A DNA-Modified Live Vaccine Prime–Boost Strategy Broadens the T-Cell Response and Enhances the Antibody Response against the Porcine Reproductive and Respiratory Syndrome Virus
Abstract
:1. Introduction
2. Material and Methods
2.1. Antibodies
2.2. Modified Live Virus
2.3. Constructions of Vectors Expressing PRRSV-AGs
2.4. 293T Transfection
2.5. Detection of VB by ELISA
2.6. Interaction of VB with Pig Low-Density Peripheral Blood Mononuclear Cells (PBMCs)
2.7. Cationic Poly-Lactide Co-Glycolide Acid (PLGA) Nanoparticles (NPs)
2.8. Immunization of Pigs
2.9. Overlapping Peptides
2.10. ELISPOTS on PBMCs from the DNA 3X and DNA + MLV Groups
2.11. Collection of Serum and Nasal Fluid and Evaluation of Their Content in Anti-N IgG by ELISA
2.12. Viral Detection by Specific qRT-PCR
2.13. Statistical and Correlation Analysis
3. Results
3.1. Production and Biochemical Characterization of Vaccibodies (VB) for Targeting PRRSV-Ags to Pig XCR1 and CD11c
3.2. Interaction of VB with Pig Cells
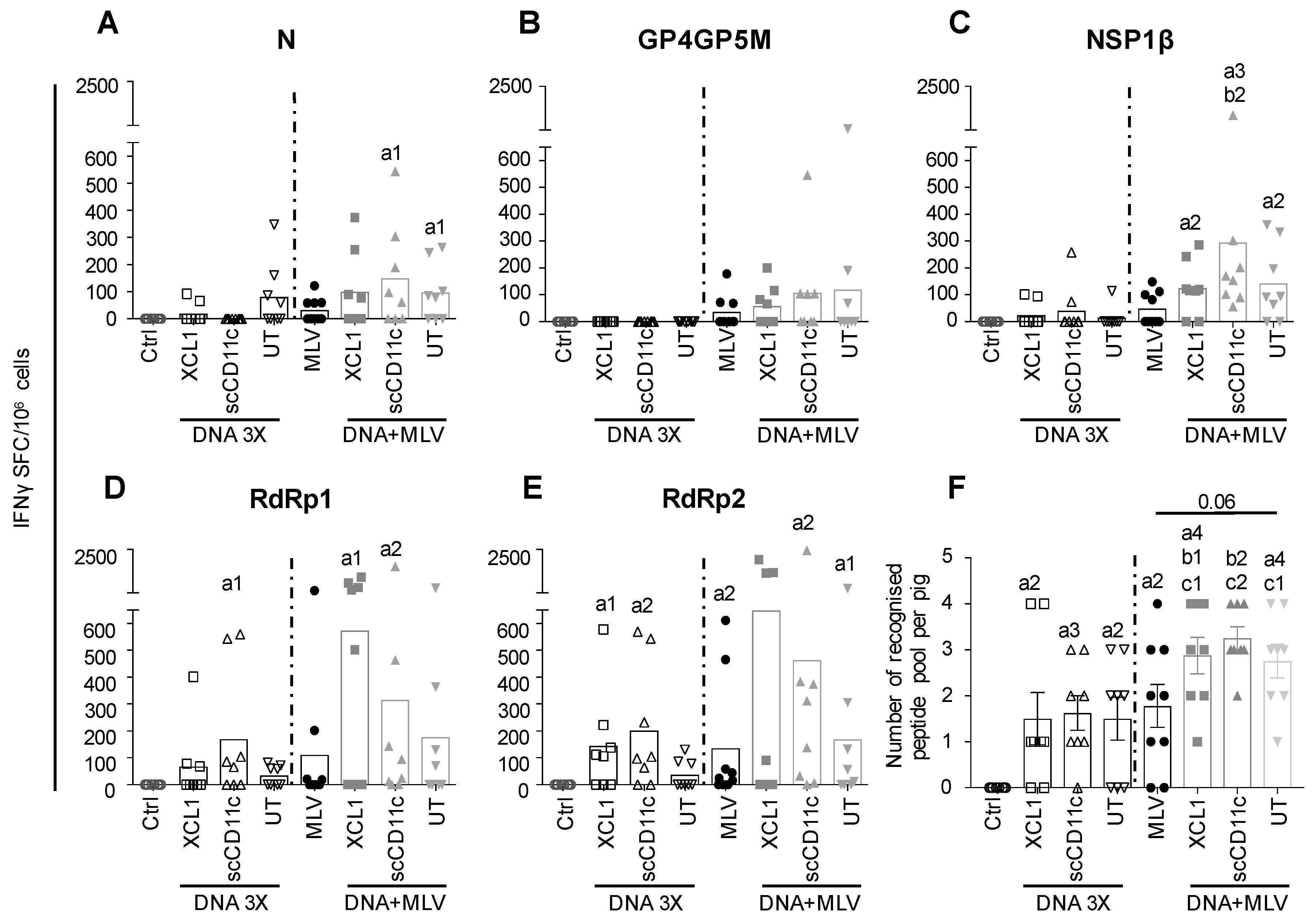
3.3. DNA Vectors Encoding PRRSV-AGs in Native Forms or Targeted to Pig APC Induce IFNγ T-Cell Responses in Pigs (DNA 3X Groups)
3.4. A DNA-MLV Prime–Boost (DNA + MLV Groups) Broadens the IFNγ T-Cell Response as Compared to MLV-Only, Induces Higher T-Cell Responses as Compared to DNA 3X, and Does not Show a Benefit of APC-Targeting on the IFNγ T-Cell Response
3.5. A DNA-MLV Prime–Boost Potentiates the Anti-N Igg Response as Compared to an MLV-Only Vaccine or to A DNA-Stand-Alone Regimen and Shows a Benefit of XCR1 and CD11c-Targeting
3.6. Anti-N IgG Response in Nasal Secretions
3.7. A DNA Prime Effect on MLV Replication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Lauck, M.; Bailey, A.L.; Shchetinin, A.M.; Vishnevskaya, T.V.; Bao, Y.; Ng, T.F.; LeBreton, M.; Schneider, B.S.; Gillis, A.; et al. Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 2016, 161, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Darwich, L.; Gimeno, M.; Sibila, M.; Diaz, I.; de la Torre, E.; Dotti, S.; Kuzemtseva, L.; Martin, M.; Pujols, J.; Mateu, E. Genetic and immunobiological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet. Microbiol. 2011, 150, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Kimman, T.G.; Cornelissen, L.A.; Moormann, R.J.; Rebel, J.M.; Stockhofe-Zurwieden, N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 2009, 27, 3704–3718. [Google Scholar] [CrossRef] [PubMed]
- Renukaradhya, G.J.; Meng, X.J.; Calvert, J.G.; Roof, M.; Lager, K.M. Live porcine reproductive and respiratory syndrome virus vaccines: Current status and future direction. Vaccine 2015, 33, 4069–4080. [Google Scholar] [CrossRef] [PubMed]
- Mateu, E.; Diaz, I. The challenge of PRRS immunology. Vet. J. 2008, 177, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Rahe, M.C.; Murtaugh, M.P. Mechanisms of Adaptive Immunity to Porcine Reproductive and Respiratory Syndrome Virus. Viruses 2017, 9, 148. [Google Scholar] [CrossRef]
- Lowe, J.E.; Husmann, R.; Firkins, L.D.; Zuckermann, F.A.; Goldberg, T.L. Correlation of cell-mediated immunity against porcine reproductive and respiratory syndrome virus with protection against reproductive failure in sows during outbreaks of porcine reproductive and respiratory syndrome in commercial herds. J. Am. Vet. Med. Assoc. 2005, 226, 1707–1711. [Google Scholar] [CrossRef] [Green Version]
- Salguero, F.J.; Frossard, J.P.; Rebel, J.M.; Stadejek, T.; Morgan, S.B.; Graham, S.P.; Steinbach, F. Host-pathogen interactions during porcine reproductive and respiratory syndrome virus 1 infection of piglets. Virus Res. 2015, 202, 135–143. [Google Scholar] [CrossRef]
- Park, C.; Seo, H.W.; Han, K.; Kang, I.; Chae, C. Evaluation of the efficacy of a new modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine (Fostera PRRS) against heterologous PRRSV challenge. Vet. Microbiol. 2014, 172, 432–442. [Google Scholar] [CrossRef]
- Meier, W.A.; Galeota, J.; Osorio, F.A.; Husmann, R.J.; Schnitzlein, W.M.; Zuckermann, F.A. Gradual development of the interferon-gamma response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology 2003, 309, 18–31. [Google Scholar] [CrossRef]
- Bautista, E.M.; Molitor, T.W. IFN gamma inhibits porcine reproductive and respiratory syndrome virus replication in macrophages. Arch. Virol. 1999, 144, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Welsh, R.M.; Selin, L.K. No one is naive: The significance of heterologous T-cell immunity. Nat. Rev. Immunol. 2002, 2, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, H.; Pedrera, M.; Frossard, J.P.; Biffar, L.; Hammer, S.E.; Kvisgaard, L.K.; Larsen, L.E.; Stewart, G.R.; Somavarapu, S.; Steinbach, F.; et al. The Non-structural Protein 5 and Matrix Protein Are Antigenic Targets of T Cell Immunity to Genotype 1 Porcine Reproductive and Respiratory Syndrome Viruses. Front. Immunol. 2016, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, H.; Eck, M.; Morgan, S.B.; Essler, S.E.; Frossard, J.P.; Ruggli, N.; Graham, S.P. Proteome-wide screening of the European porcine reproductive and respiratory syndrome virus reveals a broad range of T cell antigen reactivity. Vaccine 2014, 32, 6828–6837. [Google Scholar] [CrossRef] [PubMed]
- Burgara-Estrella, A.; Diaz, I.; Rodriguez-Gomez, I.M.; Essler, S.E.; Hernandez, J.; Mateu, E. Predicted peptides from non-structural proteins of porcine reproductive and respiratory syndrome virus are able to induce IFN-gamma and IL-10. Viruses 2013, 5, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Tregoning, J.S.; Kinnear, E. Using Plasmids as DNA Vaccines for Infectious Diseases. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Shortman, K.; Lahoud, M.H.; Caminschi, I. Improving vaccines by targeting antigens to dendritic cells. Exp. Mol. Med. 2009, 41, 61–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trumpfheller, C.; Longhi, M.P.; Caskey, M.; Idoyaga, J.; Bozzacco, L.; Keler, T.; Schlesinger, S.J.; Steinman, R.M. Dendritic cell-targeted protein vaccines: A novel approach to induce T-cell immunity. J. Intern. Med. 2012, 271, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Radford, K.J.; Caminschi, I. New generation of dendritic cell vaccines. Hum. Vaccin. Immunother. 2013, 9, 259–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gornati, L.; Zanoni, I.; Granucci, F. Dendritic Cells in the Cross Hair for the Generation of Tailored Vaccines. Front. Immunol. 2018, 9, 1484. [Google Scholar] [CrossRef] [PubMed]
- Nchinda, G.; Kuroiwa, J.; Oks, M.; Trumpfheller, C.; Park, C.G.; Huang, Y.; Hannaman, D.; Schlesinger, S.J.; Mizenina, O.; Nussenzweig, M.C.; et al. The efficacy of DNA vaccination is enhanced in mice by targeting the encoded protein to dendritic cells. J. Clin. Invest. 2008, 118, 1427–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranowska, M.; Hauge, A.G.; Hoornaert, C.; Bogen, B.; Grodeland, G. Targeting of nucleoprotein to chemokine receptors by DNA vaccination results in increased CD8(+)-mediated cross protection against influenza. Vaccine 2015, 33, 6988–6996. [Google Scholar] [CrossRef] [PubMed]
- Fossum, E.; Grodeland, G.; Terhorst, D.; Tveita, A.A.; Vikse, E.; Mjaaland, S.; Henri, S.; Malissen, B.; Bogen, B. Vaccine molecules targeting Xcr1 on cross-presenting DCs induce protective CD8(+) T-cell responses against influenza virus. Eur. J. Immunol. 2015, 45, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Grodeland, G.; Mjaaland, S.; Tunheim, G.; Fredriksen, A.B.; Bogen, B. The Specificity of Targeted Vaccines for APC Surface Molecules Influences the Immune Response Phenotype. PLoS ONE 2013, 8, e80008. [Google Scholar] [CrossRef] [PubMed]
- Crozat, K.; Guiton, R.; Contreras, V.; Feuillet, V.; Dutertre, C.A.; Ventre, E.; Vu Manh, T.P.; Baranek, T.; Storset, A.K.; Marvel, J.; et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010, 207, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Contreras, V.; Urien, C.; Guiton, R.; Alexandre, Y.; Vu Manh, T.P.; Andrieu, T.; Crozat, K.; Jouneau, L.; Bertho, N.; Epardaud, M.; et al. Existence of CD8 α-Like Dendritic Cells with a Conserved Functional Specialization and a Common Molecular Signature in Distant Mammalian Species. J. Immunol. 2010, 185, 3313–3325. [Google Scholar] [CrossRef] [PubMed]
- Maisonnasse, P.; Bouguyon, E.; Piton, G.; Ezquerra, A.; Urien, C.; Deloizy, C.; Bourge, M.; Leplat, J.J.; Simon, G.; Chevalier, C.; et al. The respiratory DC/macrophage network at steady-state and upon influenza infection in the swine biomedical model. Mucosal Immunol. 2016, 9, 835–849. [Google Scholar] [CrossRef]
- Deloizy, C.; Bouguyon, E.; Fossum, E.; Sebo, P.; Osicka, R.; Bole, A.; Pierres, M.; Biacchesi, S.; Dalod, M.; Bogen, B.; et al. Expanding the tools for identifying mononuclear phagocyte subsets in swine: Reagents to porcine CD11c and XCR1. Dev. Comp. Immunol. 2016, 65, 31–40. [Google Scholar] [CrossRef]
- Kurts, C. CD11c: Not merely a murine DC marker, but also a useful vaccination target. Eur. J. Immunol. 2008, 38, 2072–2075. [Google Scholar] [CrossRef]
- Castro, F.V.; Tutt, A.L.; White, A.L.; Teeling, J.L.; James, S.; French, R.R.; Glennie, M.J. CD11c provides an effective immunotarget for the generation of both CD4 and CD8 T cell responses. Eur. J. Immunol. 2008, 38, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Cohn, L.; Chatterjee, B.; Esselborn, F.; Smed-Sorensen, A.; Nakamura, N.; Chalouni, C.; Lee, B.C.; Vandlen, R.; Keler, T.; Lauer, P.; et al. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J. Exp. Med. 2013, 210, 1049–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernelin-Cottet, C.; Deloizy, C.; Stanek, O.; Barc, C.; Bouguyon, E.; Urien, C.; Boulesteix, O.; Pezant, J.; Richard, C.A.; Moudjou, M.; et al. A Universal Influenza Vaccine Can Lead to Disease Exacerbation or Viral Control Depending on Delivery Strategies. Front. Immunol. 2016, 7, 641. [Google Scholar] [CrossRef] [PubMed]
- Barbon, C.M.; Baker, L.; Lajoie, C.; Ramstedt, U.; Hedley, M.L.; Luby, T.M. In vivo electroporation enhances the potency of poly-lactide co-glycolide (PLG) plasmid DNA immunization. Vaccine 2010, 28, 7852–7864. [Google Scholar] [CrossRef] [PubMed]
- Bernelin-Cottet, C.; Urien, C.; McCaffrey, J.; Collins, D.; Donadei, A.; McDaid, D.; Jakob, V.; Barnier-Quer, C.; Collin, N.; Bouguyon, E.; et al. Assessing the immunogenic properties of DNA vaccines delivered with nanoparticles, electroporation and dissolvable microneedle patches in pig skin. 2019; Submitted. [Google Scholar]
- Todorova, B.; Adam, L.; Culina, S.; Boisgard, R.; Martinon, F.; Cosma, A.; Ustav, M.; Kortulewski, T.; Le Grand, R.; Chapon, C. Electroporation as a vaccine delivery system and a natural adjuvant to intradermal administration of plasmid DNA in macaques. Sci. Rep. 2017, 7, 4122. [Google Scholar] [CrossRef] [PubMed]
- Amante, D.H.; Smith, T.R.; Mendoza, J.M.; Schultheis, K.; McCoy, J.R.; Khan, A.S.; Sardesai, N.Y.; Broderick, K.E. Skin Transfection Patterns and Expression Kinetics of Electroporation-Enhanced Plasmid Delivery Using the CELLECTRA-3P, a Portable Next-Generation Dermal Electroporation Device. Hum. Gene Ther. Methods 2015, 26, 134–146. [Google Scholar] [CrossRef]
- Frydas, I.S.; Trus, I.; Kvisgaard, L.K.; Bonckaert, C.; Reddy, V.R.; Li, Y.; Larsen, L.E.; Nauwynck, H.J. Different clinical, virological, serological and tissue tropism outcomes of two new and one old Belgian type 1 subtype 1 porcine reproductive and respiratory virus (PRRSV) isolates. Vet. Res. 2015, 46, 37. [Google Scholar] [CrossRef]
- Subramaniam, S.; Pineyro, P.; Tian, D.; Overend, C.; Yugo, D.M.; Matzinger, S.R.; Rogers, A.J.; Haac, M.E.; Cao, Q.; Heffron, C.L.; et al. In vivo targeting of porcine reproductive and respiratory syndrome virus antigen through porcine DC-SIGN to dendritic cells elicits antigen-specific CD4T cell immunity in pigs. Vaccine 2014, 32, 6768–6775. [Google Scholar] [CrossRef]
- Fredriksen, A.B.; Bogen, B. Chemokine-idiotype fusion DNA vaccines are potentiated by bivalency and xenogeneic sequences. Blood 2007, 110, 1797–1805. [Google Scholar] [CrossRef] [Green Version]
- Ooi, H.S.; Kwo, C.Y.; Wildpaner, M.; Sirota, F.L.; Eisenhaber, B.; Maurer-Stroh, S.; Wong, W.C.; Schleiffer, A.; Eisenhaber, F.; Schneider, G. ANNIE: Integrated de novo protein sequence annotation. Nucleic Acids Res. 2009, 37, W435–W440. [Google Scholar] [CrossRef]
- Vu Manh, T.P.; Elhmouzi-Younes, J.; Urien, C.; Ruscanu, S.; Jouneau, L.; Bourge, M.; Moroldo, M.; Foucras, G.; Salmon, H.; Marty, H.; et al. Defining Mononuclear Phagocyte Subset Homology Across Several Distant Warm-Blooded Vertebrates Through Comparative Transcriptomics. Front. Immunol. 2015, 6, 299. [Google Scholar] [CrossRef] [PubMed]
- Biernacka, K.; Podgorska, K.; Tyszka, A.; Stadejek, T. Comparison of six commercial ELISAs for the detection of antibodies against porcine reproductive and respiratory syndrome virus (PRRSV) in field serum samples. Res. Vet. Sci. 2018, 121, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Sattler, T.; Wodak, E.; Revilla-Fernandez, S.; Schmoll, F. Comparison of different commercial ELISAs for detection of antibodies against porcine respiratory and reproductive syndrome virus in serum. BMC Vet. Res. 2014, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.; Pineyro, P.; Derscheid, R.J.; Madson, D.M.; Magstadt, D.R.; Meng, X.J. Dendritic cell-targeted porcine reproductive and respiratory syndrome virus (PRRSV) antigens adjuvanted with polyinosinic-polycytidylic acid (poly (I:C)) induced non-protective immune responses against heterologous type 2 PRRSV challenge in pigs. Vet. Immunol. Immunopathol. 2017, 190, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, B.; Poderoso, T.; Alonso, F.; Ezquerra, A.; Dominguez, J.; Revilla, C. Antigen targeting to APC: From mice to veterinary species. Dev. Comp. Immunol. 2013, 41, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Kreutz, M.; Tacken, P.J.; Figdor, C.G. Targeting dendritic cells--why bother? Blood 2013, 121, 2836–2844. [Google Scholar] [CrossRef] [PubMed]
- Kastenmuller, W.; Kastenmuller, K.; Kurts, C.; Seder, R.A. Dendritic cell-targeted vaccines—hope or hype? Nat. Rev. Immunol. 2014, 14, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Amorim, K.N.; Rampazo, E.V.; Antonialli, R.; Yamamoto, M.M.; Rodrigues, M.M.; Soares, I.S.; Boscardin, S.B. The presence of T cell epitopes is important for induction of antibody responses against antigens directed to DEC205+ dendritic cells. Sci. Rep. 2016, 6, 39250. [Google Scholar] [CrossRef]
- Losikoff, P.T.; Mishra, S.; Terry, F.; Gutierrez, A.; Ardito, M.T.; Fast, L.; Nevola, M.; Martin, W.D.; Bailey-Kellogg, C.; De Groot, A.S.; et al. HCV epitope, homologous to multiple human protein sequences, induces a regulatory T cell response in infected patients. J. Hepatol. 2015, 62, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Tenbusch, M.; Ignatius, R.; Nchinda, G.; Trumpfheller, C.; Salazar, A.M.; Topfer, K.; Sauermann, U.; Wagner, R.; Hannaman, D.; Tenner-Racz, K.; et al. Immunogenicity of DNA vaccines encoding simian immunodeficiency virus antigen targeted to dendritic cells in rhesus macaques. PLoS ONE 2012, 7, e39038. [Google Scholar] [CrossRef]
- Park, H.Y.; Tan, P.S.; Kavishna, R.; Ker, A.; Lu, J.; Chan, C.E.Z.; Hanson, B.J.; MacAry, P.A.; Caminschi, I.; Shortman, K.; et al. Enhancing vaccine antibody responses by targeting Clec9A on dendritic cells. NPJ Vaccines 2017, 2, 31. [Google Scholar] [CrossRef] [PubMed]
- Gudjonsson, A.; Lysen, A.; Balan, S.; Sundvold-Gjerstad, V.; Arnold-Schrauf, C.; Richter, L.; Baekkevold, E.S.; Dalod, M.; Bogen, B.; Fossum, E. Targeting Influenza Virus Hemagglutinin to Xcr1+ Dendritic Cells in the Absence of Receptor-Mediated Endocytosis Enhances Protective Antibody Responses. J. Immunol. 2017, 198, 2785–2795. [Google Scholar] [CrossRef] [PubMed]
- Deloizy, C.; Fossum, E.; Barnier-Quer, C.; Urien, C.; Chrun, T.; Duval, A.; Codjovi, M.; Bouguyon, E.; Maisonnasse, P.; Herve, P.L.; et al. The anti-influenza M2e antibody response is promoted by XCR1 targeting in pig skin. Sci. Rep. 2017, 7, 7639. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.R.; Rahe, M.C.; Gray, D.K.; Martins, K.V.; Murtaugh, M.P. Porcine reproductive and respiratory syndrome virus neutralizing antibodies provide in vivo cross-protection to PRRSV1 and PRRSV2 viral challenge. Virus Res. 2018, 248, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Parida, R.; Choi, I.S.; Peterson, D.A.; Pattnaik, A.K.; Laegreid, W.; Zuckermann, F.A.; Osorio, F.A. Location of T-cell epitopes in nonstructural proteins 9 and 10 of type-II porcine reproductive and respiratory syndrome virus. Virus Res. 2012, 169, 13–21. [Google Scholar] [CrossRef]
- Suradhat, S.; Wongyanin, P.; Sirisereewan, C.; Nedumpun, T.; Lumyai, M.; Triyarach, S.; Chaturavittawong, D.; Paphavasit, T.; Panyatong, R.; Thanawongnuwech, R. Transdermal delivery of plasmid encoding truncated nucleocapsid protein enhanced PRRSV-specific immune responses. Vaccine 2015, 34, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.J.; Cha, S.H.; Grimm, A.L.; Ajithdoss, D.; Rzepka, J.; Chung, G.; Yu, J.; Davis, W.C.; Ho, C.S. Pigs that recover from porcine reproduction and respiratory syndrome virus infection develop cytotoxic CD4+CD8+ and CD4+CD8- T-cells that kill virus infected cells. PLoS ONE 2018, 13, e0203482. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.M.; Moonen-Leusen, H.W.; de Visser, Y.E.; Middel, W.G.; Boersma, W.J.; Bianchi, A.T. A DNA vaccine coding for gB and gD of pseudorabies virus (suid herpes type 1) primes the immune system in the presence of maternal immunity more efficiently than conventional vaccines. Vaccine 2006, 24, 1264–1273. [Google Scholar] [CrossRef]
- Fablet, C.; Renson, P.; Eono, F.; Mahe, S.; Eveno, E.; Le Dimna, M.; Normand, V.; Lebret, A.; Rose, N.; Bourry, O. Maternally-derived antibodies (MDAs) impair piglets’ humoral and cellular immune responses to vaccination against porcine reproductive and respiratory syndrome (PRRS). Vet. Microbiol. 2016, 192, 175–180. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernelin-Cottet, C.; Urien, C.; Stubsrud, E.; Jakob, V.; Bouguyon, E.; Bordet, E.; Barc, C.; Boulesteix, O.; Contreras, V.; Barnier-Quer, C.; et al. A DNA-Modified Live Vaccine Prime–Boost Strategy Broadens the T-Cell Response and Enhances the Antibody Response against the Porcine Reproductive and Respiratory Syndrome Virus. Viruses 2019, 11, 551. https://doi.org/10.3390/v11060551
Bernelin-Cottet C, Urien C, Stubsrud E, Jakob V, Bouguyon E, Bordet E, Barc C, Boulesteix O, Contreras V, Barnier-Quer C, et al. A DNA-Modified Live Vaccine Prime–Boost Strategy Broadens the T-Cell Response and Enhances the Antibody Response against the Porcine Reproductive and Respiratory Syndrome Virus. Viruses. 2019; 11(6):551. https://doi.org/10.3390/v11060551
Chicago/Turabian StyleBernelin-Cottet, Cindy, Céline Urien, Elisabeth Stubsrud, Virginie Jakob, Edwige Bouguyon, Elise Bordet, Céline Barc, Olivier Boulesteix, Vanessa Contreras, Christophe Barnier-Quer, and et al. 2019. "A DNA-Modified Live Vaccine Prime–Boost Strategy Broadens the T-Cell Response and Enhances the Antibody Response against the Porcine Reproductive and Respiratory Syndrome Virus" Viruses 11, no. 6: 551. https://doi.org/10.3390/v11060551
APA StyleBernelin-Cottet, C., Urien, C., Stubsrud, E., Jakob, V., Bouguyon, E., Bordet, E., Barc, C., Boulesteix, O., Contreras, V., Barnier-Quer, C., Collin, N., Trus, I., Nauwynck, H., Bertho, N., & Schwartz-Cornil, I. (2019). A DNA-Modified Live Vaccine Prime–Boost Strategy Broadens the T-Cell Response and Enhances the Antibody Response against the Porcine Reproductive and Respiratory Syndrome Virus. Viruses, 11(6), 551. https://doi.org/10.3390/v11060551




