Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Viruses
2.3. Human Airway Epithelial Cell (hAEC) Culture
2.4. Viral Replication in Well-Differentiated hAEC Cultures
2.5. Virus Titration by Tissue Culture Infectious Dosis 50 (TCID50)
2.6. Hemaglutination Assay
2.7. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (PCR)
2.8. Antibody Specificity of Influenza D Virus (IDV) Nucleoprotein (NP) Antibody
2.9. Immunofluorescence of Well-Differentiated hAEC Cultures
2.10. Deep Sequencing Analysis of IDV Passaged upon Well-Differentiated hAEC Cultures
2.11. Data Presentation
3. Results
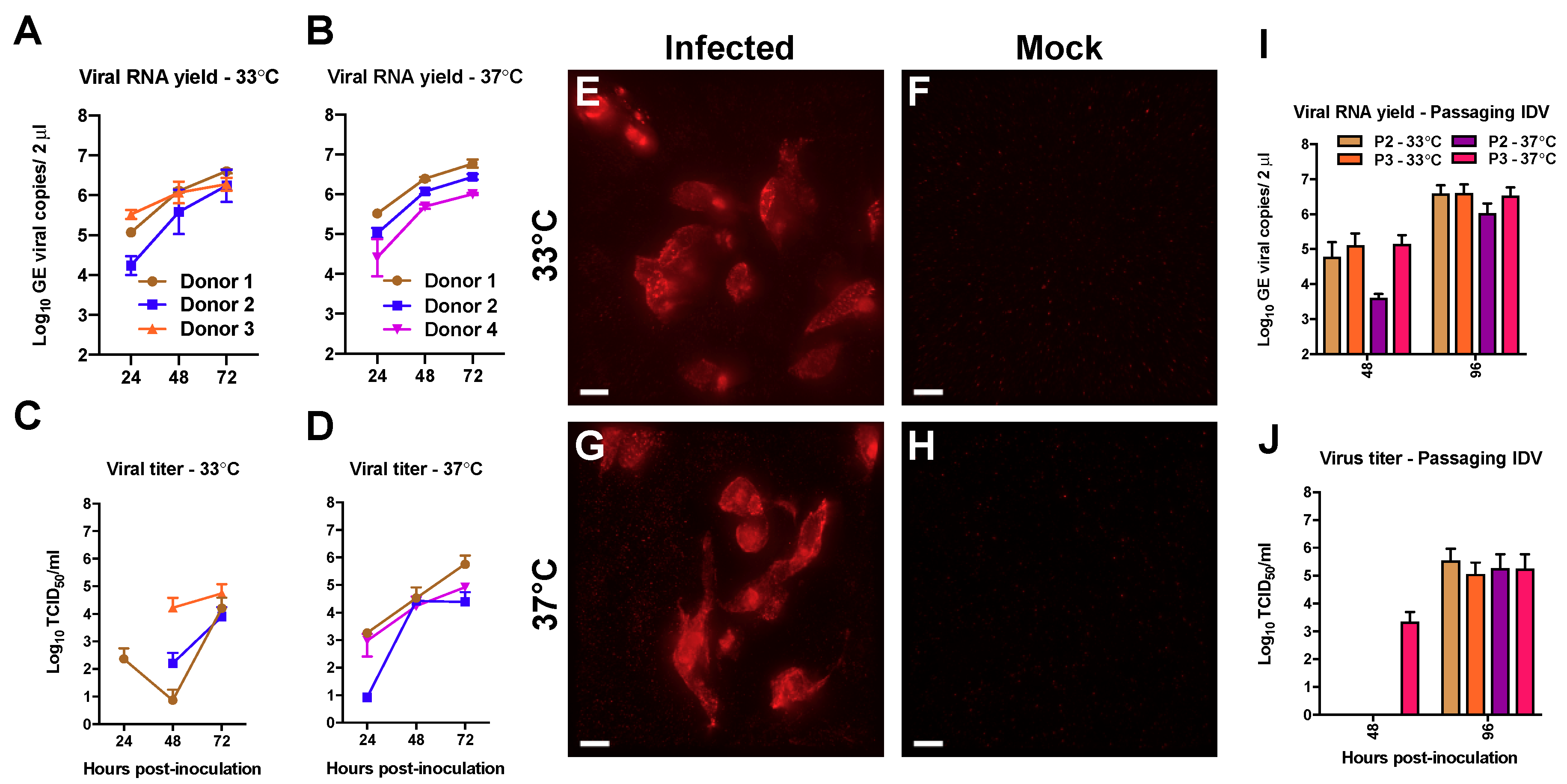
3.1. Efficient Replication of IDV in Well-Differentiated hAEC Cultures
3.2. Deep Sequencing Analysis of IDV Passaged upon Well-Differentiated hAEC Cultures
3.3. Comparison of Influenza C Virus (ICV) and IDV Infection in Well-Differentiated hAEC Cultures
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a Novel Swine Influenza Virus from Oklahoma in 2011 Which Is Distantly Related to Human Influenza C Viruses. PLoS Pathog. 2013, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5, e00031-14. [Google Scholar] [CrossRef]
- Foni, E.; Chiapponi, C.; Baioni, L.; Zanni, I.; Merenda, M.; Rosignoli, C.; Kyriakis, C.S.; Luini, M.V.; Mandola, M.L.; Nigrelli, A.D.; et al. Influenza D in Italy: towards a better understanding of an emerging viral infection in swine. Sci. Rep. 2017, 7, 11660. [Google Scholar] [CrossRef] [Green Version]
- Zhai, S.; Zhang, H.; Chen, S.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.; Wen, X.; Wei, W.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Horimoto, T.; Hiono, T.; Mekata, H.; Odagiri, T.; Lei, Z.; Kobayashi, T.; Norimine, J.; Inoshima, Y.; Hikono, H.; Murakami, K.; et al. Nationwide distribution of bovine influenza D virus infection in Japan. PLoS ONE 2016, 11, 1–7. [Google Scholar] [CrossRef]
- Luo, J.; Ferguson, L.; Smith, D.R.; Woolums, A.R.; Epperson, W.B.; Wan, X.F. Serological evidence for high prevalence of Influenza D Viruses in Cattle, Nebraska, United States, 2003–2004. Virology 2017, 501, 88–91. [Google Scholar] [CrossRef] [Green Version]
- Ducatez, M.F.; Pelletier, C.; Meyer, G. Influenza d virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef]
- Flynn, O.; Gallagher, C.; Mooney, J.; Irvine, C.; Ducatez, M.; Hause, B.; Mcgrath, G.; Ryan, E. Influenza D Virus in Cattle, Ireland. Emerg. Infect. Dis. 2018, 24, 2016–2018. [Google Scholar] [CrossRef]
- Salem, E.; Cook, E.A.J.; Lbacha, H.A.; Oliva, J.; Awoume, F.; Aplogan, G.L.; Hymann, E.C.; Muloi, D.; Deem, S.L.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017, 23, 2015–2018. [Google Scholar] [CrossRef] [PubMed]
- Collin, E.A.; Sheng, Z.; Lang, Y.; Ma, W.; Hause, B.M.; Li, F. Cocirculation of Two Distinct Genetic and Antigenic Lineages of Proposed Influenza D Virus in Cattle. J. Virol. 2015, 89, 1036–1042. [Google Scholar] [CrossRef]
- Ferguson, L.; Olivier, A.K.; Genova, S.; Epperson, W.B.; Smith, D.R.; Schneider, L.; Barton, K.; McCuan, K.; Webby, R.J.; Wan, X.-F. Pathogenesis of Influenza D virus in Cattle. J. Virol. 2016, 90, 5636–5642. [Google Scholar] [CrossRef]
- Ferguson, L.; Luo, K.; Olivier, A.K.; Cunningham, F.L.; Blackmon, S.; Hanson-dorr, K.; Sun, H.; Baroch, J.; Lutman, M.W.; Quade, B.; et al. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg. Infect. Dis. 2018, 24, 1020–1028. [Google Scholar] [CrossRef]
- Nedland, H.; Wollman, J.; Sreenivasan, C.; Quast, M.; Singrey, A.; Fawcett, L.; Christopher-Hennings, J.; Nelson, E.; Kaushik, R.S.; Wang, D.; et al. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 2017, 65, e148–e154. [Google Scholar] [CrossRef] [Green Version]
- Quast, M.; Sreenivasan, C.; Sexton, G.; Nedland, H.; Singrey, A.; Fawcett, L.; Miller, G.; Lauer, D.; Voss, S.; Pollock, S.; et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015, 180, 281–285. [Google Scholar] [CrossRef] [Green Version]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, G.C.; Lednicky, J.A. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef]
- Song, H.; Qi, J.; Khedri, Z.; Diaz, S.; Yu, H.; Chen, X.; Varki, A.; Shi, Y.; Gao, G.F. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS Pathog. 2016, 12, e1005411. [Google Scholar]
- Rogers, G.N.; Herrler, G.; Paulson, J.C.; Klenk, H.D. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 1986, 261, 5947–5951. [Google Scholar] [PubMed]
- Borkenhagen, L.K.; Mallinson, K.A.; Tsao, R.W.; Ha, S.J.; Lim, W.H.; Toh, T.H.; Anderson, B.D.; Fieldhouse, J.K.; Philo, S.E.; Chong, K.-S.; et al. Surveillance for respiratory and diarrheal pathogens at the human-pig interface in Sarawak, Malaysia. PLoS ONE 2018, 13, 1–14. [Google Scholar] [CrossRef]
- Smith, D.B.; Gaunt, E.R.; Digard, P.; Templeton, K.; Simmonds, P. Detection of influenza C virus but not influenza D virus in Scottish respiratory samples. J. Clin. Virol. 2016, 74, 50–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, E.S.; Choi, J.Y.; Zemke, J.; Yondon, M.; Gray, G.C. Molecular surveillance of respiratory viruses with bioaerosol sampling in an airport. Trop. Dis. Travel Med. Vaccines 2018, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Sims, A.C.; Baric, R.S.; Yount, B.; Burkett, S.E.; Collins, P.L.; Pickles, R.J. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 2005, 79, 15511–15524. [Google Scholar] [CrossRef]
- Kindler, E.; Jónsdóttir, H.R.; Muth, D.; Hamming, O.J.; Hartmann, R.; Rodriguez, R.; Geffers, R.; Fouchier, R.A.M.; Drosten, C.; Müller, M.A.; et al. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio 2013, 4, e00611-12. [Google Scholar] [CrossRef]
- Menachery, V.D.; Yount, B.L.; Sims, A.C.; Debbink, K.; Agnihothram, S.S.; Gralinski, L.E.; Graham, R.L.; Scobey, T.; Plante, J.A.; Royal, S.R.; et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA 2016, 113, 3048–3053. [Google Scholar] [CrossRef]
- Huang, D.T.N.; Lu, C.Y.; Chi, Y.H.; Li, W.L.; Chang, L.Y.; Lai, M.J.; Chen, J.S.; Hsu, W.M.; Huang, L.M. Adaptation of influenza A (H7N9) virus in primary human airway epithelial cells. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.-Y.; Chu, H.; Poon, V.K.-M.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827. [Google Scholar] [CrossRef]
- Dijkman, R.; Koekkoek, S.M.; Molenkamp, R.; Schildgen, O.; van der Hoek, L. Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 2009, 83, 7739–7748. [Google Scholar] [CrossRef]
- Jonsdottir, H.R.; Dijkman, R. Characterization of human coronaviruses on well-differentiated human airway epithelial cell cultures. Methods Mol. Biol. 2015, 1282, 73–87. [Google Scholar]
- Kaerber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn. Schmiedebergs. Arch. Exp. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar] [CrossRef]
- WHO. Global Influenza Surveillance Network Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; WHO Press: Geneva, Switzerland, 2011; ISBN 978-92-4-154809-0. [Google Scholar]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilm, A.; Aw, P.P.K.; Bertrand, D.; Yeo, G.H.T.; Ong, S.H.; Wong, C.H.; Khor, C.C.; Petric, R.; Hibberd, M.L.; Nagarajan, N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Palese, P.; O’Neill, R.E. The NPI-1/NPI-3 Nucleoprotein, (karyopherin α) binding site on the influenza a virus NP is a nonconventional nuclear localization signal. J. Virol. 1997, 71, 1850–1856. [Google Scholar]
- Ozawa, M.; Fujii, K.; Muramoto, Y.; Yamada, S.; Yamayoshi, S.; Takada, A.; Goto, H.; Horimoto, T.; Kawaoka, Y. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 2007, 81, 30–41. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Katsushima, N.; Nagai, Y.; Shoji, M.; Sakamoto, M.; Kitaoka, S.; Mizuta, K.; Nishimura, H.; The, S.; Diseases, I.; et al. Clinical Features of Influenza C Virus Infection in Children. J. Infect. Dis. 2006, 193, 1229–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salez, N.; Mélade, J.; Pascalis, H.; Aherfi, S.; Dellagi, K.; Charrel, R.N.; Carrat, F.; de Lamballerie, X. Influenza C virus high seroprevalence rates observed in 3 different population groups. J. Infect. 2014, 69, 182–189. [Google Scholar] [CrossRef]
- Kuiken, T.; Holmes, E.C.; McCauley, J.; Rimmelzwaan, G.F.; Williams, C.S.; Grenfell, B.T. Host species barriers to influenza virus infections. Science 2006, 312, 394–397. [Google Scholar] [CrossRef]
- Herfst, S.; Schrauwen, E.J.A.; Linster, M.; Chutinimitkul, S.; De, E.; Munster, V.J.; Sorrell, E.M.; Bestebroer, T.M.; Burke, D.F.; Derek, J.; et al. Airborne Transmission of Influenza A/H5N1 Virus Between Ferrets. Science 2012, 336, 1534–1541. [Google Scholar] [CrossRef] [Green Version]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar] [CrossRef]
- Sreenivasan, C.; Thomas, M.; Sheng, Z.; Hause, B.M.; Collin, E.A.; Knudsen, D.E.B.; Pillatzki, A.; Nelson, E.; Wang, D.; Kaushik, R.S.; et al. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J. Virol. 2015, 89, 11990–12001. [Google Scholar] [CrossRef] [Green Version]
- Maines, T.R.; Jayaraman, A.; Belser, J.A.; Wadford, D.A.; Pappas, C.; Zeng, H.; Gustin, K.M.; Pearce, M.B.; Viswanathan, K.; Shriver, Z.H.; et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 2009, 325, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Munster, V.J.; De Wit, E.; Van Den Brand, J.M.A.; Herfst, S.; Schrauwen, E.J.A.; Bestebroer, T.M.; De Van Vijver, D.; Boucher, C.A.; Koopmans, M.; Rimmelzwaan, G.F.; et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 2009, 325, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Principi, N.; Scala, A.; Daleno, C.; Esposito, S. Influenza C virus–associated community-acquired pneumonia in children. Influenza Other Respi. Viruses 2013, 7, 999–1003. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Thomas, M.; Antony, L.; Wormstadt, T.; Hildreth, M.B.; Wang, D.; Hause, B.; Francis, D.H.; Li, F.; Kaushik, R.S. Development and characterization of swine primary respiratory epithelial cells and their susceptibility to infection by four influenza virus types. Virology 2019, 528, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Porter, E.; Lohman, M.; Lu, N.; Peddireddi, L.; Hanzlicek, G.; Marthaler, D.; Liu, X.; Bai, J. Influenza C virus in cattle with respiratory disease, United States, 2016–2018. Emerg. Infect. Dis. 2018, 24, 1926–1929. [Google Scholar] [CrossRef] [PubMed]
- Yuanji (Kuo Yuanchi), G.; Fengen, J.; Ping, W.; Min, W.; Jiming (Chu Chinming), Z. Isolation of Influenza C Virus from Pigs and Experimental Infection of Pigs with Influenza C Virus. J. Gen. Virol. 1983, 64, 177–182. [Google Scholar] [CrossRef]
- Dijkman, R.; Jebbink, M.F.; Koekkoek, S.M.; Deijs, M.; Jónsdóttir, H.R.; Molenkamp, R.; Ieven, M.; Goossens, H.; Thiel, V.; van der Hoek, L. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 2013, 87, 6081–6090. [Google Scholar] [CrossRef]
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]


| Name Primer | Oligonucleotide Sequence (from 5′ to 3′) |
|---|---|
| Segment 1 Fw | CGG GTT ATT AGC AGT AGC AAG AGG ATT TTT TCA ATG TGC TTC AAA C |
| Segment 1 Rv | GGG GGG GCA TAA GCA GAG GAT GTC AC |
| Segment 2 Fw | CGG GTT ATT AGC AGT AGC AAG AGG ATT TTT CTG TTA TTA AAC AAC GC |
| Segment 2 Rv | GGG GGG GCA TAA GCA GAG GAT TTT ATA ACA ATG G |
| Segment 3 Fw | CGG GTT ATT AGC AGT AGC AAG GAG ATT TTT AAC ATT ACA AG |
| Segment 3 Rv | GGG GGG GCA TAA GCA GGA GAT TTA AAA ATG T |
| Segment 4 Fw | CGG GTT ATT AGC AGT AGC AAG GAG ATT TTT TCT AA |
| Segment 4 Rv | GGG GGA GCA TAA GCA GGA GAT TTT CAA AGA TG |
| Segment 5 Fw | CGG GTT ATT AGC AGT AGC AAG GAG ATT TTT TGT TAA ACA AGA CAA ACC AA |
| Segment 5 Rv | GGG GGG GCA TAA GCA GGA GAT TAT TAA GCA ATA |
| Segment 6 Fw | CGG GTT ATT AGC AGT AGC AAG AGG ATT TTT TCG |
| Segment 6 Rv | GGG GGG GCA TAA GCA GAG GAT ATT TTT GAC GC |
| Segment 7 Fw | CGG GTT ATT AGC AGT AGC AAG GGG TTT TTT C |
| Segment 7 Rv | GGG GGA GCA TAA GCA GGG GTG TAC AAT TTC AAT |
| Variant Detection Fraction in Total Number of Reads | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Position | Nucleotide Change | Variant | Amino Acid | Donor 1 at 33 °C | Donor 1 at 37 °C | Donor 2 at 33 °C | Donor 2 at 37 °C | Donor 4 at 33 °C | Donor 4 at 37 °C | Input |
| PB2 | 232 | T > C | missense | Ile73Thr | 0.030 | ||||||
| PB2 | 867 | G > A | missense | Ala285Thr | 0.995 | 0.521 | |||||
| PB2 | 989 | G > A | synonymous | Ala325Ala | 0.031 | ||||||
| PB2 | 2106 | G > A | missense | Asp698Asn | 0.087 | ||||||
| PB2 | 2273 | C > T | synonymous | Ser753Ser | 0.675 | ||||||
| PB1 | 325 | G > A | synonymous | Lys100Lys | 0.027 | ||||||
| PB1 | 328 | G > A | missense | Met101Ile | 0.023 | ||||||
| PB1 | 842 | T > C | synonymous | Leu273Leu | 0.142 | ||||||
| PB1 | 1144 | A > G | synonymous | Lys373Lys | 0.066 | ||||||
| PB1 | 2002 | G > C | synonymous | Thr659Thr | 0.050 | ||||||
| PB1 | 2006 | T > A | missense | Ser661Thr | 0.054 | ||||||
| PB1 | 2033 | T > C | synonymous | Leu670Leu | 0.132 | ||||||
| PB1 | 2053 | G > A | synonymous | Arg676Arg | 0.083 | ||||||
| P3 | 385 | C > A | missense | Phe121Leu | 0.023 | ||||||
| P3 | 454 | C > A | synonymous | Pro144Pro | 0.021 | ||||||
| P3 | 538 | C > A | missense | Phe172Leu | 0.020 | ||||||
| P3 | 1615 | G > A | synonymous | Val531Val | 0.184 | 0.151 | |||||
| HE | 276 | G > A | synonymous | Ala84Ala | 0.029 | ||||||
| HE | 770 | A > G | missense | Asn249Ser | 0.265 | 0.995 | 0.683 | 0.917 | 0.997 | 0.107 | |
| HE | 841 | R > A | missense | Val273Ile | 0.793 | ||||||
| HE | 841 | R > G | synonymous | Val273Val | 0.998 | 0.999 | 0.203 | ||||
| HE | 947 | G > A | missense | Arg308Lys | 0.036 | ||||||
| HE | 1145 | A > G | missense | Asn374Ser | 0.074 | ||||||
| HE | 1585 | G > A | missense | Gly521Ser | 0.032 | ||||||
| HE | 1635 | A > T | synonymous | Ala537Ala | 0.067 | ||||||
| NP | 432 | C > A | missense | Phe135Leu | 0.024 | ||||||
| NP | 891 | G > T | missense | Met288Ile | 0.021 | ||||||
| NP | 938 | T > C | missense | Val304Ala | 0.021 | ||||||
| NP | 1335 | G > A | synonymous | Glu436Glu | 0.175 | ||||||
| NP | 1446 | C > A | missense | Phe473Leu | 0.022 | ||||||
| NP | 1596 | G > T | synonymous | Val523Val | 0.994 | 0.047 | |||||
| P42 | 108 | G > A | missense | Arg27Lys | 0.036 | ||||||
| P42 | 772 | G > T | missense | Leu248Phe | 0.176 | ||||||
| Influenza C Virus | Influenza D Virus | |||
|---|---|---|---|---|
| Total number of cells | Infected (n = 6) | Mock (n = 3) | Infected (n = 6) | Mock (n = 3) |
| Ciliated cells | 1078 (42.7%) | 458 (49.1%) | 1099 (48.3%) | 584 (48.1%) |
| Non-ciliated cells | 1448 (57.3%) | 474 (50.9%) | 1174 (51.7%) | 629 (51.9%) |
| Total | 2526 (100%) | 932 (100%) | 2273 (100%) | 1213 (100%) |
| Total number of infected cells | ||||
| Ciliated cells | 82 (97.6%) | 0 (0%) | 94 (96.8%) | 0 (0%) |
| Non-ciliated cells | 2 (2.4%) | 0 (0%) | 3 (3.2%) | 0 (0%) |
| Total | 84 (100%) | 0 (0%) | 97 (100%) | 0 (0%) |
| Percentage infected cells | 3.3 | 0.0 | 4.3 | 0.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holwerda, M.; Kelly, J.; Laloli, L.; Stürmer, I.; Portmann, J.; Stalder, H.; Dijkman, R. Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells. Viruses 2019, 11, 377. https://doi.org/10.3390/v11040377
Holwerda M, Kelly J, Laloli L, Stürmer I, Portmann J, Stalder H, Dijkman R. Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells. Viruses. 2019; 11(4):377. https://doi.org/10.3390/v11040377
Chicago/Turabian StyleHolwerda, Melle, Jenna Kelly, Laura Laloli, Isabel Stürmer, Jasmine Portmann, Hanspeter Stalder, and Ronald Dijkman. 2019. "Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells" Viruses 11, no. 4: 377. https://doi.org/10.3390/v11040377
APA StyleHolwerda, M., Kelly, J., Laloli, L., Stürmer, I., Portmann, J., Stalder, H., & Dijkman, R. (2019). Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells. Viruses, 11(4), 377. https://doi.org/10.3390/v11040377





