Inclusion of Antibodies to Cell Culture Media Preserves the Integrity of Genes Encoding RL13 and the Pentameric Complex Components During Fibroblast Passage of Human Cytomegalovirus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Subjects and Clinical Sample Collection
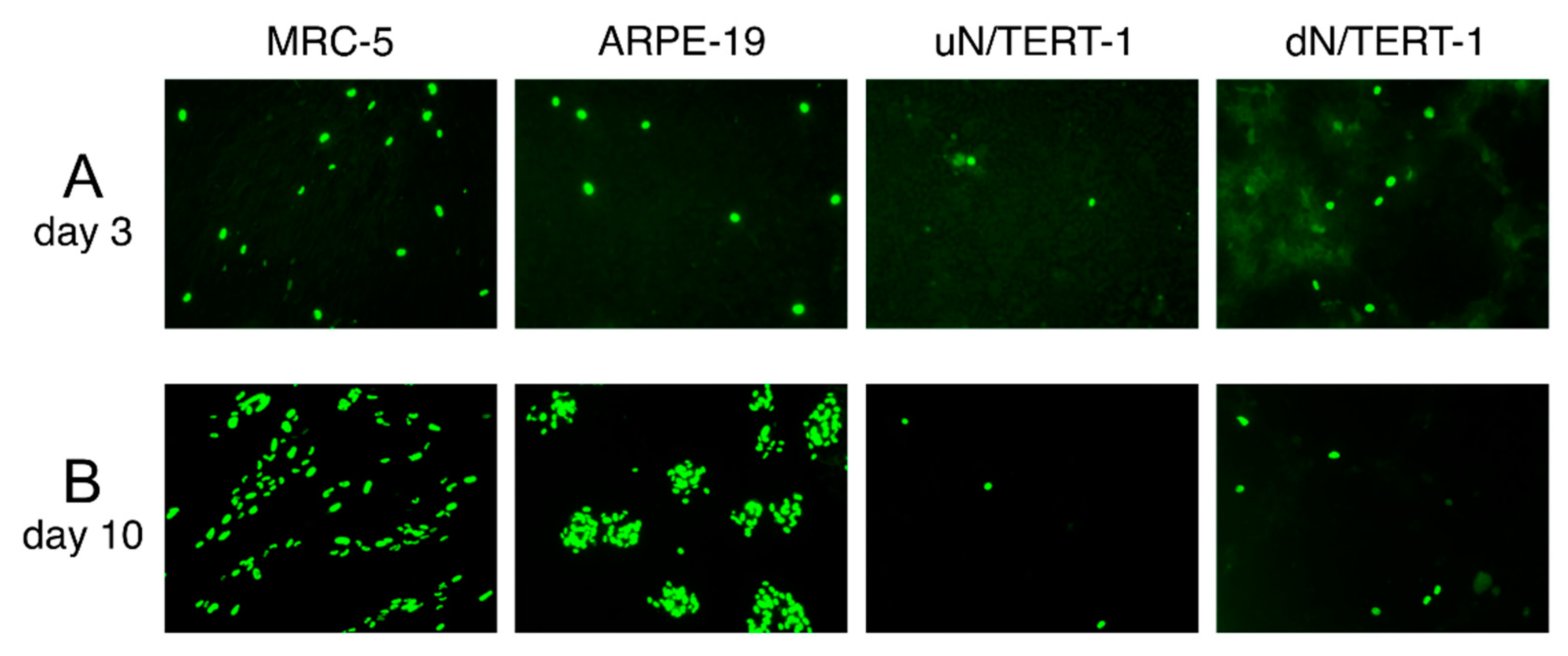
2.2. Cells
2.3. Virus
2.4. Cell Infection and Detection of Infected Cells by Immunostaining
2.5. Genetic Sequencing
2.6. Amino Acid Sequence Prediction and Alignments
3. Results
3.1. HIG in the Culture Medium Prevents Loss of Epithelial Tropism during Fibroblast Passage
3.2. Genetic Analysis of ϕ-KG and Ig-KG
3.3. Viruses Isolated and Expanded from Mixed Stocks
3.4. Experimental Use of CMV Stocks Propagated with HIG
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Craig, J.M.; Macauley, J.C.; Weller, T.H.; Wirth, P. Isolation of intranuclear inclusion producing agents from infants with illnesses resembling cytomegalic inclusion disease. Proc. Soc. Exp. Biol. Med. 1957, 94, 4–12. [Google Scholar] [PubMed]
- Dargan, D.J.; Douglas, E.; Cunningham, C.; Jamieson, F.; Stanton, R.J.; Baluchova, K.; McSharry, B.P.; Tomasec, P.; Emery, V.C.; Percivalle, E.; et al. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J. Gen. Virol. 2010, 91, 1535–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, I.; Wilkie, G.S.; Davison, A.J.; Statkute, E.; Fielding, C.A.; Tomasec, P.; Wilkinson, G.W.; Stanton, R.J. Genetic Stability of Bacterial Artificial Chromosome-Derived Human Cytomegalovirus during Culture In Vitro. J. Virol. 2016, 90, 3929–3943. [Google Scholar] [CrossRef] [PubMed]
- Murrell, I.; Tomasec, P.; Wilkie, G.S.; Dargan, D.J.; Davison, A.J.; Stanton, R.J. Impact of sequence variation in the UL128 locus on production of human cytomegalovirus in fibroblast and epithelial cells. J. Virol. 2013, 87, 10489–10500. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.A.; Hahn, W.C.; Ino, Y.; Ronfard, V.; Wu, J.Y.; Weinberg, R.A.; Louis, D.N.; Li, F.P.; Rheinwald, J.G. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell Biol. 2000, 20, 1436–1447. [Google Scholar] [CrossRef] [PubMed]
- Adler, S.P. Molecular epidemiology of cytomegalovirus: viral transmission among children attending a day care center, their parents, and caretakers. J. Pediatr. 1988, 112, 366–372. [Google Scholar] [CrossRef]
- Cui, X.; Adler, S.P.; Davison, A.J.; Smith, L.; Habib el, S.E.; McVoy, M.A. Bacterial artificial chromosome clones of viruses comprising the towne cytomegalovirus vaccine. J. Biomed. Biotechnol. 2012, 2012, 428498. [Google Scholar] [CrossRef] [PubMed]
- Mehta, K.; Laimins, L. Human Papillomaviruses Preferentially Recruit DNA Repair Factors to Viral Genomes for Rapid Repair and Amplification. MBio 2018, 9, e00064-18. [Google Scholar] [CrossRef] [PubMed]
- Coronel, R.; Takayama, S.; Juwono, T.; Hertel, L. Dynamics of Human Cytomegalovirus Infection in CD34+ Hematopoietic Cells and Derived Langerhans-Type Dendritic Cells. J. Virol. 2015, 89, 5615–5632. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Tamburro, K.; Dittmer, D.; Cui, X.; McVoy, M.A.; Hernandez-Alvarado, N.; Schleiss, M.R. Complete genome sequence of pathogenic Guinea pig cytomegalovirus from salivary gland homogenates of infected animals. Genome Announc. 2013, 1, e0005413. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.D.; Lee, G.C.; Kim, Y.Y.; Lee, C.H. A Comparison between Low- and High-Passage Strains of Human Cytomegalovirus. J. Microbiol. Biotechnol. 2016, 26, 1800–1807. [Google Scholar] [CrossRef] [PubMed]
- Hahn, G.; Revello, M.G.; Patrone, M.; Percivalle, E.; Campanini, G.; Sarasini, A.; Wagner, M.; Gallina, A.; Milanesi, G.; Koszinowski, U.; et al. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 2004, 78, 10023–10033. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shenk, T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA 2005, 102, 18153–18158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, B.; Scrivano, L.; Ruzcics, Z.; Rupp, B.; Sinzger, C.; Koszinowski, U. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 2006, 87, 2451–2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryckman, B.J.; Rainish, B.L.; Chase, M.C.; Borton, J.A.; Nelson, J.A.; Jarvis, M.A.; Johnson, D.C. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 2008, 82, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Freed, D.C.; Tang, Q.; Tang, A.; Li, F.; He, X.; Huang, Z.; Meng, W.; Xia, L.; Finnefrock, A.C.; Durr, E.; et al. Pentameric complex of viral glycoprotein H is the primary target for potent neutralization by a human cytomegalovirus vaccine. Proc. Natl. Acad. Sci. USA 2013, 110, E4997–E5005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McVoy, M. Analysis of Cytomegalovirus Sequences Obtained from the NCBI Database; Virginia Commonwealth University: Richmond, VA, USA, 2019. [Google Scholar]
- Ourahmane, A.; McVoy, M. Observations of Cell Cultures Infected with Different Cytomegalovirus Strain Variants; Virginia Commonwealth University: Richmond, VA, USA, 2018. [Google Scholar]
- Stanton, R.J.; Baluchova, K.; Dargan, D.J.; Cunningham, C.; Sheehy, O.; Seirafian, S.; McSharry, B.P.; Neale, M.L.; Davies, J.A.; Tomasec, P.; et al. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J. Clin. Invest. 2010, 120, 3191–3208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, I.; Bedford, C.; Ladell, K.; Miners, K.L.; Price, D.A.; Tomasec, P.; Wilkinson, G.W.G.; Stanton, R.J. The pentameric complex drives immunologically covert cell-cell transmission of wild-type human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2017, 114, 6104–6109. [Google Scholar] [CrossRef] [PubMed]
- Cha, T.A.; Tom, E.; Kemble, G.W.; Duke, G.M.; Mocarski, E.S.; Spaete, R.R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 1996, 70, 78–83. [Google Scholar] [PubMed]
- Prichard, M.N.; Penfold, M.E.; Duke, G.M.; Spaete, R.R.; Kemble, G.W. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 2001, 11, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Gerna, G.; Percivalle, E.; Lilleri, D.; Lozza, L.; Fornara, C.; Hahn, G.; Baldanti, F.; Revello, M.G. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol. 2005, 86, 275–284. [Google Scholar] [CrossRef] [PubMed]
- MacCormac, L.P.; Grundy, J.E. Two clinical isolates and the Toledo strain of cytomegalovirus contain endothelial cell tropic variants that are not present in the AD169, Towne, or Davis strains. J. Med. Virol. 1999, 57, 298–307. [Google Scholar] [CrossRef]
- Kari, B.; Gehrz, R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J. Virol. 1992, 66, 1761–1764. [Google Scholar] [PubMed]
- Mach, M.; Kropff, B.; Dal Monte, P.; Britt, W. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 2000, 74, 11881–11892. [Google Scholar] [CrossRef] [PubMed]
- Navarro, D.; Paz, P.; Tugizov, S.; Topp, K.; La Vail, J.; Pereira, L. Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology 1993, 197, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Alberola, J.; Tamarit, A.; Navarro, D. Functional antibody response to human cytomegalovirus in immunocompetent and HIV-1 infected individuals with antibodies that inhibit virus penetration into cells and intercellular transmission of viral infection. J. Med. Microbiol. 1999, 48, 947–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinzger, C.; Mangin, M.; Weinstock, C.; Topp, M.S.; Hebart, H.; Einsele, H.; Jahn, G. Effect of serum and CTL on focal growth of human cytomegalovirus. J. Clin. Virol. 2007, 38, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.J.; Adler, B.; Sampaio, K.L.; Digel, M.; Jahn, G.; Ettischer, N.; Stierhof, Y.D.; Scrivano, L.; Koszinowski, U.; Mach, M.; et al. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J. Virol. 2008, 82, 2802–2812. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.M.; Shenk, T. Human cytomegalovirus pUS27 G protein-coupled receptor homologue is required for efficient spread by the extracellular route but not for direct cell-to-cell spread. J. Virol. 2011, 85, 3700–3707. [Google Scholar] [CrossRef] [PubMed]
- Scrivano, L.; Sinzger, C.; Nitschko, H.; Koszinowski, U.H.; Adler, B. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog. 2011, 7, e1001256. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lee, R.; Adler, S.P.; McVoy, M.A. Antibody inhibition of human cytomegalovirus spread in epithelial cell cultures. J. Virol. Methods 2013, 192, 44–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, X.; Adler, S.P.; Schleiss, M.R.; Arav-Boger, R.; Demmler Harrison, G.J.; McVoy, M.A. Cytomegalovirus Virions Shed in Urine Have a Reversible Block to Epithelial Cell Entry and Are Highly Resistant to Antibody Neutralization. Clin. Vaccine Immunol. 2017, 24, e00024-17. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Freed, D.C.; Wang, D.; Qiu, P.; Li, F.; Fu, T.M.; Kauvar, L.M.; McVoy, M.A. Impact of Antibodies and Strain Polymorphisms on Cytomegalovirus Entry and Spread in Fibroblasts and Epithelial Cells. J. Virol. 2017, 91, e01650-16. [Google Scholar] [CrossRef] [PubMed]
- Maidji, E.; Percivalle, E.; Gerna, G.; Fisher, S.; Pereira, L. Transmission of human cytomegalovirus from infected uterine microvascular endothelial cells to differentiating/invasive placental cytotrophoblasts. Virology 2002, 304, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Gerna, G.; Sarasini, A.; Patrone, M.; Percivalle, E.; Fiorina, L.; Campanini, G.; Gallina, A.; Baldanti, F.; Revello, M.G. Human cytomegalovirus serum neutralizing antibodies block virus infection of endothelial/epithelial cells, but not fibroblasts, early during primary infection. J. Gen. Virol. 2008, 89, 853–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilleri, D.; Kabanova, A.; Revello, M.G.; Percivalle, E.; Sarasini, A.; Genini, E.; Sallusto, F.; Lanzavecchia, A.; Corti, D.; Gerna, G. Fetal human cytomegalovirus transmission correlates with delayed maternal antibodies to gH/gL/pUL128-130-131 complex during primary infection. PLoS ONE 2013, 8, e59863. [Google Scholar] [CrossRef] [PubMed]
- Chiuppesi, F.; Wussow, F.; Johnson, E.; Bian, C.; Zhuo, M.; Rajakumar, A.; Barry, P.A.; Britt, W.J.; Chakraborty, R.; Diamond, D.J. Vaccine-Derived Neutralizing Antibodies to the Human Cytomegalovirus gH/gL Pentamer Potently Block Primary Cytotrophoblast Infection. J. Virol. 2015, 89, 11884–11898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.A.; Pari, G.S. Human cytomegalovirus UL102 gene. J. Virol. 1995, 69, 1734–1740. [Google Scholar]
- Stinski, M.F.; Petrik, D.T. Functional roles of the human cytomegalovirus essential IE86 protein. Curr. Top. Microbiol. Immunol. 2008, 325, 133–152. [Google Scholar] [PubMed]
- Kagele, D.; Rossetto, C.C.; Tarrant, M.T.; Pari, G.S. Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt. Virology 2012, 424, 106–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pari, G.S. Nuts and bolts of human cytomegalovirus lytic DNA replication. Curr. Top. Microbiol. Immunol. 2008, 325, 153–166. [Google Scholar] [PubMed]
- Spector, D.J. UL84-independent replication of human cytomegalovirus strains conferred by a single codon change in UL122. Virology 2015, 476, 345–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, C.; Lee, D.; Gelbmann, C.B.; Van Sciver, N.; Nawandar, D.M.; Kenney, S.C.; Kalejta, R.F. Human Cytomegalovirus Productively Replicates In Vitro in Undifferentiated Oral Epithelial Cells. J. Virol. 2018, 92, e00903-18. [Google Scholar] [CrossRef] [PubMed]
- He, L.; McVoy, M. Observations of Cultured Keratinocytes Infected with Cytomegalovirus Strain AD169 Variant BADrUL131-Y4; Virginia Commonwealth University: Richmond, VA, USA, 2018. [Google Scholar]



| Locus | Lineage | Mutation a | Consequence | Passage Range in Which Mutations Arose b |
|---|---|---|---|---|
| RL13 | ϕ-KG | delc | frameshift after aa 164 | 6–10 |
| UL131A | ϕ-KG | insd | frameshift after aa 27 | 6–10 |
| UL100 | Ig-KG | G44T | S15I | 13–22 |
| G858T | Q286H | 13–22 | ||
| AG901C G902T | S301L | 13–22 | ||
| G1086C | E362D | 13–22 | ||
| UL102 | Ig-KG | C67G | L23V | 13–22 |
| C1033G | L345V | 13–22 | ||
| C1812T | silent | 13–22 | ||
| UL122 | Ig-KG | C1127A | S376Y | 6–10 |
| ϕ-KG | C1152G | F384L | 6–10 |
| Virus | Stock | Type | Culture Size | Titer (pfu/mL) | Volume (mL) | Total Yield (pfu) |
|---|---|---|---|---|---|---|
| ϕ-KG-B5 | B5 | medium a | 2× T75 | 6.3 × 105 | 30 | 1.89 ×107 |
| Ig-KG-H2 | H2a | lysate b | 1× T75 | 6.9 × 104 | 2 | 1.38 × 105 |
| H2b | lysate b,c | 2× T75 | 4.9 × 104 | 6 | 2.94 × 105 | |
| H2c | medium c | 2× T75 | 5.0 × 102 | 30 | 1.5 × 103 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ourahmane, A.; Cui, X.; He, L.; Catron, M.; Dittmer, D.P.; Al Qaffasaa, A.; Schleiss, M.R.; Hertel, L.; McVoy, M.A. Inclusion of Antibodies to Cell Culture Media Preserves the Integrity of Genes Encoding RL13 and the Pentameric Complex Components During Fibroblast Passage of Human Cytomegalovirus. Viruses 2019, 11, 221. https://doi.org/10.3390/v11030221
Ourahmane A, Cui X, He L, Catron M, Dittmer DP, Al Qaffasaa A, Schleiss MR, Hertel L, McVoy MA. Inclusion of Antibodies to Cell Culture Media Preserves the Integrity of Genes Encoding RL13 and the Pentameric Complex Components During Fibroblast Passage of Human Cytomegalovirus. Viruses. 2019; 11(3):221. https://doi.org/10.3390/v11030221
Chicago/Turabian StyleOurahmane, Amine, Xiaohong Cui, Li He, Meaghan Catron, Dirk P. Dittmer, Ahmed Al Qaffasaa, Mark R. Schleiss, Laura Hertel, and Michael A. McVoy. 2019. "Inclusion of Antibodies to Cell Culture Media Preserves the Integrity of Genes Encoding RL13 and the Pentameric Complex Components During Fibroblast Passage of Human Cytomegalovirus" Viruses 11, no. 3: 221. https://doi.org/10.3390/v11030221
APA StyleOurahmane, A., Cui, X., He, L., Catron, M., Dittmer, D. P., Al Qaffasaa, A., Schleiss, M. R., Hertel, L., & McVoy, M. A. (2019). Inclusion of Antibodies to Cell Culture Media Preserves the Integrity of Genes Encoding RL13 and the Pentameric Complex Components During Fibroblast Passage of Human Cytomegalovirus. Viruses, 11(3), 221. https://doi.org/10.3390/v11030221





