Transcriptome Analysis Shows That IFN-I Treatment and Concurrent SAV3 Infection Enriches MHC-I Antigen Processing and Presentation Pathways in Atlantic Salmon-Derived Macrophage/Dendritic Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Virus Infection, and IFN-I Treatment
2.2. Total RNA Isolation
2.3. Library Construction, Sequencing, and Data Analysis for RNA-Seq
2.4. De Novo Assembly, Functional Annotation, and Gene Ontology Classification
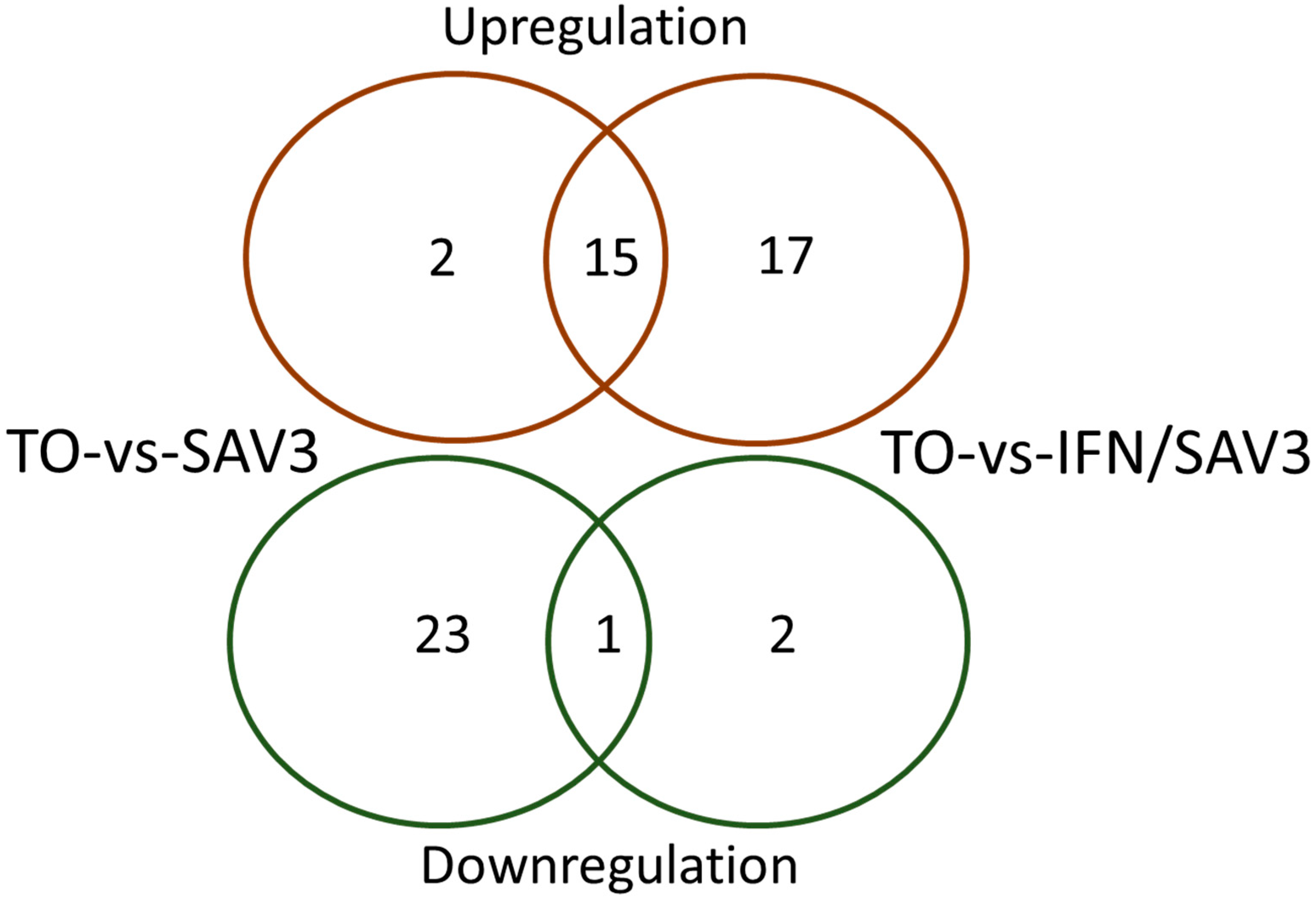
2.5. Identification of Differentially Expressed Genes
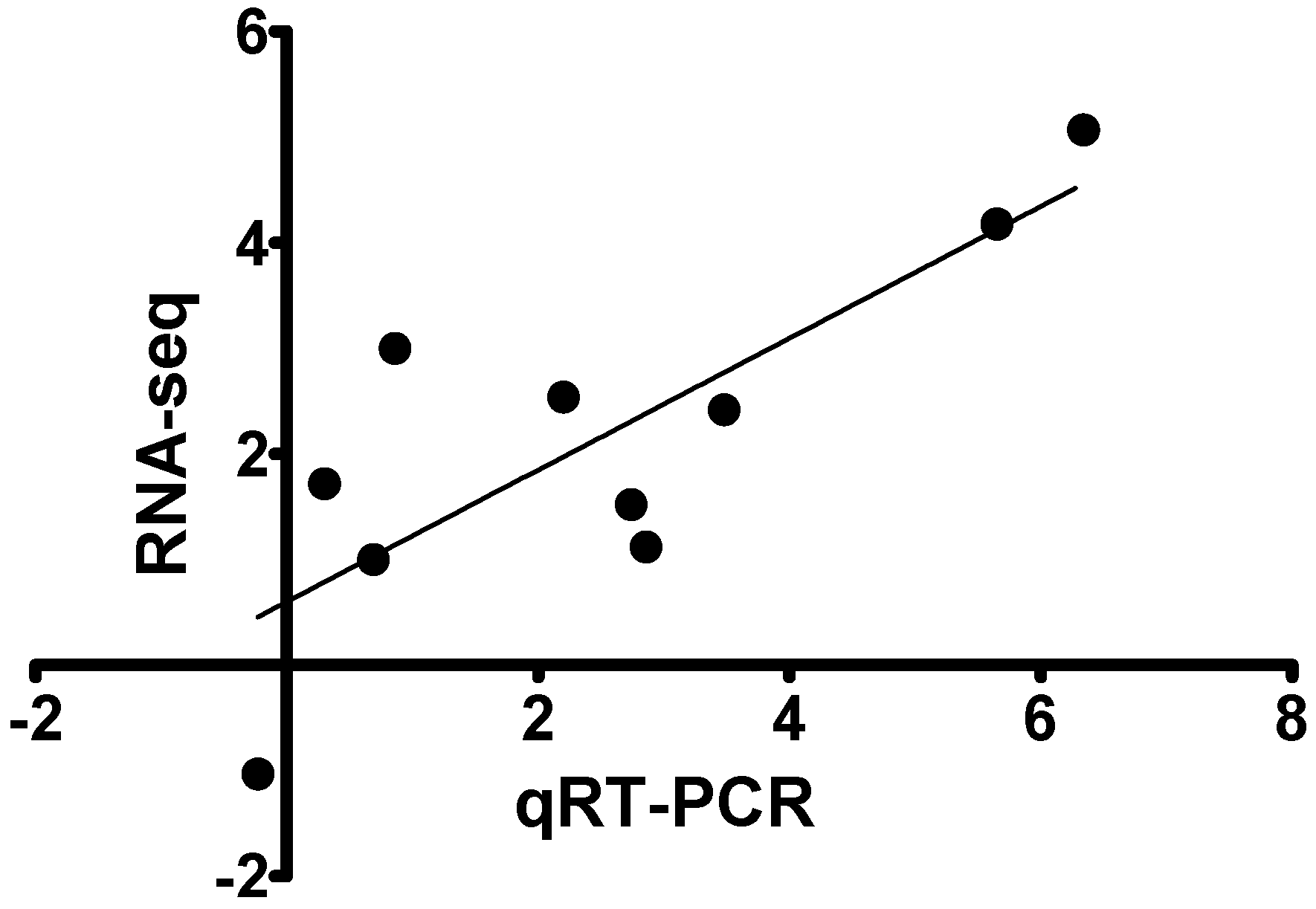
2.6. Validation of RNA-Seq Data
3. Results
3.1. KEGG Pathway Analysis
3.2. Major Histocompatibility Complex I Pathway
3.2.1. Proteasome Genes
3.2.2. Endoplasmic Reticulum Genes
3.3. Major Histocompatibility Complex II Pathway
3.3.1. Endosomal Compartment Genes
3.3.2. Endolysosomal Compartment Genes
3.4. Other Genes
3.5. Quantitative Real Time PCR
4. Discussion
4.1. KEGG Pathway Analysis
4.2. MHC-I Pathway Analysis
4.3. MHC-II Pathway Analysis
5. Conclusions
6. Data Access
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Vilcek, J. Fifty years of interferon research: Aiming at a moving target. Immunity 2006, 25, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Belardelli, F.; Gresser, I. The neglected role of type I interferon in the T-cell response: Implications for its clinical use. Immunol. Today 1996, 17, 369–372. [Google Scholar] [CrossRef]
- Belardelli, F.; Ferrantini, M. Cytokines as a link between innate and adaptive antitumor immunity. Trends Immunol. 2002, 23, 201–208. [Google Scholar] [CrossRef]
- Proietti, E.; Bracci, L.; Puzelli, S.; Di Pucchio, T.; Sestili, P.; De Vincenzi, E.; Venditti, M.; Capone, I.; Seif, I.; De Maeyer, E.; et al. Type I IFN as a natural adjuvant for a protective immune response: Lessons from the influenza vaccine model. J. Immunol. 2002, 169, 375–383. [Google Scholar] [CrossRef]
- Bracci, L.; Schumacher, R.; Provenzano, M.; Adamina, M.; Rosenthal, R.; Groeper, C.; Zajac, P.; Lezzi, G.; Proietti, E.; Belardelli, F.; Spagnoli, G.C. Efficient stimulation of T cell responses by human IFN-alpha-induced dendritic cells does not require toll-like receptor triggering. J. Immunother. 2008, 31, 466–474. [Google Scholar] [CrossRef]
- Tough, D.F. Modulation of T-cell function by type I interferon. Immunol. Cell Biol. 2012, 90, 492–497. [Google Scholar] [CrossRef]
- Hervas-Stubbs, S.; Perez-Gracia, J.L.; Rouzaut, A.; Sanmamed, M.F.; Le Bon, A.; Melero, I. Direct Effects of Type I Interferons on Cells of the Immune System. Clin. Cancer Res. 2011, 17, 2619–2627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Bon, A.; Etchart, N.; Rossmann, C.; Ashton, M.; Hou, S.; Gewert, D.; Borrow, P.; Tough, D.F. Cross-priming of CD8(+) T cells stimulated by virus-induced type I interferon. Nat. Immunol. 2003, 4, 1009–1015. [Google Scholar] [CrossRef]
- Le Bon, A.; Thompson, C.; Kamphuis, E.; Durand, V.; Rossmann, C.; Kalinke, U.; Tough, D.F. Cutting edge: Enhancement of antibody responses through direct stimulation of B and T cells by type IIFN. J. Immunol. 2006, 176, 2074–2078. [Google Scholar] [CrossRef] [PubMed]
- Gessani, S.; Conti, L.; Del, C.M.; Belardelli, F. Type I interferons as regulators of human antigen presenting cell functions. Toxins (Basel) 2014, 6, 1696–1723. [Google Scholar] [CrossRef]
- Lapenta, C.; Santini, S.M.; Spada, M.; Donati, S.; Urbani, F.; Accapezzato, D.; Franceschini, D.; Andreotti, M.; Barnaba, V.; Belardelli, F. IFN-alpha-conditioned dendritic cells are highly efficient in inducing cross-priming CD8(+) T cells against exogenous viral antigens. Eur. J. Immunol. 2006, 36, 2046–2060. [Google Scholar] [CrossRef]
- Lattanzi, L.; Rozera, C.; Marescotti, D.; D’Agostino, G.; Santodonato, L.; Cellini, S.; Belardelli, F.; Gavioli, R.; Ferrantini, M. IFN-alpha boosts epitope cross-presentation by dendritic cells via modulation of proteasome activity. Immunobiology 2011, 216, 537–547. [Google Scholar] [CrossRef]
- Mohty, M.; Vialle-Castellano, A.; Nunes, J.A.; Isnardon, D.; Olive, D.; Gaugler, W. IFN-alpha skews monocyte differentiation into toll-like receptor 7-expressing dendritic cells with potent functional activities. J. Immunol. 2003, 171, 3385–3393. [Google Scholar] [CrossRef]
- Parlato, S.; Romagnoli, G.; Spadaro, F.; Canini, I.; Sirabella, P.; Borghi, P.; Ramoni, C.; Filesi, I.; Biocca, S.; Gabriele, L.; et al. LOX-1 as a natural IFN-alpha-mediated signal for apoptotic cell uptake and antigen presentation in dendritic cells. Blood 2010, 115, 1554–1563. [Google Scholar] [CrossRef]
- Santini, S.M.; Lapenta, C.; Logozzi, M.; Parlato, S.; Spada, M.; Di Pucchio, T.; Belardelli, F. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J. Exp. Med. 2000, 191, 1777–1788. [Google Scholar] [CrossRef]
- Tosello, V.; Zamarchi, R.; Merlo, A.; Gorza, M.; Piovan, E.; Mandruzzato, S.; Bronte, V.; Wang, X.H.; Ferrone, S.; Amadori, A.; et al. Differential expression of constitutive and inducible proteasome subunits in human monocyte-derived DC differentiated in the presence of IFN-alpha or IL-4. Eur. J. Immunol. 2009, 39, 56–66. [Google Scholar] [CrossRef]
- Spadaro, F.; Lapenta, C.; Donati, S.; Abalsamo, L.; Barnaba, V.; Belardelli, F.; Santini, S.M.; Ferrantini, M. IFN-alpha enhances cross-presentation in human dendritic cells by modulating antigen survival, endocytic routing, and processing. Blood 2012, 119, 1407–1417. [Google Scholar] [CrossRef]
- Ito, T.; Amakawa, R.; Inaba, M.; Ikehara, S.; Inaba, K.; Fukuhara, S. Differential regulation of human blood dendritic cell subsets by IFNs. J. Immunol. 2001, 166, 2961–2969. [Google Scholar] [CrossRef]
- Radvanyi, L.G.; Banerjee, A.; Weir, M.; Messner, H. Low levels of interferon-alpha induce CD86 (B7.2) expression and accelerates dendritic cell maturation from human peripheral blood mononuclear cells. Scand. J. Immunol. 1999, 50, 499–509. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Ingerslev, H.C.; Stavang, V.; Egenberg, M.; Wergeland, H.I. A highly phagocytic cell line TO from Atlantic salmon is CD83 positive and M-CSFR negative, indicating a dendritic-like cell type. Fish Shellfish Immunol. 2008, 25, 809–819. [Google Scholar] [CrossRef]
- Wergeland, H.I.; Jakobsen, R.A. A salmonid cell line (TO) for production of infectious salmon anaemia virus (ISAV). Dis. Aquat. Organ. 2001, 44, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Guo, T.C.; Mutoloki, S.; Haugland, O.; Marjara, I.S.; Evensen, O. Alpha Interferon and Not Gamma Interferon Inhibits Salmonid Alphavirus Subtype 3 Replication In Vitro. J. Virol. 2010, 84, 8903–8912. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Evensen, O.; Munang’andu, H.M. De novo assembly and transcriptome analysis of Atlantic salmon macrophage/dendritic-like TO cells following type I IFN treatment and Salmonid alphavirus subtype-3 infection. BMC Genom. 2015, 16, 96. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.P.; Zhang, Y.; Song, J.; Zhao, L.J.; Wang, Z.Z. De novo transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients. Genomics 2011, 98, 272–279. [Google Scholar] [Green Version]
- Munang’andu, H.M.; Evensen, O. A Review of Intra- and Extracellular Antigen Delivery Systems for Virus Vaccines of Finfish. J. Immunol. Res. 2015, 2015. [Google Scholar] [CrossRef]
- Rock, K.L.; Goldberg, A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999, 17, 739–779. [Google Scholar] [CrossRef]
- Tanaka, K.; Chiba, T. The proteasome: A protein-destroying machine. Genes Cells 1998, 3, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K. Molecular biology of the proteasome. Biochem. Biophys. Res. Commun. 1998, 247, 537–541. [Google Scholar] [CrossRef]
- Tanaka, K.; Kasahara, M. The MHC class I ligand-generating system: Roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol. Rev. 1998, 163, 161–176. [Google Scholar] [CrossRef]
- Yamano, T.; Mizukami, S.; Murata, S.; Chiba, T.; Tanaka, K.; Udono, H. Hsp90-mediated assembly of the 26 S proteasome is involved in major histocompatibility complex class I antigen processing. J. Biol. Chem. 2008, 283, 28060–28065. [Google Scholar] [CrossRef]
- Kasthuri, S.R.; Umasuthan, N.; Whang, I.; Kim, E.; Park, H.C.; Lee, J. Genomic structural characterization and transcriptional expression analysis of proteasome activator PA28 alpha and PA28 beta subunits from Oplegnathus fasciatus. Fish Shellfish Immunol. 2013, 35, 1224–1234. [Google Scholar] [CrossRef]
- Kim, D.H.; Lee, S.M.; Hong, B.Y.; Kim, Y.T.; Choi, T.J. Cloning and sequence analysis of cDNA for the proteasome activator PA28-beta subunit of flounder (Paralichthys olivaceus). Mol. Immunol. 2003, 40, 611–616. [Google Scholar] [CrossRef]
- Liu, G.Z.; Zheng, W.B.; Chen, X.H. Molecular cloning of proteasome activator PA28-beta subunit of large yellow croaker (Pseudosciana crocea) and its coordinated up-regulation with MHC class I alpha-chain and beta(2)-microglobulin in poly I: C-treated fish. Mol. Immunol. 2007, 44, 1190–1197. [Google Scholar] [CrossRef]
- Pan, F.; Zarate, J.M.; Tremblay, G.C.; Bradley, T.M. Cloning and characterization of salmon hsp90 cDNA: Upregulation by thermal and hyperosmotic stress. J. Exp. Zool. 2000, 287, 199–212. [Google Scholar] [CrossRef]
- Yamano, T.; Murata, S.; Shimbara, N.; Tanaka, N.; Chiba, T.; Tanaka, K.; Yui, K.; Udono, H. Two distinct pathways mediated by PA28 and hsp90 in major histocompatibility complex class I antigen processing. J. Exp. Med. 2002, 196, 185–196. [Google Scholar] [CrossRef]
- Binder, R.J.; Blachere, N.E.; Srivastava, P.K. Heat shock protein-chaperoned peptides but not free peptides introduced into the cytosol are presented efficiently by major histocompatibility complex I molecules. J. Biol. Chem. 2001, 276, 17163–17171. [Google Scholar] [CrossRef]
- Peaper, D.R.; Cresswell, P. Regulation of MHC Class I Assembly and Peptide Binding. Annu. Rev. Cell Dev. Biol. 2008, 24, 343–368. [Google Scholar] [CrossRef]
- Mantegazza, A.R.; Magalhaes, J.G.; Amigorena, S.; Marks, M.S. Presentation of Phagocytosed Antigens by MHC Class I and II. Traffic 2013, 14, 135–152. [Google Scholar] [CrossRef]
- Landis, E.D.; Palti, Y.; Dekoning, J.; Drew, R.; Phillips, R.B.; Hansen, J.D. Identification and regulatory analysis of rainbow trout tapasin and tapasin-related genes. Immunogenetics 2006, 58, 56–69. [Google Scholar] [CrossRef]
- Hansen, J.D.; La Patra, S. Induction of the rainbow trout MHC class I pathway during acute IHNV infection. Immunogenetics 2002, 54, 654–661. [Google Scholar] [CrossRef]
- Jorgensen, S.M.; Lyng-Syvertsen, B.; Lukacs, M.; Grimholt, U.; Gjoen, T. Expression of MHC class I pathway genes in response to infectious salmon anaemia virus in Atlantic salmon (Salmo salar L.) cells. Fish Shellfish Immunol. 2006, 21, 548–560. [Google Scholar] [CrossRef]
- Jorgensen, S.M.; Hetland, D.L.; Press, C.M.; Grimholt, U.; Gjoen, T. Effect of early infectious salmon anaemia virus (ISAV) infection on expression of MHC pathway genes and type I and II interferon in Atlantic salmon (Salmo salar L.) tissues. Fish Shellfish Immunol. 2007, 23, 576–588. [Google Scholar] [CrossRef]
- Deussing, J.; Roth, W.; Saftig, P.; Peters, C.; Ploegh, H.L.; Villadangos, J.A. Cathepsins B and D are dispensable for major histocompatibility complex class II-mediated antigen presentation. Proc. Natl. Acad. Sci. USA 1998, 95, 4516–4521. [Google Scholar] [CrossRef] [Green Version]
- Roche, P.A.; Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015, 15, 203–216. [Google Scholar] [CrossRef]
- Cresswell, P.; Androlewicz, M.J.; Ortmann, B. Assembly and Transport of Class-I Mhc-Peptide Complexes. Vaccines Against Virally Induced Cancers 1994, 187, 150–169. [Google Scholar]
- Cresswell, P. Assembly, Transport, and Function of Mhc Class-Ii Molecules. Annu. Rev. Immunol. 1994, 12, 259–293. [Google Scholar] [CrossRef]
- Avva, R.R.; Cresswell, P. Alpha-Beta-Clip—An Intermediate in Mhc Class-Ii Processing. J. Cell. Biochem. 1994, 330. [Google Scholar]
- Avva, R.R.; Cresswell, P. In-Vivo and In-Vitro Formation and Dissociation of Hla-Dr Complexes with Invariant Chain-Derived Peptides. Immunity 1994, 1, 763–774. [Google Scholar] [CrossRef]
- Katz, J.F.; Stebbins, C.; Appella, E.; Sant, A.J. Invariant chain and DM edit self-peptide presentation by major histocompatibility complex (MHC) class II molecules. J. Exp. Med. 1996, 184, 1747–1753. [Google Scholar] [CrossRef]
- Kropshofer, H.; Hammerling, G.J.; Vogt, A.B. How HLA-DM edits the MHC class II peptide repertoire: Survival of the fittest? Immunol. Today 1997, 18, 77–82. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Grimholt, U.; Leong, J.; Koop, B.F.; Hashimoto, K. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol. Biol. 2013, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.M.; Kiryu, I.; Kollner, B.; Yoshiura, Y.; Ototake, M. MHC class II invariant chain homologues in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2003, 15, 91–105. [Google Scholar] [CrossRef]
- Yoder, J.A.; Haire, R.N.; Litman, G.W. Cloning of two zebrafish cDNAs that share domains with the MHC class II-associated invariant chain. Immunogenetics 1999, 50, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Hetland, D.L.; Jorgensen, S.M.; Skjodt, K.; Dale, O.B.; Falk, K.; Xu, C.; Mikalsen, A.B.; Grimholt, U.; Gjoen, T.; Press, C.M. In situ localisation of major histocompatibility complex class I and class II and CD8 positive cells in infectious salmon anaemia virus (ISAV)-infected Atlantic salmon. Fish Shellfish Immunol. 2010, 28, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Jiang, L.X.; Han, J.B.; Su, H.; Yang, Q.; He, C.B. Major histocompatibility complex class IIA and IIB genes of the spotted halibut Verasper variegatus: Genomic structure, molecular polymorphism, and expression analysis. Fish Physiol. Biochem. 2011, 37, 767–780. [Google Scholar] [CrossRef]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrom, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Star, B.; Jentoft, S. Why does the immune system of Atlantic cod lack MHC II? Bioessays 2012, 34, 648–651. [Google Scholar] [CrossRef] [Green Version]


| Primer Name | Sequence | |
|---|---|---|
| HSP90 | F | CCACCATGGGCTACATGATG |
| R | CCTTCACCGCCTTGTCATTC | |
| TAP1 | F | ACGAGCCTGAAGCCTTTAC |
| R | TCACACACAAACTCACACAC | |
| TAP2 | F | GGGAAACAGAAGACACAGAAG |
| R | ATGCCCCAACCAAAAGGAG | |
| β2M | F | TCGTTGTACTTGTGCTCATTTACAGC |
| R | CAGGGTATTCTTATCTCCAAAGTTGC | |
| MHCI | F | CTGCATTGAGTGGCTGAAGA |
| R | GGTGATCTTGTCCGTCTTTC | |
| MHCII | F | TCTCCAGTCTGCCCTTCACC |
| R | GAACACAGCAGGACCCACAC | |
| CathepsinS | F | CGAAGGGAGGTCTGGGAGAGGAAT |
| R | GCCCAGGTCATAGGTGTGCATGTC | |
| CathepsinB | F | TGTGAGACTGGATACACACCTGGCTAC |
| R | GCTCCTTCCACAGGTCCGTTCTTC | |
| β-actin | F | CCAGTCCTGCTCACTGAGGC |
| R | GGTCTCAAACATGATCTGGGTCA |
| Parameters | TO-vs.-SAV3 | TO-vs.-SAV3/IFN |
|---|---|---|
| Antigen presentation pathway ID | K04612 | K04612 |
| DEG with pathway annotation of antigen presentation | 41 (0.44%) | 35 (2.15%) |
| Total DEG with pathway annotations | 9315 | 1629 |
| All genes with pathway annotation of antigen presentation | 145 | 145 |
| Q-value | 1.0000e+00 | 1.2654e−07 |
| p-value | 0.9999 | 3.2132e−09 |
| Number of upregulated unigenes | 17 | 32 |
| Number of downregulated unigenes | 24 | 3 |
| Percentage of upregulated unigenes | 41.46% | 91.43% |
| Percentage of downregulated unigenes | 58.44% | 8.57% |
| Gene Name | Abbrev. | KEGG | Up/Down | TO-vs.-SAV3 (Unigenes) | TO-vs.-SAV3/IFN (Unigenes) |
|---|---|---|---|---|---|
| Cathepsin L | CTSL | K01365 | Up | 3 | |
| Class II transactivator | CIITA | K08060 | Up | 3 | 3 |
| Transport associated protein 2 | TAP2 | K05654 | Up | 2 | 4 |
| Transport associated protein 1 | TAP1 | K05653 | Up | 2 | 2 |
| TAP binding protein (tapasin) | TAPBP | K08058 | Up | 2 | 3 |
| Heat shock protein 90 | HSP90 | K04090 | Up | 5 | 5 |
| Major histocompatibility complex I | MHC-I | K06751 | Up | 1 | 12 |
| Lysophosphatidic acid receptor 5 | LPAR5 | K08390 | Up | 1 | |
| Proteasome activator 28 | PA28 | K06697 | Up | 1 | |
| Beta-2-microglobulin | β2M | K08055 | Up | 1 | |
| Major histocompatibility complex, class I | MHC-I | K06751 | Down | 1 | |
| Cyclic AMP response element-binding protein 1 | CREB-1 | K05870 | Down | 5 | |
| Heat shock protein 70 | HSP70 | K09489 | Down | 7 | |
| PAX-interacting protein 1 | PAXIP1 | K14972 | Down | 1 | |
| Recyclin-1 | RCY1 | K15071 | Down | 1 | |
| Non-specific serine/threonine protein kinase | STK | K08282 | Down | 1 | |
| Calreticulin | CALR | K08057 | Down | 2 | 1 |
| Nuclear factor Y | NFY | K08066 | Down | 1 | |
| Filaggrin | FLG | K10384 | Down | 1 | |
| Regulatory factor X-associated protein | RFXAP | K08063 | Down | 1 | |
| Calnexin | CANX | K08054 | Down | 2 | |
| Gamma IFN inducible lysosome thiol | GILT | K08059 | Down | 1 | |
| Protein disulfide isomerase family A, member 3 | PDIA3 | K08056 | Down | 1 | |
| Total unigenes expressed | 41 | 35 |
| Gene Name | Abbrev. | KEGG | Unigene | GeneBank | Up/Down | TO-vs.-SAV3 | TO-vs.-SA-3/IFN | ||
|---|---|---|---|---|---|---|---|---|---|
| Log2 Ratio (PD/TO) | p-Value | Log2 Ratio (PD/TO) | p-Value | ||||||
| Cathepsin L | CTSL | K01365 | Unigene20896 | BT043962.1| | Up | 11.54 | 2.84E−51 | ||
| Class II transactivator | CIITA | K08060 | CL7384.1 | NP_001186995.1 | Up | 9.31 | 1.22E−17 | 9.87 | 9.83E−25 |
| Transport associated protein 2 | TAP2 | K05654 | CL7652.1 | Z83329.1 | Up | 4.18 | 1.14E−142 | 1.38 | 6.21E−08 |
| Transport associated protein 1 | TAP1 | K05653 | Unigene6385 | AF115538.1 | Up | 1.52 | 2.83E−124 | 2.34 | 0 |
| TAP binding protein (tapasin) | TAPBP | K08058 | CL1564.1 | NM_001124553.1 | Up | 1.85 | 3.36E−110 | 2.01 | 8.00E−305 |
| Heat shock protein 90 | HSP90 | K04090 | CL11346. 1 | NM_001173702.1 | Up | 1.72 | 1.62E−73 | 3.00 | 7.42E−182 |
| Lysophosphatidic acid receptor 5 | LPAR5 | K08390 | Unigene7594 | NP_001133983.1 | Up | 1.50 | 4.47E−57 | ||
| Proteasome activator 28 | PA28 | K06697 | CL10. 2 | ACM08763.1 | Up | 1.15 | 3.41E−27 | ||
| Beta-2-microglobulin | β2M | K08055 | Unigene75.1 | AF180485 | Up | 1.12 | 0 | ||
| Major histocompatibility complex, class I | MHC-I | K06751 | Unigene8180 | L63541.1 | Up | 1.10 | 3.14E−10 | 2.74 | 7.64E−155 |
| Major histocompatibility complex, class I | MHC-I | K06751 | Unigene23484 | BT072706.1 | Down | −1.11 | 3.25E−06 | ||
| Cyclic AMP response element-binding protein 1 | CREB-1 | K05870 | CL3991.2 | EMC89054.1 | Down | −2.52 | 7.25E−06 | ||
| Heat shock protein 70 | HSP70 | K09489 | CL1805. 2 | NP_999881.1 | Down | −1.03 | 6.00E−80 | ||
| PAX-interacting protein 1 | PAXIP1 | K14972 | CL5737. 1 | NM_001025462.1 | Down | −1.68 | 3.83E−07 | ||
| Recyclin-1 | RCY1 | K15071 | CL1550. 1 | NM_001165325.1 | Down | −1.65 | 4.03E−39 | ||
| Non-specific serine/threonine protein kinase | STK | K08282 | Unigene4701 | XM_004075980.1 | Down | −1.64 | 2.57E−14 | ||
| Calreticulin | CALR | K08057 | CL1908. 1 | XM_004068445.1 | Down | −1.91 | 5.93E−07 | −1.33 | 1.66E−05 |
| Nuclear factor Y | NFY | K08066 | CL10158. 1 | AC133690.1 | Down | −1.39 | 1.67E−15 | ||
| Filaggrin | FLG | K10384 | CL9396. 1 | XM_003437908.1 | Down | −1.38 | 8.33E−29 | ||
| Regulatory factor X | RFX | K08063 | Unigene16842 | XM_004319280.1 | Down | −1.17 | 5.47E−05 | ||
| Calnexin | CANX | K08054 | CL8069. 2 | AAQ18011.1 | Down | −1.16 | 4.79E−-54 | ||
| Cathepsin S | CTSS | K01365 | Unigene2181 | NP_001134963.1 | Down | −1.03 | 1.85E−61 | ||
| Interferon, gamma-inducible protein 30 | GILT | K08059 | Unigene12822 | ACI67174.1 | Down | −1.10 | 6.76E−06 | ||
| Protein disulfide isomerase family A, member 3 | PDIA3 | K08056 | Unigene19161 | NP_998070.1 | Down | −1.05 | 7.23E−-07 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Evensen, Ø.; Munang’andu, H.M. Transcriptome Analysis Shows That IFN-I Treatment and Concurrent SAV3 Infection Enriches MHC-I Antigen Processing and Presentation Pathways in Atlantic Salmon-Derived Macrophage/Dendritic Cells. Viruses 2019, 11, 464. https://doi.org/10.3390/v11050464
Xu C, Evensen Ø, Munang’andu HM. Transcriptome Analysis Shows That IFN-I Treatment and Concurrent SAV3 Infection Enriches MHC-I Antigen Processing and Presentation Pathways in Atlantic Salmon-Derived Macrophage/Dendritic Cells. Viruses. 2019; 11(5):464. https://doi.org/10.3390/v11050464
Chicago/Turabian StyleXu, Cheng, Øystein Evensen, and Hetron Mweemba Munang’andu. 2019. "Transcriptome Analysis Shows That IFN-I Treatment and Concurrent SAV3 Infection Enriches MHC-I Antigen Processing and Presentation Pathways in Atlantic Salmon-Derived Macrophage/Dendritic Cells" Viruses 11, no. 5: 464. https://doi.org/10.3390/v11050464
APA StyleXu, C., Evensen, Ø., & Munang’andu, H. M. (2019). Transcriptome Analysis Shows That IFN-I Treatment and Concurrent SAV3 Infection Enriches MHC-I Antigen Processing and Presentation Pathways in Atlantic Salmon-Derived Macrophage/Dendritic Cells. Viruses, 11(5), 464. https://doi.org/10.3390/v11050464





