homeostasis. Virology 2015, 476, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, T.; Shen, D.; Wang, J.; Ling, X.; Hu, Z.; Chen, T.; Hu, J.; Huang, J.; Yu, W.; et al. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathog. 2019, 15, e1007534. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Lin, L.; Wang, S.; Guo, Q.; Zhou, H.; Rong, L.; Li, J.; Peng, J.; Lu, Y.; Zheng, H.; et al. Identification and regulation of host genes related to rice stripe virus symptom production. New Phytol. 2016, 209, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Moyo, L.; Ramesh, S.V.; Kappagantu, M.; Mitter, N.; Sathuvalli, V.; Pappu, H.R. The effects of potato virus Y-derived virus small interfering RNAs of three biologically distinct strains on potato (Solanum tuberosum) transcriptome. Virol. J. 2017, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shibuya, M.; Taneda, A.; Kurauchi, T.; Senda, M.; Owens, R.A.; Sano, T. Accumulation of potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 2011, 413, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhou, G. Characteristics of siRNAs derived from southern rice black-streaked dwarf virus in infected rice and their potential role in host gene regulation. Virol. J. 2017, 14, 27. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Y.; Ding, B.; Fei, Z. Comprehensive transcriptome analyses reveal that potato spindle tuber viroid triggers genome-wide changes in alternative splicing, inducible trans-acting activity of phased secondary small interfering RNAs, and immune responses. J. Virol. 2017, 91, e00247-17. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, K.K.; Scholthof, K.-B.G. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 2015, 27, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, M.; Lu, L.; Zhang, X. Diverse functions of small RNAs in different plant-pathogen communications. Front. Microbiol. 2016, 7, 1552. [Google Scholar] [CrossRef] [PubMed]
- He, B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006, 13, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Toribio, R.; Ventoso, I. Inhibition of host translation by virus infection in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 9837–9842. [Google Scholar] [CrossRef] [PubMed]
- Miras, M.; Miller, W.A.; Truniger, V.; Aranda, M.A. Non-canonical translation in plant RNA viruses. Front. Plant Sci. 2017, 8, 494. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.; Lopes, K.V.G.; Apfata, J.A.C.; Fontes, E.P.B. NSP-interacting kinase, NIK: A transducer of plant defence signalling. J. Exp. Bot. 2010, 61, 3839–3845. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.P.B.; Brustolini, O.J.B.; Mendes, G.C.; Santos, A.A.; Fontes, E.P.B. NIK1, a host factor specialized in antiviral defense or a novel general regulator of plant immunity? BioEssays 2015, 37, 1236–1242. [Google Scholar] [CrossRef] [PubMed]
- Chenon, M.; Camborde, L.; Cheminant, S.; Jupin, I. A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity. EMBO J. 2012, 31, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, R.; et al. CLCuMuB βC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, e1005668. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.H.; Chen, I.H.; Baulcombe, D.C.; Tsai, C.H. The silencing suppressor P25 of potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 2010, 11, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Fusaro, A.F.; Correa, R.L.; Nakasugi, K.; Jackson, C.; Kawchuk, L.; Vaslin, M.F.S.; Waterhouse, P.M. The enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology 2012, 426, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Lozsa, R.; Hutvagner, G.; Burgyan, J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010, 62, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Tsai, C.-H.; Lie, M.; Havecker, E.; Baulcombe, D.C. The polerovirus silencing suppressor P0 targets Argonaute proteins for degradation. Curr. Biol. 2007, 17, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Derrien, B.; Baumberger, N.; Schepetilnikov, M.; Viotti, C.; De Cillia, J.; Ziegler-Graff, V.; Isono, E.; Schumacher, K.; Genschik, P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 15942–15946. [Google Scholar] [CrossRef] [PubMed]
- Aronson, M.N.; Meyer, A.D.; Györgyey, J.; Katul, L.; Vetten, H.J.; Gronenborn, B.; Timchenko, T. Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 2000, 74, 2967–2972. [Google Scholar] [PubMed]
- Aronson, M.N.; Complainville, A.; Clérot, D.; Alcalde, H.; Katul, L.; Vetten, H.J.; Gronenborn, B.; Timchenko, T. In planta protein-protein interactions assessed using a nanovirus-based replication and expression system. Plant J. 2002, 31, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Xu, Y.; Li, C.; Li, Y.; Wu, J.; Zhou, X. Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 2018, 11, 269–287. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Duran, R.; Bejarano, E.R. Geminivirus C2 protein might be the key player for geminiviral co-option of SCF-mediated ubiquitination. Plant Signal Behav. 2011, 6, 999–1001. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, A. The potyviral silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 2017, 91, e01478-16. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.B.; Kim, B.-W.; Lee, K.E.; Kim, S.W.; Jeon, H.; Kim, J.; Song, H.K. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat. Struct. Mol. Biol. 2011, 18, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, Y.; Xie, X.; Yue, N.; Li, J.; Wang, X.B.; Han, C.G.; Yu, J.L.; Liu, Y.; Li, D. Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 interaction. Plant Cell 2018, 30, 1582–1595. [Google Scholar] [CrossRef] [PubMed]
- Wileman, T. Aggresomes and autophagy generate sites for virus replication. Science 2006, 312, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Suhy, D.A.; Giddings, T.H.; Kirkegaard, K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: An autophagy-like origin for virus-induced vesicles. J. Virol. 2000, 74, 8953–8965. [Google Scholar] [CrossRef] [PubMed]
- Dreux, M.; Gastaminza, P.; Wieland, S.F.; Chisari, F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 2009, 106, 14046–14051. [Google Scholar] [CrossRef] [PubMed]
- Romero-Brey, I.; Bartenschlager, R. Membranous replication factories induced by plus-strand RNA viruses. Viruses 2014, 6, 2826. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Luo, H. Interplay between the cellular autophagy machinery and positive-stranded RNA viruses. Acta Biochim. Biophys. Sin. 2012, 44, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Boiteux, L.S.; de Noronha Fonseca, M.E.; Vieira, J.V.; de Cássia Pereira-Carvalho, R. Breeding for resistance to viral diseases. In Plant Breeding for Biotic Stress Resistance; Fritsche-Neto, R., Borém, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 57–79. [Google Scholar]
- Rey, C.; Vanderschuren, H. Cassava mosaic and brown streak diseases: Current perspectives and beyond. Annu. Rev. Virol. 2017, 4, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Singh, S.P.; Kianian, S.F. Engineering resistance to plant viruses: Present status and future prospects. In Current Developments in Biotechnology and Bioengineering; Larroche, C., Sanroman, M., Du, G., Pandey, A., Eds.; Elsevier: London, UK, 2017; pp. 75–100. [Google Scholar]
- Galvez, L.C.; Banerjee, J.; Pinar, H.; Mitra, A. Engineered plant virus resistance. Plant Sci. 2014, 228, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA based genetic engineering for plant viral resistance: Application in crop protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [PubMed]
- Chen, L.; Cheng, X.; Cai, J.; Zhan, L.; Wu, X.; Liu, Q.; Wu, X. Multiple virus resistance using artificial trans-acting siRNAs. J. Virol. Methods 2016, 228, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, F.; Cai, J.; Chen, W.; Zhao, N.; Sun, Y.; Guo, Y.; Yang, X.; Wu, X. Artificial TALE as a convenient protein platform for engineering broad-spectrum resistance to begomoviruses. Viruses 2015, 7, 4772–4782. [Google Scholar] [PubMed]
- Lin, C.Y.; Ku, H.M.; Tsai, W.S.; Green, S.K.; Jan, F.J. Resistance to a DNA and a RNA virus in transgenic plants by using a single chimeric transgene construct. Transgenic Res. 2011, 20, 261–270. [Google Scholar] [PubMed]
- Chung, B.N.; Palukaitis, P. Resistance to multiple viruses in transgenic tobacco expressing fused, tandem repeat, virus-derived double-stranded RNAs. Virus Genes 2011, 43, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Bucher, E.; Lohuis, D.; van Poppel, P.M.; Geerts-Dimitriadou, C.; Goldbach, R.; Prins, M. Multiple virus resistance at a high frequency using a single transgene construct. J. Gen. Virol. 2006, 87, 3697–3701. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. (Stuttg.) 2011, 13, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M. Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 2017, 26, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.-T.; Widyasari, K.; Park, J.Y.; Kim, K.-H. Engineering an auto-activated R protein that is in vivo activated by a viral protease. Virology 2017, 510, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Qi, D.; Ashfield, T.; Helm, M.; Innes, R.W. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 2016, 351, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Römer, P.; Recht, S.; Lahaye, T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. USA 2009, 106, 20526–20531. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, S.; Rougon-Cardoso, A.; Sherwood, E.; Peeters, N.; Dahlbeck, D.; van Esse, H.P.; Smoker, M.; Rallapalli, G.; Thomma, B.P.H.J.; Staskawicz, B.; et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 2010, 28, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]
- García-Cano, E.; Resende, R.O.; Fernández-Muñoz, R.; Moriones, E. Synergistic interaction between tomato chlorosis virus and tomato spotted wilt virus results in breakdown of resistance in tomato. Phytopathology 2006, 96, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Valli, A.; García, J.A.; Zhou, X.; Cheng, X. The Tug-of-War between Plants and Viruses: Great Progress and Many Remaining Questions. Viruses 2019, 11, 203. https://doi.org/10.3390/v11030203
Wu X, Valli A, García JA, Zhou X, Cheng X. The Tug-of-War between Plants and Viruses: Great Progress and Many Remaining Questions. Viruses. 2019; 11(3):203. https://doi.org/10.3390/v11030203
Chicago/Turabian StyleWu, Xiaoyun, Adrian Valli, Juan Antonio García, Xueping Zhou, and Xiaofei Cheng. 2019. "The Tug-of-War between Plants and Viruses: Great Progress and Many Remaining Questions" Viruses 11, no. 3: 203. https://doi.org/10.3390/v11030203
APA StyleWu, X., Valli, A., García, J. A., Zhou, X., & Cheng, X. (2019). The Tug-of-War between Plants and Viruses: Great Progress and Many Remaining Questions. Viruses, 11(3), 203. https://doi.org/10.3390/v11030203





