Understanding and Exploiting Phage–Host Interactions
Abstract
1. Introduction
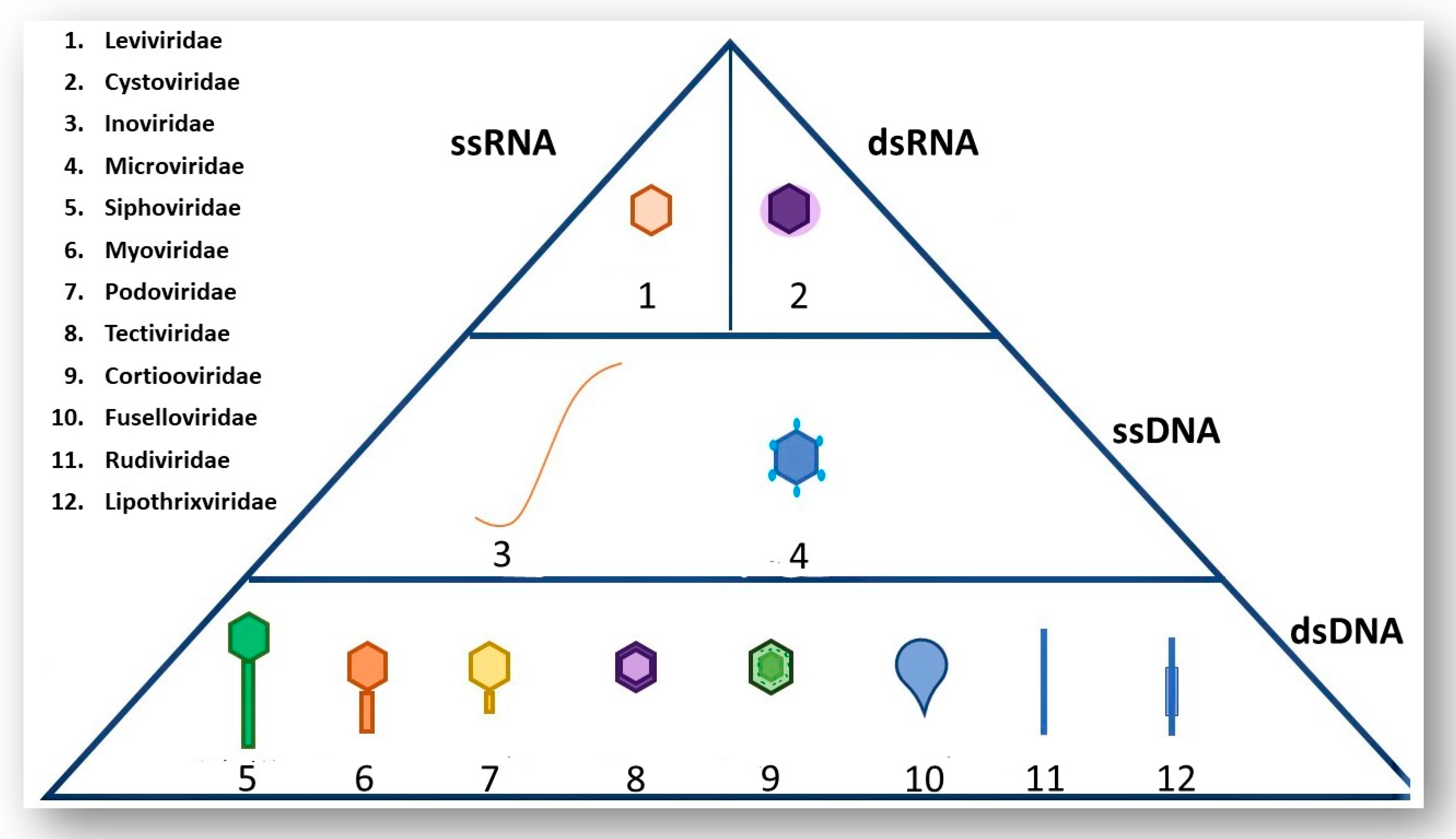
2. A Brief Overview of Phage Morphology and Classification
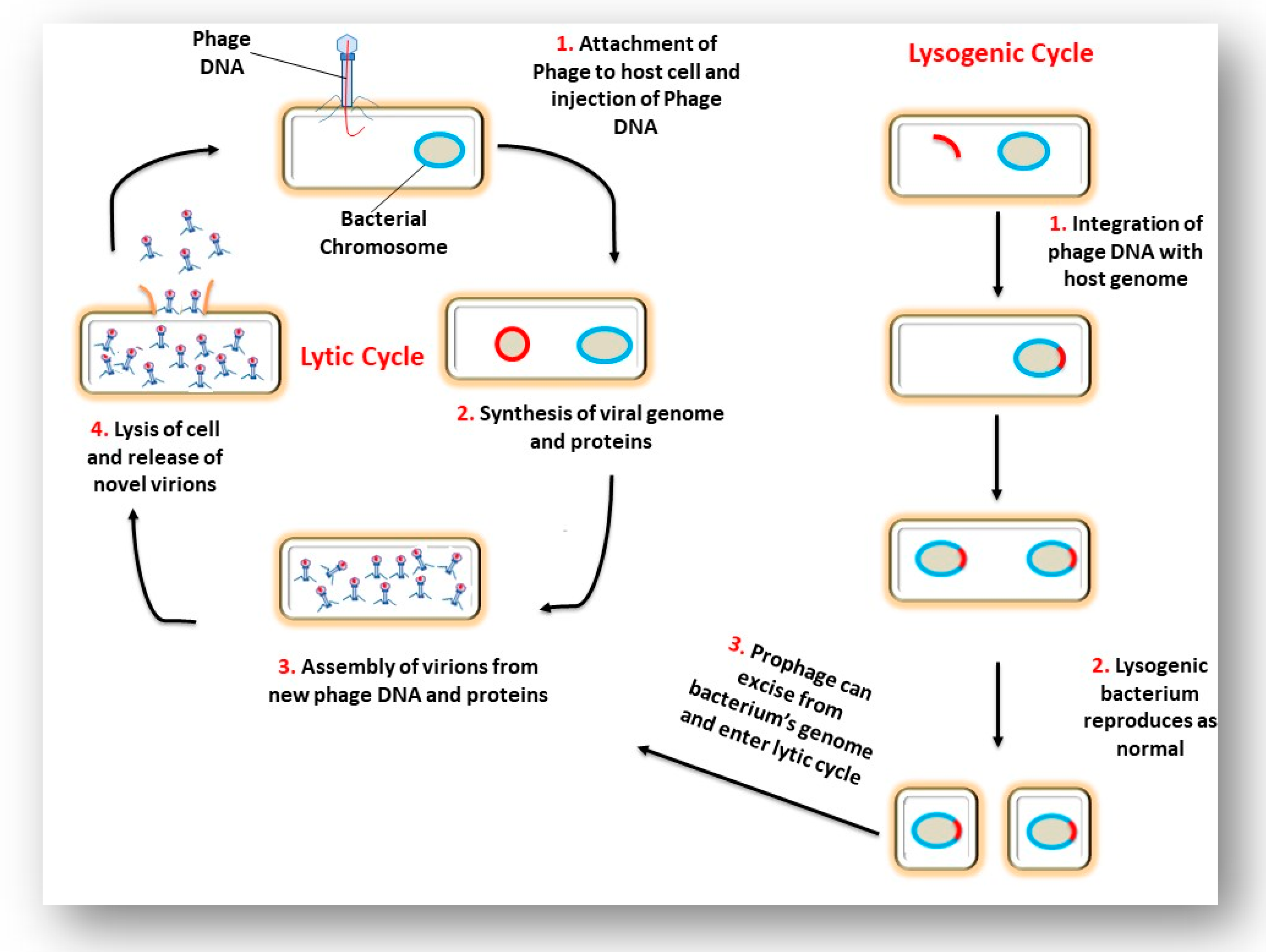
3. The Interaction of a Phage and its Bacterial Host Cell
3.1. Endolysins from Phages Infecting Gram-Negative Bacteria
3.2. Endolysins from Phages Infecting Gram-Positive Bacteria
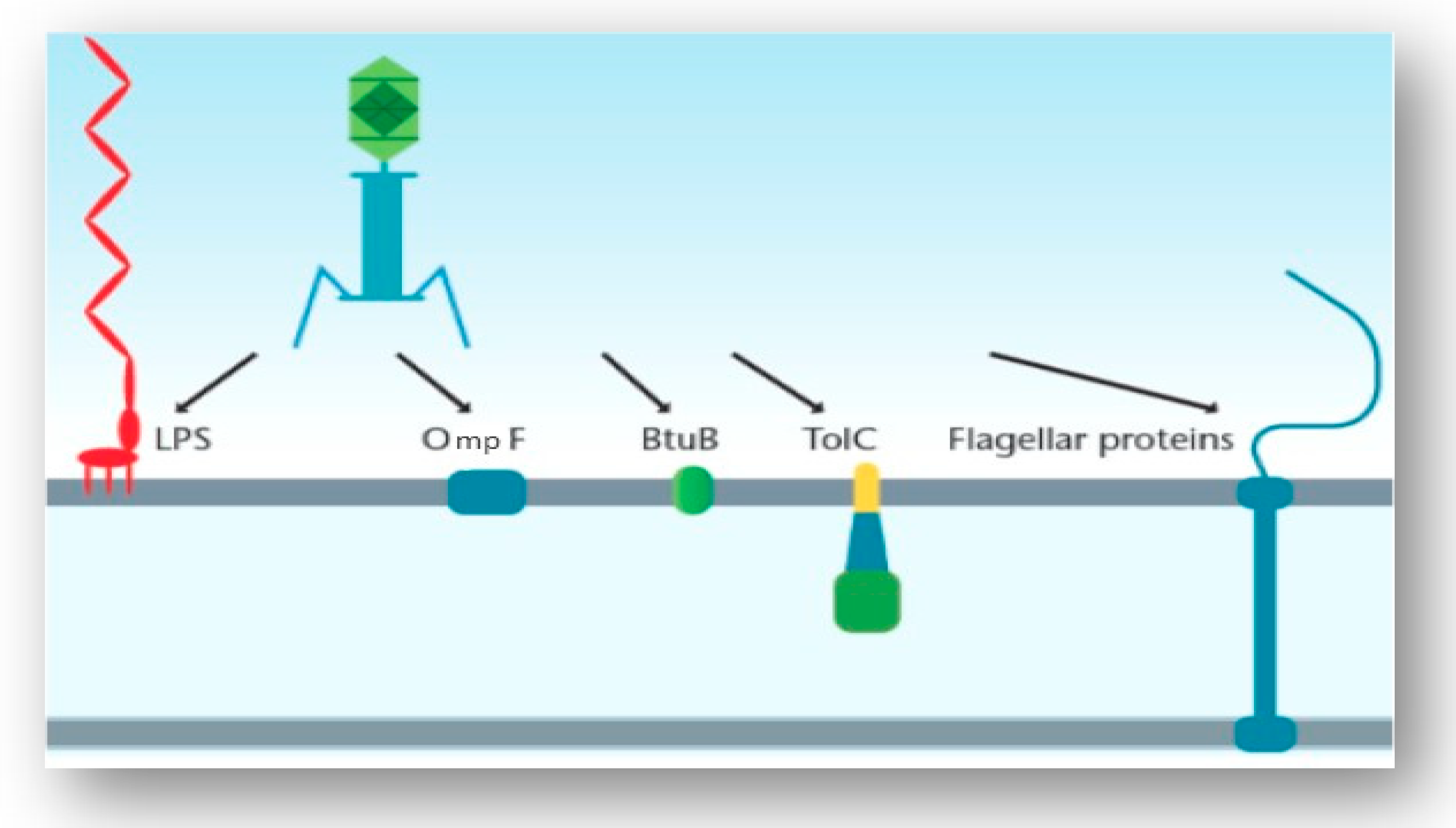
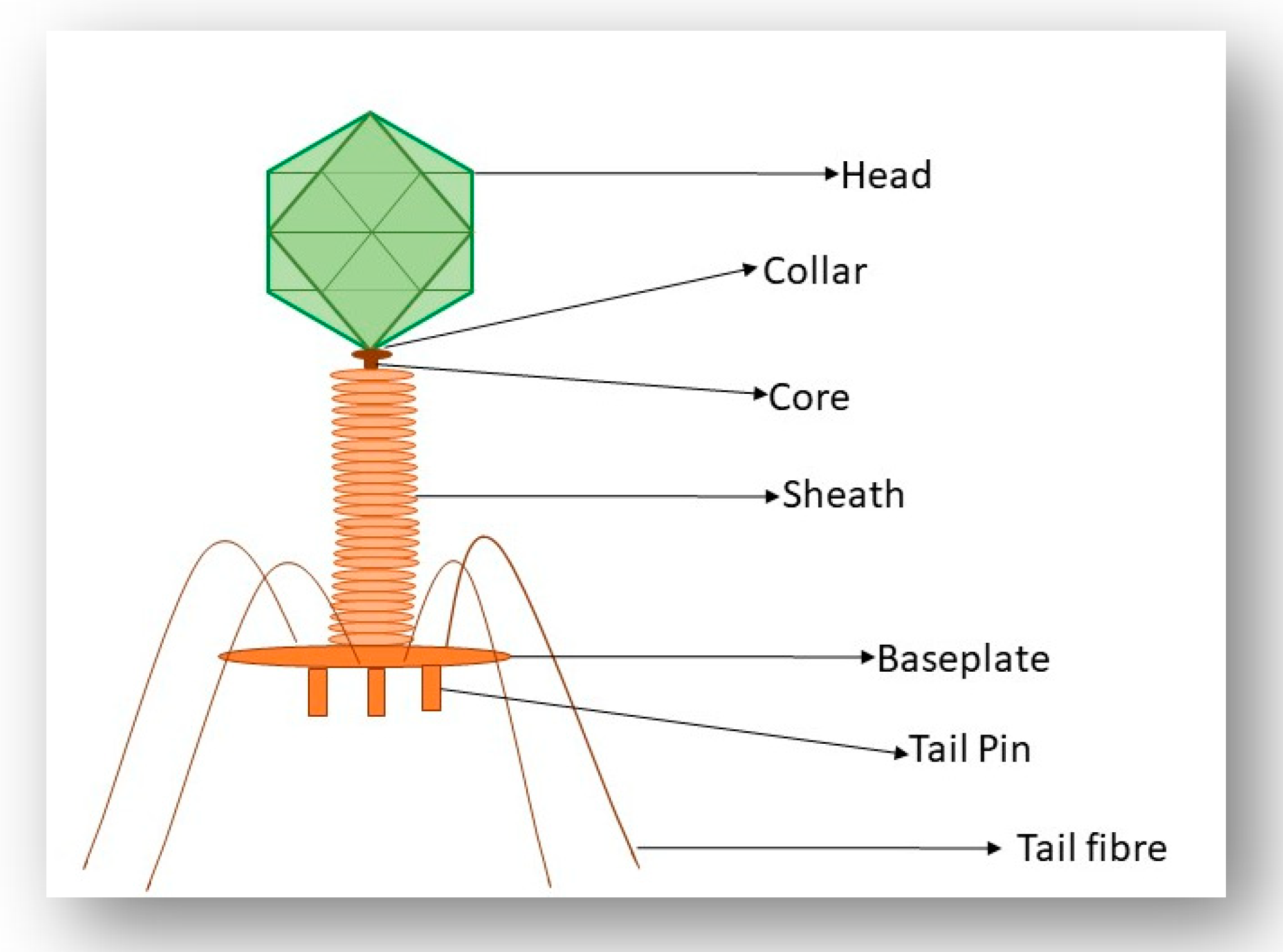
3.3. Interaction of the T4 Phage and its Bacterial Host Cell (Receptors for Attachment)
3.4. Interactions of Other Phages and Their Gram-Negative Host Bacterial Cell (Receptors for Attachment)
3.5. Phage–Host Interactions in Gram-Positive Bacteria (Receptors for Attachment)
3.5.1. Lactococcus lactis Phage–Host Interactions
3.5.2. Listeria monocytogenes Phage–Host Interactions
4. Exploitation of Phage–Host Interactions
4.1. Detection of Foodborne Pathogens
4.2. Exploitation of Phage–Host Interactions for the Detection of Foodborne Pathogens
4.2.1. Whole Phages in the Detection of Foodborne Pathogens
4.2.2. Phage-Derived Proteins for the Detection of Foodborne Pathogens
4.3. The Use of Biosensors to Detect Bacteria
5. Exploitation of Phages as Biocontrol Agents
5.1. Exploitation of Phages as Biocontrol Agents in Food
The Use of Whole Phages and Phage-Derived Proteins as Biocontrol Agents in Foods
5.2. Exploitation of Phages as Biocontrol Agents in Food Producing Plants
5.2.1. Pre-Harvest Treatment of Food Producing Plants
5.2.2. Post-Harvest Treatment of Food Producing Plants
5.3. Exploitation of Phages as Biocontrol Agents in Agricultural Animals
5.4. The Pros and Cons of Using Phages as Biocontrol Agents
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in Nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and Their Genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Summers, W.C. The Strange History of Phage Therapy. Bacteriophage 2012, 2, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Betts, A.; Gray, C.; Zelek, M.; MacLean, R.C.; King, K.C. High Parasite Diversity Accelerates Host Adaptation and Diversification. Science 2018, 360, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Rüger, W. Bacteriophage T4 genome. Microbiol. Mol. Rev. 2003, 67, 86–156. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; São-José, C.; Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef] [PubMed]
- Weitz, J.S.; Poisot, T.; Meyer, J.R.; Flores, C.O.; Valverde, S.; Sullivan, M.B.; Hochberg, M.E. Phage–bacteria Infection Networks. Trends Microbiol. 2013, 21, 82–91. [Google Scholar] [CrossRef]
- Ackermann, H.-W. 5500 Phages Examined in the Electron Microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Phage Classification and Characterization. Methods Mol. Biol. 2009, 501, 127–140. [Google Scholar] [CrossRef]
- International Committee for Virus Taxonomy. Taxonomic Information. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 29 April 2019).
- Campbell, A. Phage Evolution and Speciation. In The Bacteriophages; Plenum Press: New York, NY, USA, 1988; pp. 1–14. [Google Scholar]
- Eichhorn, I.; Heidemanns, K.; Ulrich, R.G.; Schmidt, H.; Semmler, T.; Fruth, A.; Bethe, A.; Goulding, D.; Pickard, D.; Karch, H.; et al. Lysogenic Conversion of Atypical Enteropathogenic Escherichia coli (aEPEC) from Human, Murine, and Bovine Origin with Bacteriophage Φ3538 Δstx2::cat Proves Their Enterohemorrhagic E. coli (EHEC) Progeny. Int. J. Med. Microbiol. 2018, 308, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Munson-McGee, J.; Snyder, J.; Young, M.; Munson-McGee, J.H.; Snyder, J.C.; Young, M.J. Archaeal Viruses from High-Temperature Environments. Genes 2018, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host Receptors for Bacteriophage Adsorption. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage Receptors, Mechanisms of Phage Adsorption and Penetration into Host Cel. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, R.J.; Holt, K.E. Diversity-Generating Machines: Genetics of Bacterial Sugar-Coating. Trends Microbiol. 2018, 26, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage Resistance Mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Mao, J.; Xie, J. Bacteriophage Polysaccharide Depolymerases and Biomedical Applications. BioDrugs 2014, 28, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-Encoded Virion-Associated Enzymes to Overcome the Carbohydrate Barriers during the Infection Process. Appl. Microb. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J.; Kramer, K.; Ebel, F.; Scherer, S. C-Terminal Domains of Listeria monocytogenes Bacteriophage Murein Hydrolases Determine Specific Recognition and High-Affinity Binding to Bacterial Cell Wall Carbohydrates. Mol. Microbiol. 2002, 44, 335–349. [Google Scholar] [CrossRef]
- Eugster, M.R.; Haug, M.C.; Huwiler, S.G.; Loessner, M.J. The Cell Wall Binding Domain of Listeria Bacteriophage Endolysin PlyP35 Recognizes Terminal GlcNAc Residues in Cell Wall Teichoic Acid. Mol. Microbiol. 2011, 81, 1419–1432. [Google Scholar] [CrossRef]
- Berry, J.; Rajaure, M.; Pang, T.; Young, R. The Spanin Complex Is Essential for Lambda Lysis. J. Bacteriol. 2012, 194, 5667–5674. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.; Savva, C.G.; Fleming, K.G.; Struck, D.K.; Young, R. Structure of the Lethal Phage Pinhole. Proc. Natl. Acad. Sci. USA 2009, 106, 18966–18971. [Google Scholar] [CrossRef] [PubMed]
- Emrich, J.; Streisinger, G. The Role of Phage Lysozyme in the Life Cycle of Phage T4. Virology 1968, 36, 387–391. [Google Scholar] [CrossRef]
- Moussa, S.H.; Kuznetsov, V.; Tran, T.A.T.; Sacchettini, J.C.; Young, R. Protein Determinants of Phage T4 Lysis Inhibition. Protein Sci. 2012, 21, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Lysis from Without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Arisaka, F.; Kanamaru, S.; Leiman, P.; Rossmann, M.G. The Tail Lysozyme Complex of Bacteriophage T4. Int. J. Biochem. Cell Biol. 2003, 35, 16–21. [Google Scholar] [CrossRef]
- Tarahovsky, Y.S.; Ivanitsky, G.R.; Khusainov, A.A. Lysis of Escherichia coli Cells Induced by Bacteriophage T4. FEMS Microbiol. Lett. 1994, 122, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Gerstmans, H.; Thorpe, S.; Mesnage, S.; Lavigne, R.; Briers, Y. DUF3380 Domain from a Salmonella Phage Endolysin Shows Potent N-Acetylmuramidase Activity. Appl. Environ. Microbiol. 2016, 82, 4975–4981. [Google Scholar] [CrossRef]
- Hu, S.; Kong, J.; Kong, W.; Guo, T.; Ji, M. Characterization of a Novel LysM Domain from Lactobacillus fermentum Bacteriophage Endolysin and Its Use as an Anchor To Display Heterologous Proteins on the Surfaces of Lactic Acid Bacteria. Appl. Environ. Microbiol. 2010, 76, 2410–2418. [Google Scholar] [CrossRef]
- Mahony, J.; van Sinderen, D. Gram-Positive Phage-Host Interactions. Front. Microbiol. 2015, 6, 61. [Google Scholar] [CrossRef]
- Marti, R.; Zurfluh, K.; Hagens, S.; Pianezzi, J.; Klumpp, J.; Loessner, M.J. Long Tail Fibres of the Novel Broad-Host-Range T-Even Bacteriophage S16 Specifically Recognize Salmonella OmpC. Mol. Microbiol. 2013, 87, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; van Raaij, M. Bacteriophage T4 Long Tail Fiber Domains. Biophys. Rev. 2018, 10, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Washizaki, A.; Yonesaki, T.; Otsuka, Y. Characterization of the Interactions between Escherichia coli Receptors, LPS and OmpC, and Bacteriophage T4 Long Tail Fibers. Microbiologyopen 2016, 5, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Prehm, P.; Jann, B.; Jann, K.; Schmidt, G.; Stirm, S. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coli B. J. Mol. Biol. 1976, 101, 277–281. [Google Scholar] [CrossRef]
- Leiman, P.G.; Chipman, P.R.; Kostyuchenko, V.A.; Mesyanzhinov, V.V.; Rossmann, M.G. Three-Dimensional Rearrangement of Proteins in the Tail of Bacteriophage T4 on Infection of Its Host. Cell 2004, 118, 419–429. [Google Scholar] [CrossRef]
- Dunne, M.; Denyes, J.M.; Arndt, H.; Loessner, M.J.; Leiman, P.G.; Klumpp, J. Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition. Structure 2018, 26, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Lee, J.-H.; Kim, H.; Choi, Y.; Heu, S.; Ryu, S. Receptor Diversity and Host Interaction of Bacteriophages Infecting Salmonella enterica Serovar Typhimurium. PLoS ONE 2012, 7, e43392. [Google Scholar] [CrossRef]
- Choi, Y.; Shin, H.; Lee, J.-H.; Ryu, S. Identification and Characterization of a Novel Flagellum-Dependent Salmonella-Infecting Bacteriophage, iEPS5. Appl. Environ. Microbiol. 2013, 79, 4829–4837. [Google Scholar] [CrossRef]
- Kojima, S.; Furukawa, Y.; Matsunami, H.; Minamino, T.; Namba, K. Characterization of the Periplasmic Domain of MotB and Implications for Its Role in the Stator Assembly of the Bacterial Flagellar Motor. J. Bacteriol. 2008, 190, 3314–3322. [Google Scholar] [CrossRef]
- Pickard, D.; Toribio, A.L.; Petty, N.K.; van Tonder, A.; Yu, L.; Goulding, D.; Barrell, B.; Rance, R.; Harris, D.; Wetter, M.; et al. A Conserved Acetyl Esterase Domain Targets Diverse Bacteriophages to the Vi Capsular Receptor of Salmonella enterica Serovar Typhi. J. Bacteriol. 2010, 192, 5746–5754. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochima et Biophysia Acta 2016, 1858, 936–946. [Google Scholar] [CrossRef] [PubMed]
- InjectionChapot-Chartier, M.-P. Interactions of the Cell-Wall Glycopolymers of Lactic Acid Bacteria with Their Bacteriophages. Front. Microbiol. 2014, 5, 236. [Google Scholar] [CrossRef]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.-P.; van Sinderen, D. Differences in Lactococcal Cell Wall Polysaccharide Structure Are Major Determining Factors in Bacteriophage Sensitivity. MBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Monteville, M.R.; Ardestani, B.; Geller, B.L. Lactococcal Bacteriophages Require a Host Cell Wall Carbohydrate and a Plasma Membrane Protein for Adsorption and Injection of DNA. Appl. Environ. Microbiol. 1994, 60, 3204–3211. [Google Scholar] [PubMed]
- Millen, A.M.; Romero, D.A. Genetic Determinants of Lactococcal c2 viruses for Host Infection and Their Role in Phage Evolution. J. Gen. Virol. 2016, 97, 1998–2007. [Google Scholar] [CrossRef] [PubMed]
- Bielmann, R.; Habann, M.; Eugster, M.R.; Lurz, R.; Calendar, R.; Klumpp, J.; Loessner, M.J. Receptor Binding Proteins of Listeria monocytogenes Bacteriophages A118 and P35 Recognize Serovar-Specific Teichoic Acids. Virology 2015, 477, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Parent, K.N.; Erb, M.L.; Cardone, G.; Nguyen, K.; Gilcrease, E.B.; Porcek, N.B.; Pogliano, J.; Baker, T.S.; Casjens, S.R. OmpA and OmpC Are Critical Host Factors for Bacteriophage Sf6 Entry in Shigella. Mol. Microbiol. 2014, 92, 47–60. [Google Scholar] [CrossRef]
- Jakhetia, R.; Verma, N.K. Identification and Molecular Characterisation of a Novel Mu-Like Bacteriophage, SfMu, of Shigella flexneri. PLoS ONE 2015, 10, e0124053. [Google Scholar] [CrossRef]
- Faruque, S.M.; Bin Naser, I.; Fujihara, K.; Diraphat, P.; Chowdhury, N.; Kamruzzaman, M.; Qadri, F.; Yamasaki, S.; Ghosh, A.N.; Mekalanos, J.J. Genomic Sequence and Receptor for the Vibrio cholerae Phage KSF-1: Evolutionary Divergence among Filamentous Vibriophages Mediating Lateral Gene Transfer. J. Bacteriol. 2005, 187, 4095–4103. [Google Scholar] [CrossRef]
- Seed, K.D.; Faruque, S.M.; Mekalanos, J.J.; Calderwood, S.B.; Qadri, F. Phase Variable O Antigen Biosynthetic Genes Control Expression of the Major Protective Antigen and Bacteriophage Receptor in Vibrio Cholerae O1. PLoS Pathog. 2012, 8, 1002917. [Google Scholar] [CrossRef]
- Morita, M.; Tanji, Y.; Mizoguchi, K.; Akitsu, T.; Kijima, N.; Unno, H. Characterization of a Virulent Bacteriophage Specific for Escherichia coli O157:H7 and Analysis of Its Cellular Receptor and Two Tail Fiber Genes. FEMS Microbiol. Lett. 2002, 211, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Perry, L.L.; SanMiguel, P.; Minocha, U.; Terekhov, A.I.; Shroyer, M.L.; Farris, L.A.; Bright, N.; Reuhs, B.L.; Applegate, B.M. Sequence Analysis of Escherichia coli O157:H7 Bacteriophage ΦV10 and Identification of a Phage-Encoded Immunity Protein That Modifies the O157 Antigen. FEMS Microbiol. Lett. 2009, 292, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Baxa, U.; Steinbacher, S.; Miller, S.; Weintraub, A.; Huber, R.; Seckler, R. Interactions of Phage P22 Tails with Their Cellular Receptor, Salmonella O-Antigen Polysaccharide. Biophys. J. 1996, 71, 2040–2048. [Google Scholar] [CrossRef]
- Schmidt, A.; Rabsch, W.; Broeker, N.K.; Barbirz, S. Bacteriophage Tailspike Protein Based Assay to Monitor Phase Variable Glucosylations in Salmonella O-Antigens. BMC Microbiol. 2016, 16, 207. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.C.H.; Van Alphen, L.B.; Harboe, A.; Li, J.; Christensen, B.B.; Szymanski, C.M.; Brøndsted, L. Bacteriophage F336 Recognizes the Capsular Phosphoramidate Modification of Campylobacter jejuni NCTC11168 #. J. Bacteriol. 2011, 193, 6742–6749. [Google Scholar] [CrossRef] [PubMed]
- Baldvinsson, S.B.; Sørensen, M.C.H.; Vegge, C.S.; Clokie, M.R.J.; Brøndsted, L. Campylobacter Jejuni Motility Is Required for Infection of the Flagellotropic Bacteriophage F341. Appl. Environ. Microbiol. 2014, 80, 7096–7106. [Google Scholar] [CrossRef]
- Le, S.; He, X.; Tan, Y.; Huang, G.; Zhang, L.; Lux, R.; Shi, W.; Hu, F. Mapping the Tail Fiber as the Receptor Binding Protein Responsible for Differential Host Specificity of Pseudomonas aeruginosa Bacteriophages PaP1 and JG004. PLoS ONE 2013, 8, e68562. [Google Scholar] [CrossRef]
- Mcshan, W.M.; Lam, J.S.; Van Nguyen, S.; Yang, H.; Pan, X.; Cui, X.; Zhang, F.; He, Y.; Li, L. Genetic Evidence for O-Specific Antigen as Receptor of Pseudomonas aeruginosa Phage K8 and Its Genomic Analysis. Front. Microbiol. 2016, 7, 252. [Google Scholar] [CrossRef]
- Gillis, A.; Mahillon, J. Phages Preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: Past, Present and Future. Viruses 2014, 6, 2623–2672. [Google Scholar] [CrossRef]
- Bishop-Lilly, K.A.; Plaut, R.D.; Chen, P.E.; Akmal, A.; Willner, K.M.; Butani, A.; Dorsey, S.; Mokashi, V.; Mateczun, A.J.; Chapman, C.; et al. Whole Genome Sequencing of Phage Resistant Bacillus anthracis Mutants Reveals an Essential Role for Cell Surface Anchoring Protein CsaB in Phage AP50c Adsorption. Virol. J. 2012, 9, 246. [Google Scholar] [CrossRef]
- Xia, G.; Corrigan, R.M.; Winstel, V.; Goerke, C.; Gründling, A.; Peschel, A. Wall Teichoic Acid-Dependent Adsorption of Staphylococcal Siphovirus and Myovirus. J. Bacteriol. 2011, 193, 4006–4009. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, J.; Narita-Yamada, S.; Wakabayashi, Y.; Kamio, Y. Identification of ORF636 in Phage SLT Carrying Panton-Valentine Leukocidin Genes, Acting as an Adhesion Protein for a Poly(Glycerophosphate) Chain of Lipoteichoic Acid on the Cell Surface of Staphylococcus aureus. J. Bacteriol. 2009, 191, 4674–4680. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.A.; Shah, M.A.; Mir, M.M.; Dar, B.N.; Greiner, R.; Roohinejad, S. Microbiological Contamination of Ready-to-Eat Vegetable Salads in Developing Countries and Potential Solutions in the Supply Chain to Control Microbial Pathogens. Food Control 2018, 85, 235–244. [Google Scholar] [CrossRef]
- Rohde, A.; Hammerl, J.A.; Boone, I.; Jansen, W.; Fohler, S.; Klein, G.; Dieckmann, R.; Al Dahouk, S. Overview of Validated Alternative Methods for the Detection of Foodborne Bacterial Pathogens. Trends Food Sci. Technol. 2017, 62, 113–118. [Google Scholar] [CrossRef]
- Mandal, P.K.; Biswas, A.K.; Choi, K.; Pal, U.K. Methods for Rapid Detection of Foodborne Pathogens: An Overview. Am. J. Food Technol. 2011, 6, 87–102. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.-W.; Wang, J.; Oh, D.H. Advances in Rapid Detection Methods for Foodborne Pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.; Campbell, K.; McAuliffe, O. Bacteriophages and Rapid Detection of Bacterial Pathogens: A Novel Approach. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Kim, S.Y.; Ko, G. Using Propidium Monoazide to Distinguish between Viable and Nonviable Bacteria, MS2 and Murine Norovirus. Lett. Appl. Microbiol. 2012, 55, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Vesper, S.; McKinstry, C.; Hartmann, C.; Neace, M.; Yoder, S.; Vesper, A. Quantifying Fungal Viability in Air and Water Samples Using Quantitative PCR after Treatment with Propidium Monoazide (PMA). J. Microbiol. Methods 2008, 72, 180–184. [Google Scholar] [CrossRef]
- Petty, N.K.; Evans, T.J.; Fineran, P.C.; Salmond, G.P.C. Biotechnological Exploitation of Bacteriophage Research. Trends Biotechnol. 2007, 25, 7–15. [Google Scholar] [CrossRef]
- O’Sullivan, L.; Bolton, D.; McAuliffe, O.; Coffey, A. Bacteriophages in Food Applications: From Foe to Friend. Annu. Rev. Food Sci. Technol. 2019, 10, 151–172. [Google Scholar] [CrossRef]
- Jung, L.-S.; Ahn, J. Evaluation of Bacteriophage Amplification Assay for Rapid Detection of Shigella boydii in Food Systems. Ann. Microbiol. 2016, 66. Available online: https://link.springer.com/content/pdf/10.1007%2Fs13213-015-1178-y.pdf (accessed on 29 April 2019). [CrossRef]
- Garrido-Maestu, A.; Fuciños, P.; Azinheiro, S.; Carvalho, C.; Carvalho, J.; Prado, M. Specific Detection of Viable Salmonella Enteritidis by Phage Amplification Combined with qPCR (PAA-qPCR) in Spiked Chicken Meat Samples. Food Control 2019, 99, 79–83. [Google Scholar] [CrossRef]
- Cox, R.C.; Jensen, R.K.; Mondesire, R.R.; Voorhees, J.K. Rapid detection of Bacillus anthracis by γ phage amplification and lateral flow immunochromatography. J. Microbiol. Methods 2015, 118, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Coronel-Aguilera, C.P.; Romero, P.L.; Perry, L.; Minocha, U.; Rosenfield, C.; Gehring, A.G.; Paoli, G.C.; Bhunia, A.K.; Applegate, B. The Use of a Novel NanoLuc-Based Reporter Phage for the Detection of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 33235. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, M.; Ryu, S. Development of an Engineered Bioluminescent Reporter Phage for the Sensitive Detection of Viable Salmonella Typhimurium. Anal. Chem. 2014, 86, 5858–5864. [Google Scholar] [CrossRef]
- Smartt, A.E.; Xu, T.; Jegier, P.; Carswell, J.J.; Blount, S.A.; Sayler, G.S.; Ripp, S. Pathogen Detection Using Engineered Bacteriophages. Anal. Bioanal. Chem. 2012, 402, 3127–3146. [Google Scholar] [CrossRef]
- Hagens, S.; de Wouters, T.; Vollenweider, P.; Loessner, M.J. Reporter Bacteriophage A511::celB Transduces a Hyperthermostable Glycosidase from Pyrococcus furiosus for Rapid and Simple Detection of Viable Listeria Cells. Bacteriophage 2011, 1, 143–151. [Google Scholar] [CrossRef][Green Version]
- Javed, M.A.; Poshtiban, S.; Arutyunov, D.; Evoy, S.; Szymanski, C.M. Bacteriophage Receptor Binding Protein Based Assays for the Simultaneous Detection of Campylobacter jejuni and Campylobacter coli. PLoS ONE 2013, 8, e69770. [Google Scholar] [CrossRef]
- Denyes, J.M.; Dunne, M.; Steiner, S.; Mittelviefhaus, M.; Weiss, A.; Schmidt, H.; Klumpp, J.; Loessner, M.J. Modified bacteriophage S16 long tail fiber proteins for rapid and specific immobilization and detection of Salmonella cells. Appl. Environ. Microbiol. 2017, 83, e00277-17. [Google Scholar] [CrossRef]
- Junillon, T.; Vimont, A.; Mosticone, D.; Mallen, B.; Baril, F.; Rozand, C.; Flandrois, J.P. Simplified Detection of Food-Borne Pathogens: An in Situ High Affinity Capture and Staining Concept. J. Microbiol. Methods 2012, 91, 501–505. [Google Scholar] [CrossRef]
- Niyomdecha, S.; Limbut, W.; Numnuam, A.; Kanatharana, P.; Charlermroj, R.; Karoonuthaisiri, N.; Thavarungkul, P. Phage-Based Capacitive Biosensor for Salmonella Detection. Talanta 2018, 188, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Hamo, H.B.; Kushmaro, A.; Marks, R.S.; Grüner, C.; Rauschenbach, B.; Abdulhalim, I. Highly Sensitive and Specific Detection of E. coli by a SERS Nanobiosensor Chip Utilizing Metallic Nanosculptured Thin Films. Analyst 2015, 140, 3201–3209. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface Plasmon Resonance Detection of E. coli and Methicillin-Resistant S. aureus Using Bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Sorokulova, I.B.; Vodyanoy, V.J.; Simonian, A.L. Lytic Phage as a Specific and Selective Probe for Detection of Staphylococcus aureus—A Surface Plasmon Resonance Spectroscopic Study. Biosens. Bioelectron. 2007, 22, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arya, S.K.; Glass, N.; Hanifi-Moghaddam, P.; Naidoo, R.; Szymanski, C.M.; Tanha, J.; Evoy, S. Bacteriophage Tailspike Proteins as Molecular Probes for Sensitive and Selective Bacterial Detection. Biosens. Bioelectron. 2010, 26, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Chen, W.; Pelton, R.; Griffiths, M.W. Biocontrol of Listeria Monocytogenes and Escherichia coli O157:H7 in Meat by Using Phages Immobilized on Modified Cellulose Membranes. Appl. Environ. Microbiol. 2011, 77, 6379–6387. Available online: https://aem.asm.org/content/aem/77/18/6379.full.pdf (accessed on 2 May 2019). [CrossRef] [PubMed]
- Singh, A.; Arutyunov, D.; McDermott, M.T.; Szymanski, C.M.; Evoy, S. Specific Detection of Campylobacterjejuni Using the Bacteriophage NCTC 12673 Receptor Binding Protein as a Probe. Analyst 2011, 136, 4780. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C. The Problem of Antimicrobial Resistance in the Foodchain. Reports. 2010. Available online: https://www.safefood.eu/SafeFood/files/8a/8abb9354-4cc2-49a4-b586-2bf0008eb8cf.pdf (accessed on 15 March 2019).
- Mahony, J.; McAuliffe, O.; Ross, R.P.; van Sinderen, D. Bacteriophages as Biocontrol Agents of Food Pathogens. Curr. Opin. Biotechnol. 2011, 22, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Herzig, O.; Fieseler, L.; Klumpp, J.; Loessner, M.J. Biocontrol of Salmonella Typhimurium in RTE Foods with the Virulent Bacteriophage FO1-E2. Int. J. Food Microbiol. 2012, 154, 66–72. [Google Scholar] [CrossRef]
- Tanji, Y.; Shimada, T.; Yoichi, M.; Miyanaga, K.; Hori, K.; Unno, H. Toward Rational Control of Escherichia coli O157:H7 by a Phage Cocktail. Appl. Microbiol. Biotechnol. 2004, 64, 270–274. [Google Scholar] [CrossRef]
- Fischer, S.; Kittler, S.; Klein, G.; Glünder, G. Impact of a Single Phage and a Phage Cocktail Application in Broilers on Reduction of Campylobacter jejuni and Development of Resistance. PLoS ONE 2013, 8, e78543. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Jeon, B.; Ryu, S. Effective Inhibition of Salmonella Typhimurium in Fresh Produce by a Phage Cocktail Targeting Multiple Host Receptors. Food Microbiol. 2019, 77, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Coffey, B.; Rivas, L.; Duffy, G.; Coffey, A.; Ross, R.P.; McAuliffe, O. Assessment of Escherichia coli O157:H7-Specific Bacteriophages e11/2 and e4/1c in Model Broth and Hide Environments. Int. J. Food Microbiol. 2011, 147, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Bao, H.; Billington, C.; Hudson, J.A.; Wang, R. Isolation and Lytic Activity of the Listeria Bacteriophage Endolysin LysZ5 against Listeria monocytogenes in Soya Milk. Food Microbiol. 2012, 31, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Martínez, B.; Donovan, D.M.; García, P.; Rodríguez, A. Potential of the Virion-Associated Peptidoglycan Hydrolase HydH5 and Its Derivative Fusion Proteins in Milk Biopreservation. PLoS ONE 2013, 8, e54828. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Bhowmick, T.S.; Ahern, S.J.; Young, R.; Gonzalez, C.F. Control of Pierce’s Disease by Phage. PLoS ONE 2015, 10, e0128902. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, A.; Fujisawa, M.; Hamasaki, R.; Kawasaki, T.; Fujie, M.; Yamada, T. Biocontrol of Ralstonia solanacearum by Treatment with Lytic Bacteriophages †. Appl. Environ. Microbiol. 2011, 77, 4155–4162. [Google Scholar] [CrossRef]
- Oliveira, M.; Viñas, I.; Colàs, P.; Anguera, M.; Usall, J.; Abadias, M. Effectiveness of a Bacteriophage in Reducing Listeria Monocytogenes on Fresh-Cut Fruits and Fruit Juices. Food Microbiol. 2014, 38, 137–142. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Gannon, B.W.; Halfhide, D.E.; Santos, S.B.; Hayes, C.M.; Roe, J.M.; Azeredo, J. The in Vivo Efficacy of Two Administration Routes of a Phage Cocktail to Reduce Numbers of Campylobacter coli and Campylobacter jejuni in Chickens. BMC Microbiol. 2010, 10, 232. [Google Scholar] [CrossRef]
- Callaway, T.R.; Edrington, T.S.; Brabban, A.; Kutter, B.; Karriker, L.; Stahl, C.; Wagstrom, E.; Anderson, R.; Poole, T.L.; Genovese, K.; et al. Evaluation of Phage Treatment as a Strategy to Reduce Salmonella Populations in Growing Swine. Foodborne Pathogens and Disease 2011, 8. [Google Scholar] [CrossRef]
- Schmelcher, M.; Powell, A.M.; Becker, S.C.; Camp, M.J.; Donovan, D.M. Chimeric Phage Lysins Act Synergistically with Lysostaphin To Kill Mastitis-Causing Staphylococcus Aureus in Murine Mammary Glands. Appl. Environ. Microbiol. 2012, 78, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Loc-Carrillo, C.; Abedon, S.T. Pros and Cons of Phage Therapy. No. 2, 111–114. Available online: https://www.tandfonline.com/doi/pdf/10.4161/bact.1.2.14590?needAccess=true (accessed on 2 May 2019).
- Kazi, M.; Annapure, U.S. Bacteriophage Biocontrol of Foodborne Pathogens. J. Food Sci. Technol. 2016, 53, 1355–1362. [Google Scholar] [CrossRef] [PubMed]


| Receptors Localized on the Surface of Gram-Negative Bacteria | |||
|---|---|---|---|
| Phage | Host Bacterial Cell | Receptor(s) | Reference |
| Sf6 | Shigella flexneri (found in contaminated food and water) | OmpA OmpC | [49] |
| SfMu | Shigella flexneri | O-antigen of lipopolysaccharide (LPS) | [50] |
| KSF-1 | Vibrio Cholera (found in contaminated food and water) | Mannose-sensitive hemagglutinin type IV pilus | [51] |
| ICP1 | Vibrio Cholera | O1 antigen | [52] |
| PP01 | Escherichia coli O157:H7 (carried by some amphibians, fish and invertebrates) | OmpC | [53] |
| ᶲV10 | Escherichia coli O157:H7 | O157 antigen | [54] |
| P22 | Salmonella Typhimurium (found in intestinal tract of humans) | O-antigen of LPS | [55] |
| 9NA | Salmonella Typhimurium | O-antigen of LPS | [56] |
| F336 | Campylobacter jejuni (found in contaminated food and water) | O-methyl phosphoramidate (MeOPN) | [57] |
| F341 | Campylobacter jejuni | Flagellum | [58] |
| JG004 | Pseudomonas aeruginosa (found in soil and contaminated water) | O-antigen of LPS | [59] |
| Phage K8 | Pseudomonas aeruginosa | O-antigen of LPS | [60] |
| Receptors Localized on the Surface of Gram-Positive Bacteria | |||
| Gamma Phage | Bacillusanthracis (found in soil and often infects livestock) | GamR (LPXTG-harboring protein) | [61] |
| AP50c | Bacillusanthracis | CsaB | [62] |
| ᶲ11 | Staphylococcus aureus (found on skin and mucous layers of human and animals) | wall teichoic acids (WTA) | [63] |
| ᶲSLT | Staphylococcus aureus | lipoteichoic acids (LTA) | [64] |
| A118 | Listeria monocytogenes | Rhamnose residues in WTA | [48] |
| P35 | Listeria monocytogenes | Rhamnose and N-acetylglucosamine | [48] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stone, E.; Campbell, K.; Grant, I.; McAuliffe, O. Understanding and Exploiting Phage–Host Interactions. Viruses 2019, 11, 567. https://doi.org/10.3390/v11060567
Stone E, Campbell K, Grant I, McAuliffe O. Understanding and Exploiting Phage–Host Interactions. Viruses. 2019; 11(6):567. https://doi.org/10.3390/v11060567
Chicago/Turabian StyleStone, Edel, Katrina Campbell, Irene Grant, and Olivia McAuliffe. 2019. "Understanding and Exploiting Phage–Host Interactions" Viruses 11, no. 6: 567. https://doi.org/10.3390/v11060567
APA StyleStone, E., Campbell, K., Grant, I., & McAuliffe, O. (2019). Understanding and Exploiting Phage–Host Interactions. Viruses, 11(6), 567. https://doi.org/10.3390/v11060567






