Rice Dwarf Virus Small RNA Profiles in Rice and Leafhopper Reveal Distinct Patterns in Cross-Kingdom Hosts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rice Plant Growth, Insect Raising, and Cell Culture
2.2. Virus Inoculation
2.3. Small RNA Deep Sequencing
2.4. Bioinformatics Analysis
2.5. RNA Northern Blotting
2.6. Accession Numbers
2.7. RNA Preparation and qPCR Analysis
3. Results
3.1. RDV Infection Triggers Abundant Production of vsiRNAs in Rice and Leafhopper
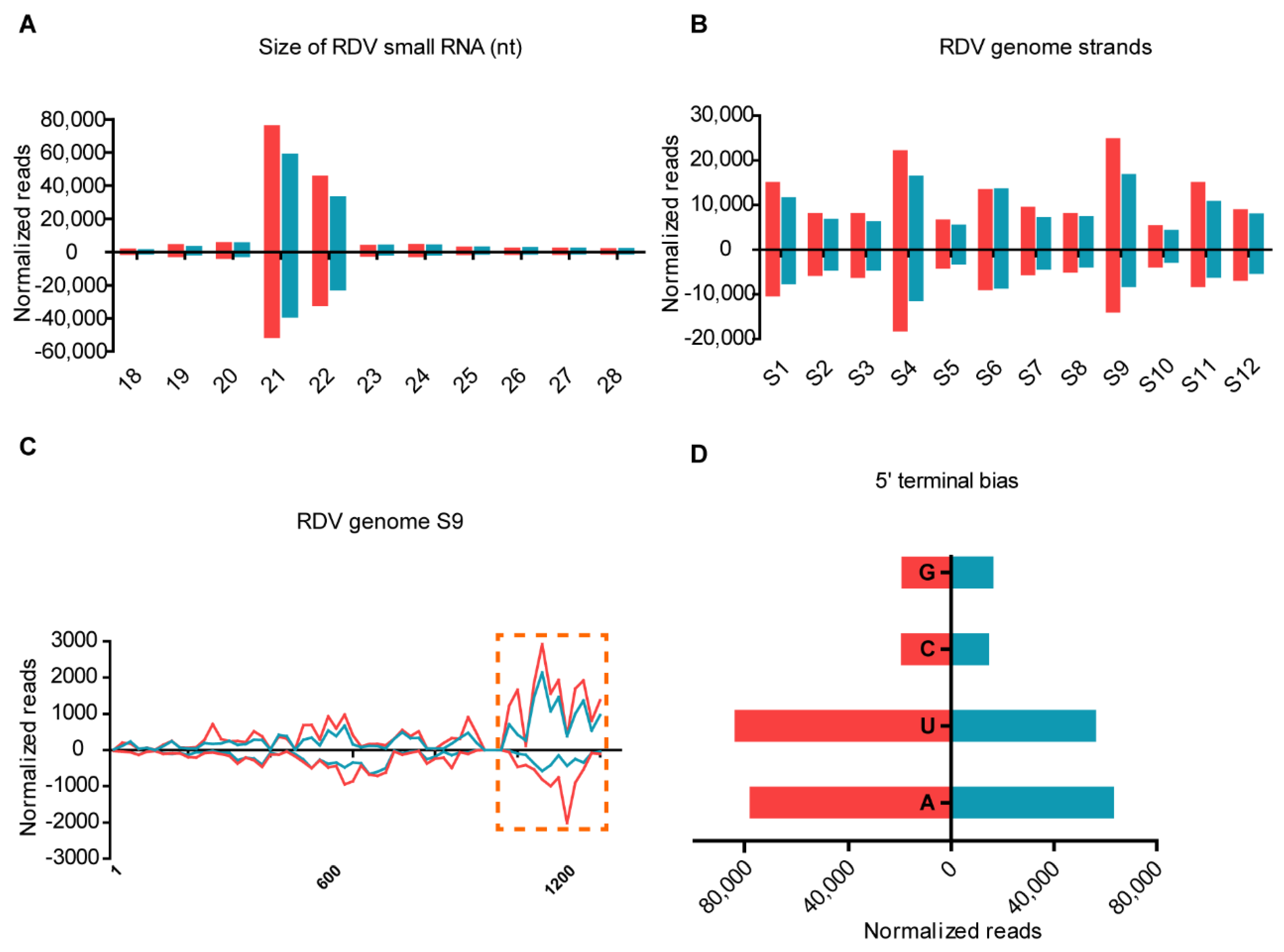
3.2. vsiRNAs Are Differentially Produced in Leafhopper and Rice Response to RDV Infection
3.3. Distribution of vsiRNAs on RDV Genomic RNAs from RDV-Infected Leafhopper and Rice Hosts
3.4. Characterization of Virus-Derived Small RNAs in Cross-Kingdom Hosts Infected with RDV
3.5. OsRDR6 Is Involved in RDV-Derived Small RNAs Production in Rice Plants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, Z.; Li, Y.; Ding, S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, Y.; Zhang, Y.; Zhou, H.; Deng, Y.Q.; Li, X.F.; Miao, M.; Zhang, Q.; Zhong, B.; Hu, Y.; et al. Human Virus-Derived Small RNAs Can Confer Antiviral Immunity in Mammals. Immunity 2018, 49, 780–781. [Google Scholar] [CrossRef]
- Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef]
- Llave, C. Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 2010, 15, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.F.; Wang, X.B.; Ding, S.W. Viral suppressors of RNA-based viral immunity: Host targets. Cell Host Microbe 2010, 8, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.S.; Zhou, P.; Zhang, X.M.; Zhu, S.F.; Zhong, X.H.; Xiao, Q.; Ding, B.; Li, Y. Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J. Virol. 2005, 79, 13018–13027. [Google Scholar] [CrossRef]
- Li, H.; Li, W.X.; Ding, S.W. Induction and suppression of RNA silencing by an animal virus. Science 2002, 296, 1319–1321. [Google Scholar] [CrossRef]
- Li, Y.; Lu, J.; Han, Y.; Fan, X.; Ding, S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013, 342, 231–234. [Google Scholar] [CrossRef]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T.; et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants 2017, 3, 16203. [Google Scholar] [CrossRef]
- Zhao, S.; Hong, W.; Wu, J.; Wang, Y.; Ji, S.; Zhu, S.; Wei, C.; Zhang, J.; Li, Y. A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Li, Y. Rice Reoviruses in Insect Vectors. Annu. Rev. Phytopathol. 2016, 54, 99–120. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Correction. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 2015, 27, 944–945. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Qian, D.; Zheng, H.; Meng, L.Y.; Chen, J.; Le, W.J.; Zhou, T.; Zhou, Y.J.; Wei, C.H.; Li, Y. RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Res. 2012, 163, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Qian, D.; Sun, R.; Jiang, L.; Wang, Y.; Wei, C.; Zhang, Z.; Li, Y. OsRDR6 plays role in host defense against double-stranded RNA virus, Rice Dwarf Phytoreovirus. Sci. Rep. 2015, 5, 11324. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gao, F.; Cao, X.; Chen, M.; Ye, G.; Wei, C.; Li, Y. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 2005, 139, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Qin, Q.; Wang, Y.; Pu, Y.; Liu, L.; Wen, X.; Ji, S.; Wu, J.; Wei, C.; Ding, B.; et al. Rice Dwarf Virus P2 Protein Hijacks Auxin Signaling by Directly Targeting the Rice OsIAA10 Protein, Enhancing Viral Infection and Disease Developmen. PLoS Pathog 2016, 12, e1005847. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Guo, Y.; Gao, F.; Zhou, P.; Wu, F.; Meng, Z.; Wei, C.; Li, Y. Multiple functions of Rice dwarf phytoreovirus Pns10 in suppressing systemic RNA silencing. J. Virol. 2010, 84, 12914–12923. [Google Scholar] [CrossRef]
- Zhou, F.; Pu, Y.; Wei, T.; Liu, H.; Deng, W.; Wei, C.; Ding, B.; Omura, T.; Li, Y. The P2 capsid protein of the nonenveloped rice dwarf phytoreovirus induces membrane fusion in insect host cells. Proc. Natl. Acad. Sci. USA 2007, 104, 19547–19552. [Google Scholar] [CrossRef] [Green Version]
- Kimura, I. A study of Rice dwarf virus in vector cell monolayers by fluorescent-antibody focus counting. J. Gen. Virol. 1986, 67, 2119–2124. [Google Scholar] [CrossRef]
- Du, P.; Wu, J.G.; Zhang, J.Y.; Zhao, S.Q.; Zheng, H.; Gao, G.; Wei, L.P.; Li, Y. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011, 7, e1002176. [Google Scholar] [CrossRef]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. Elife 2015, 4. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, S.; Navarro, B.; Gisel, A.; Pena, L.; Navarro, L.; Moreno, P.; Di Serio, F.; Flores, R. Citrus tristeza virus infection induces the accumulation of viral small RNAs (21-24-nt) mapping preferentially at the 3’-terminal region of the genomic RNA and affects the host small RNA profile. Plant Mol. Biol. 2011, 75, 607–619. [Google Scholar] [CrossRef]
- Aliyari, R.; Wu, Q.F.; Li, H.W.; Wang, X.H.; Li, F.; Green, L.D.; Han, C.S.; Li, W.X.; Ding, S.W. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 2008, 4, 387–397. [Google Scholar] [CrossRef]
- Myles, K.M.; Wiley, M.R.; Morazzani, E.M.; Adelman, Z.N. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. USA 2008, 105, 19938–19943. [Google Scholar] [CrossRef] [Green Version]
- Scott, J.C.; Brackney, D.E.; Campbell, C.L.; Bondu-Hawkins, V.; Hjelle, B.; Ebel, G.D.; Olson, K.E.; Blair, C.D. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl. Trop. Dis. 2010, 4, e848. [Google Scholar] [CrossRef]
- Lan, H.; Wang, H.; Chen, Q.; Chen, H.; Jia, D.; Mao, Q.; Wei, T. Small interfering RNA pathway modulates persistent infection of a plant virus in its insect vector. Sci. Rep. 2016, 6, 20699. [Google Scholar] [CrossRef] [Green Version]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.S.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef]
- Liu, B.; Li, P.C.; Li, X.; Liu, C.Y.; Cao, S.Y.; Chu, C.C.; Cao, X.F. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005, 139, 296–305. [Google Scholar] [CrossRef]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 2008, 9, 451. [Google Scholar] [CrossRef]
- Tomari, Y.; Du, T.; Zamore, P.D. Sorting of Drosophila small silencing RNAs. Cell 2007, 130, 299–308. [Google Scholar] [CrossRef]
- Steiner, F.A.; Hoogstrate, S.W.; Okihara, K.L.; Thijssen, K.L.; Ketting, R.F.; Plasterk, R.H.; Sijen, T. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2007, 14, 927–933. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef]
- Mourrain, P.; Beclin, C.; Elmayan, T.; Feuerbach, F.; Godon, C.; Morel, J.B.; Jouette, D.; Lacombe, A.M.; Nikic, S.; Picault, N.; et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 2000, 101, 533–542. [Google Scholar] [CrossRef]
- Qi, X.P.; Bao, F.S.; Xie, Z.X. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One 2009, 4, e4971. [Google Scholar] [CrossRef]
- Qu, F.; Ye, X.H.; Hou, G.C.; Sato, S.; Clemente, T.E.; Morris, T.J. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 2005, 79, 15209–15217. [Google Scholar] [CrossRef]
- Vaistij, F.E.; Jones, L. Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol. 2009, 149, 1399–1407. [Google Scholar] [CrossRef]
- McDonald, S.M.; Patton, J.T. Core-Associated Genome Replication Mechanisms of dsRNA Viruses. In Viral Genome Replication; Cameron, C.E., Gotte, M., Raney, K.D., Eds.; Springer Science+Business Media: New York, NY, USA, 2009; pp. 201–224. [Google Scholar]
- Susi, P.; Hohkuri, M.; Wahlroos, T.; Kilby, N.J. Characteristics of RNA silencing in plants: Similarities and differences across kingdoms. Plant Mol. Biol. 2004, 54, 157–174. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vazquez, F.; Blevins, T.; Farinelli, L.; et al. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog 2012, 8, e1002941. [Google Scholar] [CrossRef]
- Dalmay, T.; Hamilton, A.; Rudd, S.; Angell, S.; Baulcombe, D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 2000, 101, 543–553. [Google Scholar] [CrossRef]
- Xie, Z.; Fan, B.; Chen, C.; Chen, Z. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA 2001, 98, 6516–6521. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.J.; Carter, S.A.; Cole, A.B.; Cheng, N.H.; Nelson, R.S. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2004, 101, 6297–6302. [Google Scholar] [CrossRef]
- Wang, X.B.; Wu, Q.; Ito, T.; Cillo, F.; Li, W.X.; Chen, X.; Yu, J.L.; Ding, S.W. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 484–489. [Google Scholar] [CrossRef]
- Qu, F. Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. Mol. Plant Microbe Interact 2010, 23, 1248–1252. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Z.; Song, X.; Liu, C.; Cui, X.; Zhao, X.; Fang, J.; Xu, W.; Zhang, H.; Wang, X.; et al. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 2007, 19, 2705–2718. [Google Scholar] [CrossRef]
- Urayama, S.; Moriyama, H.; Aoki, N.; Nakazawa, Y.; Okada, R.; Kiyota, E.; Miki, D.; Shimamoto, K.; Fukuhara, T. Knock-down of OsDCL2 in rice negatively affects maintenance of the endogenous dsRNA virus, Oryza sativa endornavirus. Plant Cell Physiol 2010, 51, 58–67. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef]
- Pu, Y.Y.; Kikuchi, A.; Moriyasu, Y.; Tomaru, M.; Jin, Y.; Suga, H.; Hagiwara, K.; Akita, F.; Shimizu, T.; Netsu, O.; et al. Rice dwarf viruses with dysfunctional genomes generated in plants are filtered out in vector insects: Implications for the origin of the virus. J. Virol. 2011, 85, 2975–2979. [Google Scholar] [CrossRef]




| Unique Sequences a | Total Reads a | Unique RDV vsiRNA a, b | Total RDV vsiRNA Reads d (% of Total Reads) a, b | (+)-Strands (%) a, c | (−)-Strands (%) a, d | ||
|---|---|---|---|---|---|---|---|
| Leafhopper | Replicate 1 | 1,322,051 | 6,998,353 | 12,212 | 44,014 (0.63%) | 56.60% | 43.40% |
| Replicate 2 | 1,585,173 | 7,514,114 | 15,017 | 47,476 (0.63%) | 56.73% | 43.27% | |
| VCM | Replicate 1 | 1,360,138 | 8,766,394 | 9,292 | 35,897 (0.41%) | 56.34% | 43.66% |
| Replicate 2 | 1,219,667 | 7,837,090 | 8,394 | 36,411 (0.46%) | 58.83% | 41.17% | |
| Rice | Replicate 1 | 2,610,966 | 6,999,547 | 144,347 | 1,637,665 (23.40%) | 60.49% | 39.51% |
| Replicate 2 | 3,143,474 | 9,214,996 | 164,829 | 2,164,499 (23.49%) | 61.14% | 38.86% | |
| OsRDR6AS | Replicate 1 | 1,996,171 | 6,283,390 | 112,334 | 1,112,734 (17.71%) | 62.69% | 37.31% |
| Replicate 2 | 1,924,762 | 5,408,334 | 100,218 | 1,069,626 (19.78%) | 62.90% | 37.10% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Qiao, R.; Wei, C.; Li, Y. Rice Dwarf Virus Small RNA Profiles in Rice and Leafhopper Reveal Distinct Patterns in Cross-Kingdom Hosts. Viruses 2019, 11, 847. https://doi.org/10.3390/v11090847
Wang Y, Qiao R, Wei C, Li Y. Rice Dwarf Virus Small RNA Profiles in Rice and Leafhopper Reveal Distinct Patterns in Cross-Kingdom Hosts. Viruses. 2019; 11(9):847. https://doi.org/10.3390/v11090847
Chicago/Turabian StyleWang, Yu, Rui Qiao, Chunhong Wei, and Yi Li. 2019. "Rice Dwarf Virus Small RNA Profiles in Rice and Leafhopper Reveal Distinct Patterns in Cross-Kingdom Hosts" Viruses 11, no. 9: 847. https://doi.org/10.3390/v11090847
APA StyleWang, Y., Qiao, R., Wei, C., & Li, Y. (2019). Rice Dwarf Virus Small RNA Profiles in Rice and Leafhopper Reveal Distinct Patterns in Cross-Kingdom Hosts. Viruses, 11(9), 847. https://doi.org/10.3390/v11090847





