Self-Assembly of Tail Tube Protein of Bacteriophage vB_EcoS_NBD2 into Extremely Long Polytubes in E. coli and S. cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Construction of an Expression Vector in Bacteria
2.3. Construction of an Expression Vector in Yeast
2.4. Synthesis of Recombinant gp39 Protein in Bacteria and Yeast Cells
2.5.Purification of Recombinant gp39 Protein
2.6. Transmission Electron Microscopy
2.7. Stability Analysis of Yeast- and Bacteria-Expressed Tail Tube Proteins
2.8. Bioinformatics Analysis
3. Results
3.1. Bioinformatics Analysis
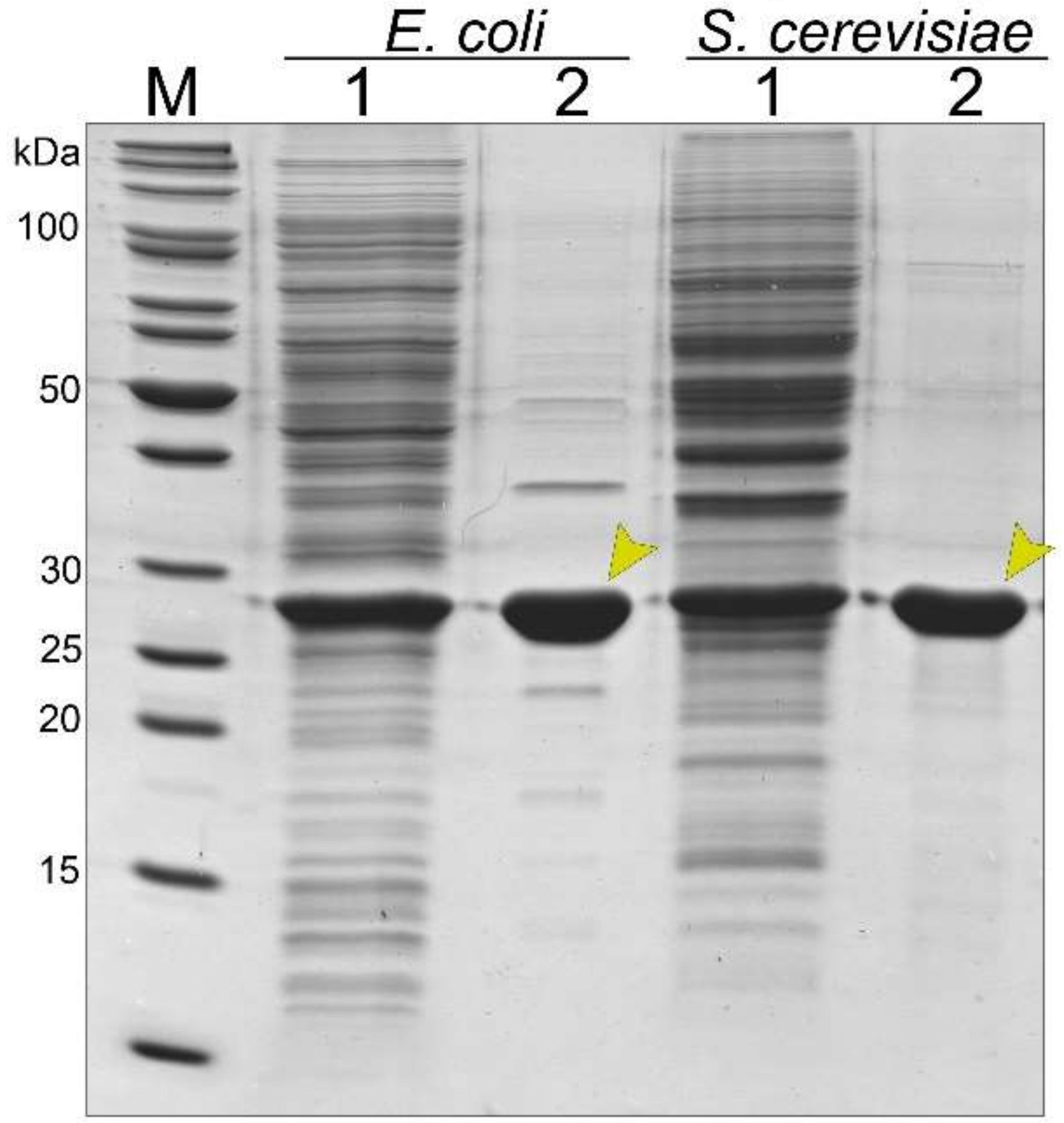
3.2. Synthesis and Purification of the Recombinant Tail Tube Protein gp39 in Bacteria and Yeast Cells
3.3. Electron Microscopy Analysis of Bacteria- and Yeast-Derived Tubular Structures
3.4. Stability of Bacteria- and Yeast-Derived Tubular Structures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lehn, J.M. Toward complex matter: Supramolecular chemistry and self-organization. Proc. Natl. Acad. Sci. USA 2002, 99, 4763–4768. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Moon, K.S.; Lee, M. Recent advances in functional supramolecular nanostructures assembled from bioactive building blocks. Chem. Soc. Rev. 2009, 38, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Webber, M.J.; Stupp, S.I. Self-assembly of peptide amphiphiles: From molecules to nanostructures to biomaterials. Biopolymers 2010, 94, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Matson, J.B.; Stupp, S.I. Self-assembling peptide scaffolds for regenerative medicine. Chem. Commun. 2012, 48, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E. Self-assembled peptide nanostructures: The design of molecular building blocks and their technological utilization. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Glover, D.J.; Giger, L.; Kim, S.S.; Naik, R.R.; Clark, D.S. Geometrical assembly of ultrastable protein templates for nanomaterials. Nat. Commun. 2016, 7, 11771. [Google Scholar] [CrossRef] [PubMed]
- McMillan, R.A.; Howard, J.; Zaluzec, N.J.; Kagawa, H.K.; Mogul, R.; Li, Y.F.; Paavola, C.D.; Trent, J.D. A self-assembling protein template for constrained synthesis and patterning of nanoparticle arrays. J. Am. Chem. Soc. 2005, 127, 2800–2801. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Nanomaterials based on DNA. Annu. Rev. Biochem. 2010, 79, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Liu, D.; Booth, G.; Gao, W.; Lu, Y. Virus-like particle engineering: From rational design to versatile applications. Biotechnol. J. 2018, 13, e1700324. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Zha, L.; Cabral-Miranda, G.; Bachmann, M.F. Major findings and recent advances in virus–like particle (VLP)-based vaccines. Semin. Immunol. 2017, 34, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, C.; Wagner, R. Virus-like particles-universal molecular toolboxes. Curr. Opin. Biotechnol. 2007, 18, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lin, T.; Tang, L.; Johnson, J.E.; Finn, M.G. Icosahedral virus particles as addressable nanoscale building blocks. Angew. Chem. Int. Ed. Engl. 2002, 41, 459–462. [Google Scholar] [CrossRef]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically engineered phages: A review of advances over the last decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef] [PubMed]
- Molek, P.; Bratkovič, T. Bacteriophages as scaffolds for bipartite display: Designing swiss army knives on a nanoscale. Bioconj. Chem. 2015, 26, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Schoonen, L.; Van Hest, J.C.M. Functionalization of protein-based nanocages for drug delivery applications. Nanoscale 2014, 6, 7124–7141. [Google Scholar] [CrossRef] [PubMed]
- Zvirbliene, A.; Kucinskaite-Kodze, I.; Razanskiene, A.; Petraityte-Burneikiene, R.; Klempa, B.; Ulrich, R.G.; Gedvilaite, A. The use of chimeric virus-like particles harbouring a segment of hantavirus Gc glycoprotein to generate a broadly-reactive hantavirus-specific monoclonal antibody. Viruses 2014, 6, 640–660. [Google Scholar] [CrossRef] [PubMed]
- Pleckaityte, M.; Bremer, C.M.; Gedvilaite, A.; Kucinskaite-Kodze, I.; Glebe, D.; Zvirbliene, A. Construction of polyomavirus-derived pseudotype virus-like particles displaying a functionally active neutralizing antibody against hepatitis B virus surface antigen. BMC Biotechnol. 2015, 15, 85. [Google Scholar] [CrossRef] [PubMed]
- Rakonjac, J.; Bennett, N.J.; Spagnuolo, J.; Gagic, D.; Russel, M. Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011, 13, 51–76. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Lee, Y.; Thomas, S.; Kohli, A.G.; Yun, D.S.; Belcher, A.M.; Kelly, K.A. M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer. Nat. Nanotechnol. 2012, 7, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Merzlyak, A.; Indrakanti, S.; Lee, S.W. Genetically engineered nanofiber-like viruses for tissue regenerating materials. Nano Lett. 2009, 9, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.; Merzlyak, A.; Lee, S. Fabrication of engineered M13 bacteriophages into liquid crystalline films and fibers for directional growth and encapsulation of fibroblasts. Soft Matter 2010, 6, 4454–4459. [Google Scholar] [CrossRef]
- Zeltins, A. Viral Nanoparticles. Principles of construction and characterization. In Viral Nanotechnology; Khudyakov, Y.E., Pumpens, P., Eds.; CRC Press, Taylor & Francis group: Boca Taon, FL, USA, 2016; pp. 93–120. ISBN 9781466583528. [Google Scholar]
- Pattenden, L.K.; Middelberg, A.P.J.; Niebert, M.; Lipin, D.I. Towards the preparative and large-scale precision manufacture of virus-like particles. Trends Biotechnol. 2005, 23, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Zeltins, A. Construction and characterization of virus-like particles: A review. Mol. Biotechnol. 2013, 53, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Kushnir, N.; Streatfield, S.; Yusibov, V. Virus-like particles as a highly efficient vaccine platform: Diversity of targets and production systems and advances in clinical development. Vaccine 2012, 31, 58–83. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Allen, M.J.; Wang, Y.; Wang, B.; Wang, N.; Shi, L.; Sitrin, R.D. Disassembly and reassembly improves morphology and thermal stability of human papillomavirus type 16 virus-like particles. Nanomedicine 2012, 8, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Sasnauskas, K.; Bulavaite, A.; Hale, A.; Jin, L.; Knowles, W.A.; Gedvilaite, A.; Dargevičiute, A.; Bartkevičiute, D.; Žvirbliene, A.; Staniulis, J.; et al. Generation of recombinant virus-like particles of human and non-human polyomaviruses in yeast Saccharomyces cerevisiae. Intervirology 2002, 45, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Slibinskas, R.; Samuel, D.; Gedvilaite, A.; Staniulis, J.; Sasnauskas, K. Synthesis of the measles virus nucleoprotein in yeast Pichia pastoris and Saccharomyces cerevisiae. J. Biotechnol. 2004, 107, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Tamošiunas, P.L.; Simutis, K.; Kodzė, I.; Firantienė, R.; Emužytė, R.; Petraitytė-Burneikienė, R.; Zvirblienė, A.; Sasnauskas, K. Production of human parvovirus 4 VP2 virus-like particles in yeast and their evaluation as an antigen for detection of virus-specific antibodies in human serum. Intervirology 2013, 56, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J. Production of biopharmaceutical proteins by yeast. Bioengineered 2013, 4, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.S.; Brown, D.R.; Bryan, J.T.; Cook, J.C.; George, H.A.; Hofmann, K.J.; Hurni, W.M.; Joyce, J.G.; Lehman, E.D.; Markus, H.Z.; et al. Human papillomavirus type II (HPV-11) neutralizing antibodies in the serum and genital mucosal secretions of African green monkeys immunized with HPV-11 virus-like particles expressed in yeast. J. Infect. Dis. 1997, 176, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Wiedermann, G.; Scheiermann, N.; Goubau, P.; Ambrosch, F.; Gesemann, M.; De Bel, C.; Kremsner, P.; Paar, D.; Kunz, C.; Hauser, P.; et al. Multicentre dose range study of a yeast-derived hepatitis B vaccine. Vaccine 1987, 5, 179–183. [Google Scholar] [CrossRef]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Katsura, I. Mechanism of length determination in bacteriophage lambda tails. Adv. Biophys. 1990, 26, 1–18. [Google Scholar] [CrossRef]
- Veesler, D.; Cambillau, C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 2011, 75, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.R.; Cardarelli, L.; Pell, L.G.; Radford, D.R.; Maxwell, K.L. Long noncontractile tail machines of bacteriophages. In Viral Molecular Machines; Rossmann, M.G., Rao, V.B., Eds.; Springer: Boston, MA, USA, 2012; pp. 115–142. ISBN 978-1-4614-0979-3. [Google Scholar]
- Zivanovic, Y.; Confalonieri, F.; Ponchon, L.; Lurz, R.; Chami, M.; Flayhan, A.; Renouard, M.; Huet, A.; Decottignies, P.; Davidson, A.R.; et al. Insights into bacteriophage T5 structure from analysis of its morphogenesis genes and protein components. J. Virol. Methods 2014, 88, 1162–1174. [Google Scholar] [CrossRef] [PubMed]
- Leonard, K.R.; Kleinschmidt, A.K.; Lake, J.A. Caulobacter crescentus Bacteriophage phiCbK: Structure and in vitro self-assembly of the tail. J. Mol. Biol. 1973, 81, 349–365. [Google Scholar] [CrossRef]
- Effantin, G.; Boulanger, P.; Neumann, E.; Letellier, L.; Conway, J.F. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 2006, 361, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Langlois, C.; Cukkemane, A.; Auzat, I.; Chagot, B.; Gilquin, B.; Ignatiou, A.; Petitpas, I.; Kasotakis, E.; Paternostre, M.; White, H.E.; et al. Bacteriophage SPP1 tail tube protein self-assembles into β-structure-rich tubes. J. Biol. Chem. 2015, 290, 3836–3849. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Valluzzi, R.; Goldberg, E. Design of protein struts for self-assembling nanoconstructs. Proc. Natl. Acad. Sci. USA 2002, 99, 8488–8493. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Walker-Kopp, N.; Wilkens, S.; Cingolani, G. Foldon-guided self-assembly of ultra-stable protein fibers. Protein Sci. 2008, 17, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Guo, P. Bacterial virus phi29 DNA-packaging motor and its potential applications in gene therapy and nanotechnology. Methods Mol. Biol. 2005, 300, 285–324. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, E.; Boy de la Tour, E. On the fine structure of normal and “Polymerized” tail sheath of phage T4. J. Ultrasructure Res. 1964, 11, 545–563. [Google Scholar] [CrossRef]
- Moody, M.F. Structure of the sheath of bacteriphage T4. Structure of the contracted sheath and polysheath. J. Mol. Biol. 1967, 25, 167–174. [Google Scholar] [CrossRef]
- Tschopp, J.; Arisaka, F.; van Driel, R.; Engel, J. Purification, characterization and reassembly of the bacteriophage T4D tail sheath protein P18. J. Mol. Biol. 1979, 128, 247–258. [Google Scholar] [CrossRef]
- Šimoliūnas, E.; Truncaitė, L.; Rutkienė, R.; Povilonienė, S.; Goda, K.; Kaupinis, A.; Valius, M.; Meškys, R. The robust self-assembling tubular nanostructures formed by gp053 from phage vB_EcoM_FV3. Viruses 2019, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Kaliniene, L.; Truncaitė, L.; Šimoliūnas, E.; Zajančkauskaitė, A.; Vilkaitytė, M.; Kaupinis, A.; Skapas, M.; Meškys, R. Molecular analysis of the low-temperature Escherichia coli phage vB_EcoS_NBD2. Arch. Virol. 2017, 163, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Sasnauskas, K.; Buzaite, O.; Vogel, F.; Jandrig, B.; Razanskas, R.; Staniulis, J.; Scherneck, S.; Krüger, D.H.; Ulrich, R. Yeast cells allow high-level expression and formation of polyomavirus-like particles. Biol. Chem. 1999, 380, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Transeq. Available online: http://www.ebi.ac.uk/Tools/st/emboss_transeq (accessed on 20 December 2018).
- Clustal Omega. Available online: http://www.ebi.ac.uk/Tools/msa/clustalo (accessed on 20 December 2018).
- PIR. Available online: http://pir.georgetown.edu/pirwww/search/comp_mw.shtml (accessed on 20 December 2018).
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A completely reimplemented mpi bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- HHpred. Available online: https://toolkit.tuebingen.mpg.de/#/tools/hhpred (accessed on 18 February 2019).
- Gedvilaite, A.; Frömmel, C.; Sasnauskas, K.; Micheel, B.; Özel, M.; Behrsing, O.; Staniulis, J.; Jandrig, B.; Scherneck, S.; Ulrich, R. Formation of immunogenic virus-like particles by inserting epitopes into surface-exposed regions of hamster polyomavirus major capsid protein. Virology 2000, 273, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, C.A.; Effantin, G.; Vives, C.; Engilberge, S.; Bacia, M.; Boulanger, P.; Girard, E.; Schoehn, G.; Breyton, C. Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection. Nat. Commun. 2017, 8, 1953. [Google Scholar] [CrossRef] [PubMed]
- Qing, G.; Ma, L.C.; Khorchid, A.; Swapna, G.V.; Mal, T.K.; Takayama, M.M.; Xia, B.; Phadtare, S.; Ke, H.; Acton, T.; et al. Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 2004, 22, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Keum, K.C.; Lee, S.Y. Production of recombinant proteins by high cell density culture of Escherichia coli. Chem. Eng. Sci. 2006, 61, 876–885. [Google Scholar] [CrossRef]
- Leavitt, A.D.; Roberts, T.M.; Garcea, R.L. Polyoma virus major capsid protein, VP1. Purification after high level expression in Escherichia coli. J. Biol. Chem. 1985, 260, 12803–12809. [Google Scholar] [PubMed]
- Buckholz, R.G.; Gleeson, M.A. Yeast systems for the commercial production of heterologous proteins. Biotechnol. 1991, 9, 1067–1072. [Google Scholar] [CrossRef]
- Boga, J.A.; Martín Alonso, J.M.; Casais, R.; Parra, F. A single dose immunization with rabbit haemorrhagic disease virus major capsid protein produced in Saccharomyces cerevisiae induces protection. J. Gen. Virol. 1997, 78, 2315–2318. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.C.; Joyce, J.G.; George, H.A.; Schultz, L.D.; Hurni, W.M.; Jansen, K.U.; Hepler, R.W.; Ip, C.; Lowe, R.S.; Keller, P.M.; et al. Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expr. Purif. 1999, 17, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Hallewell, R.; Mills, R.; Tekamp-Olson, P.; Blacher, R.; Rosenberg, S.; Ötting, F.; Masiarz, F.R.; Scandella, C.J. Amino terminal acetylation of authentic human Cu, Zn superoxide dismutase produced in yeast. Bio/Technology 1987, 5, 363–366. [Google Scholar] [CrossRef]
- Verma, R.; Boleti, E.; George, A.J. Antibody engineering: Comparison of bacterial, yeast, insect and mammalian expression systems. J. Immunol. Methods 1998, 216, 165–181. [Google Scholar] [CrossRef]
- Freivalds, J.; Dislers, A.; Ose, V.; Skrastina, D.; Cielens, I.; Pumpens, P.; Sasnauskas, K.; Kazaks, A. Assembly of bacteriophage Qβ virus-like particles in yeast Saccharomyces cerevisiae and Pichia pastoris. J. Biotechnol. 2006, 123, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Katsura, I. Tail assembly and injection. In Lambda II; Hendrix, R., Roberts, J., Stahl, F., Weisberg, R., Eds.; Cold Spring Harbor: Plainview, NY, USA, 1983; pp. 331–346. [Google Scholar]
- Katsura, I. Structure and function of the major tail protein of bacteriophage lambda. Mutants having small major tail protein molecules in their virion. J. Mol. Biol. 1981, 146, 493–512. [Google Scholar] [CrossRef]
- Pell, L.G.; Kanelis, V.; Donaldson, L.W.; Howell, P.L.; Davidson, A.R. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. USA 2009, 106, 4160–4165. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Wang, F.; Taylor, N.M.I.; Guerrero-Ferreira, R.C.; Leiman, P.G.; Egelman, E.H. Refined cryo-EM structure of the T4 tail tube: Exploring the lowest dose limit. Structure 2017, 25, 1436–1441. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.I.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Aksyuk, A.A.; Leiman, P.G.; Kurochkina, L.P.; Shneider, M.M.; Kostyuchenko, V.A.; Mesyanzhinov, V.V.; Rossmann, M.G. The tail sheath structure of bacteriophage T4: A molecular machine for infecting bacteria. EMBO J. 2009, 28, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Ge, P.; Scholl, D.; Leiman, P.G.; Yu, X.; Miller, J.F.; Zhou, Z.H. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat. Struct. Mol. Biol. 2015, 22, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Takashima, K.; Ishihara, H.; Shinomiya, T.; Kageyama, M.; Kanaya, S.; Ohnishi, M.; Murata, T.; Mori, H.; Hayashi, T. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 2000, 38, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.I.; van Raaij, M.J.; Leiman, P.G. Contractile injection systems of bacteriophages and related systems. Mol. Microbiol. 2018, 108, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Aksyuk, A.A.; Kurochkina, L.P.; Fokine, A.; Forouhar, F.; Mesyanzhinov, V.V.; Tong, L.; Rossmann, M.G. Structural conservation of the Myoviridae phage tail sheath protein fold. Structure 2011, 19, 1885–1894. [Google Scholar] [CrossRef] [PubMed]
- Jobichen, C.; Chakraborty, S.; Li, M.; Zheng, J.; Joseph, L.; Mok, Y.K.; Leung, Y.K.; Sivaraman, J. Structural basis for the secretion of EvpC: A key type VI secretion system protein from Edwardsiella tarda. PLoS ONE 2010, 5, e12910. [Google Scholar] [CrossRef] [PubMed]
- Bebeacua, C.; Lai, L.; Vegge, C.S.; Brondsted, L.; van Heel, M.; Veesler, D.; Cambillau, C. Visualizing a complete Siphoviridae member by single-particle electron microscopy: The structure of Lactococcal Phage TP901-1. J. Virol. 2013, 87, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Yui-Furihata, C. Structure of pyocin R. III. Self-assembly of sheath subunits. J. Biochem. 1971, 70, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef] [PubMed]
- Del Tordello, E.; Danilchanka, O.; McCluskey, A.J.; Mekalanos, J.J. Type VI secretion system sheaths as nanoparticles for antigen display. Proc. Natl. Acad. Sci. USA 2016, 113, 3042–3047. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, G.; Ren, X.; Herrler, G. Select what you need: A comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 2007, 127, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Kolhar, P.; Anselmo, A.C.; Gupta, V.; Pant, K.; Prabhakarpandian, B.; Ruoslahti, E.; Mitragotri, S. Using shape effects to target antibody-coated nanoparticles to lung and brain endothelium. Proc. Natl. Acad. Sci. USA 2013, 110, 10753–10758. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Ghosh, D.; Ham, M.; Qi, J.; Barone, P.W.; Strano, M.S.; Belcher, A.M. M13 Phage-Functionalized Single-Walled Carbon Nanotubes as Nanoprobes for Second Near-Infrared Window Fluorescence Imaging of Targeted Tumors. Nano Lett. 2012, 12, 1176–1183. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, N.F.; Miertens, M.E.; Taurog, R.E.; Johnson, J.E.; Commandeur, U.; Fischer, R.; Manchester, M. Potato virus X as a novel platform for potential biomedical applications. Nano Lett. 2010, 10, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Bakhshinejad, B.; Karimi, M.; Khalaj-Kondori, M. Phage display: Development of nanocarriers for targeted drug delivery to the brain. Neural Regen. Res. 2015, 10, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Zhao, X.; Chen, L.; Lan, X.; Li, Y.; Lin, Y.; Wang, Q. Viral nanoparticles as antigen carriers: Influence of shape on humoral immune responses in vivo. RSC Adv. 2014, 4, 23017–23021. [Google Scholar] [CrossRef]
- Pomerantseva, E.A.; Gerasopoulos, K.; Gnerlich, M.; Odenwald, P.; Culver, J.; Ghodssi, R. Creating high-performance microbatteries with tobacco mosaic virus. Nanotechnology 2013, 3–5. [Google Scholar] [CrossRef]


| Composition of Buffers | pH |
|---|---|
| 50 mM sodium citrate, 2 mM EDTA | 3.2 |
| 50 mM sodium acetate, 2 mM EDTA | 4.7 |
| PBS, 2 mM EDTA | 7.6 |
| 50 mM Tris, 2 mM EDTA | 8.7 |
| 50 mM NaHCO3, 2 mM EDTA, 200 mM NaCl | 9.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Špakova, A.; Šimoliūnas, E.; Batiuškaitė, R.; Pajeda, S.; Meškys, R.; Petraitytė-Burneikienė, R. Self-Assembly of Tail Tube Protein of Bacteriophage vB_EcoS_NBD2 into Extremely Long Polytubes in E. coli and S. cerevisiae. Viruses 2019, 11, 208. https://doi.org/10.3390/v11030208
Špakova A, Šimoliūnas E, Batiuškaitė R, Pajeda S, Meškys R, Petraitytė-Burneikienė R. Self-Assembly of Tail Tube Protein of Bacteriophage vB_EcoS_NBD2 into Extremely Long Polytubes in E. coli and S. cerevisiae. Viruses. 2019; 11(3):208. https://doi.org/10.3390/v11030208
Chicago/Turabian StyleŠpakova, Aliona, Eugenijus Šimoliūnas, Raminta Batiuškaitė, Simonas Pajeda, Rolandas Meškys, and Rasa Petraitytė-Burneikienė. 2019. "Self-Assembly of Tail Tube Protein of Bacteriophage vB_EcoS_NBD2 into Extremely Long Polytubes in E. coli and S. cerevisiae" Viruses 11, no. 3: 208. https://doi.org/10.3390/v11030208
APA StyleŠpakova, A., Šimoliūnas, E., Batiuškaitė, R., Pajeda, S., Meškys, R., & Petraitytė-Burneikienė, R. (2019). Self-Assembly of Tail Tube Protein of Bacteriophage vB_EcoS_NBD2 into Extremely Long Polytubes in E. coli and S. cerevisiae. Viruses, 11(3), 208. https://doi.org/10.3390/v11030208





