Interaction of a Densovirus with Glycans of the Peritrophic Matrix Mediates Oral Infection of the Lepidopteran Pest Spodoptera frugiperda
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Virus Preparation
2.2. Calcofluor, Glycans and Lectins
2.3. Insect Bioassays
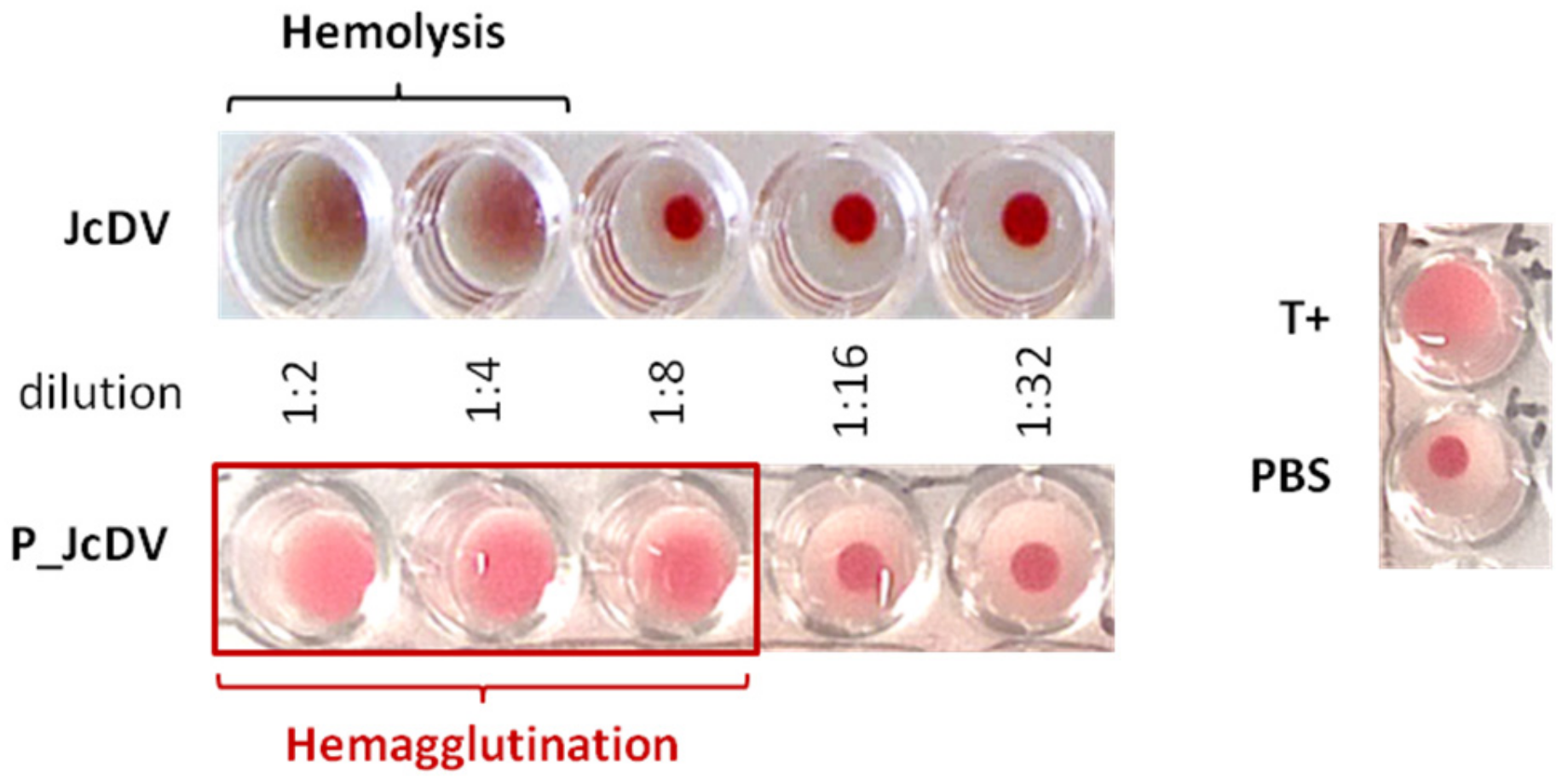
2.4. Hemagglutination Assay
2.5. PM Isolation, Ex Vivo Infection and Microscopic Observation
2.6. Midgut Semi-Thin Sections and Immunolabelling
2.7. Chitin Binding Assay and Pull Down
2.8. SDS-PAGE, PAS Staining and VOPBA
2.9. Proteomic LC-MS/MS Analysis and Data Processing
2.10. RNA Extraction, DGE Library Construction and Sequencing
2.11. Transcriptomic Analysis of the Midgut
3. Results
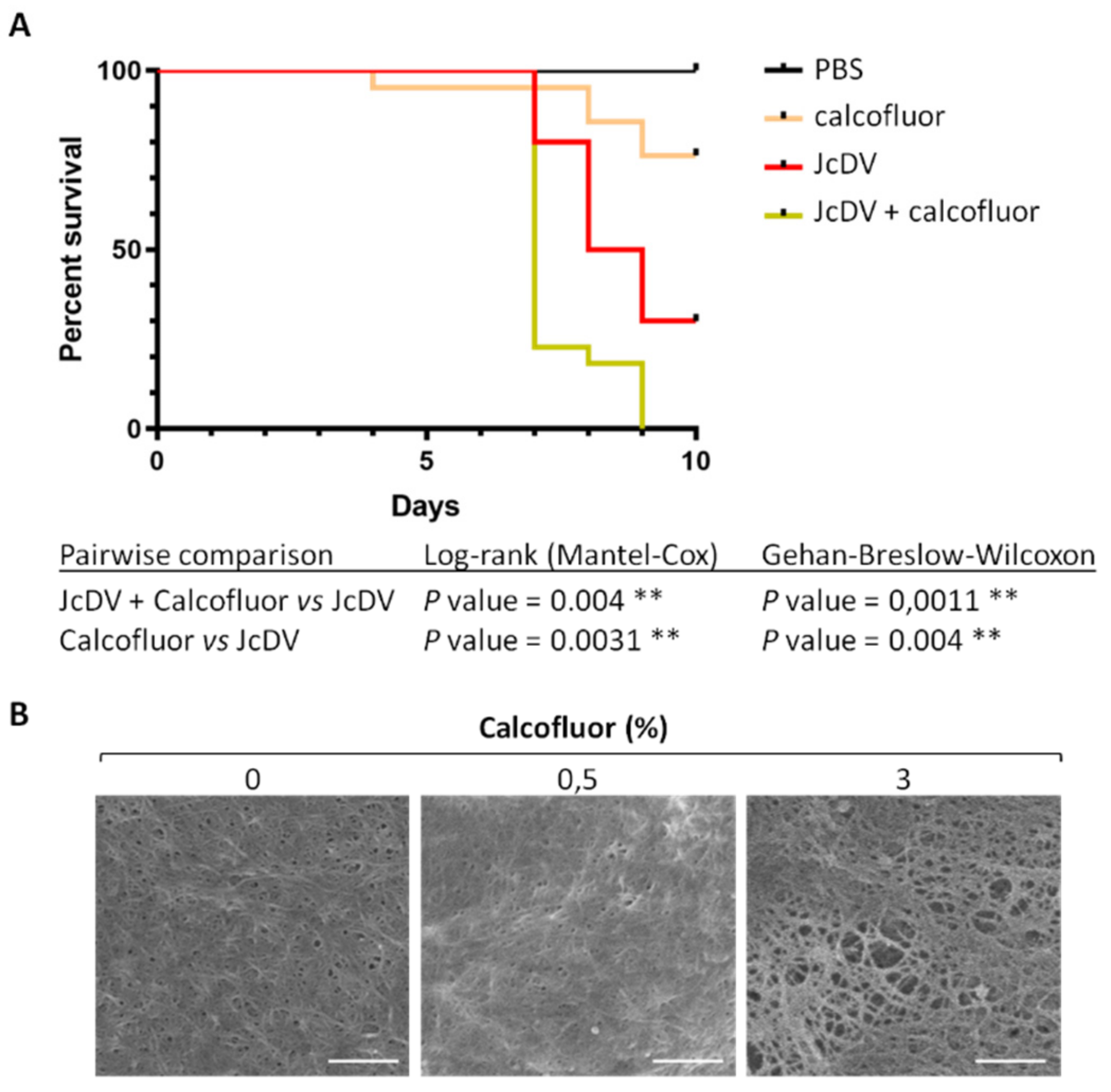
3.1. The Peritrophic Matrix of S. frugiperda is a Barrier to JcDV Infection
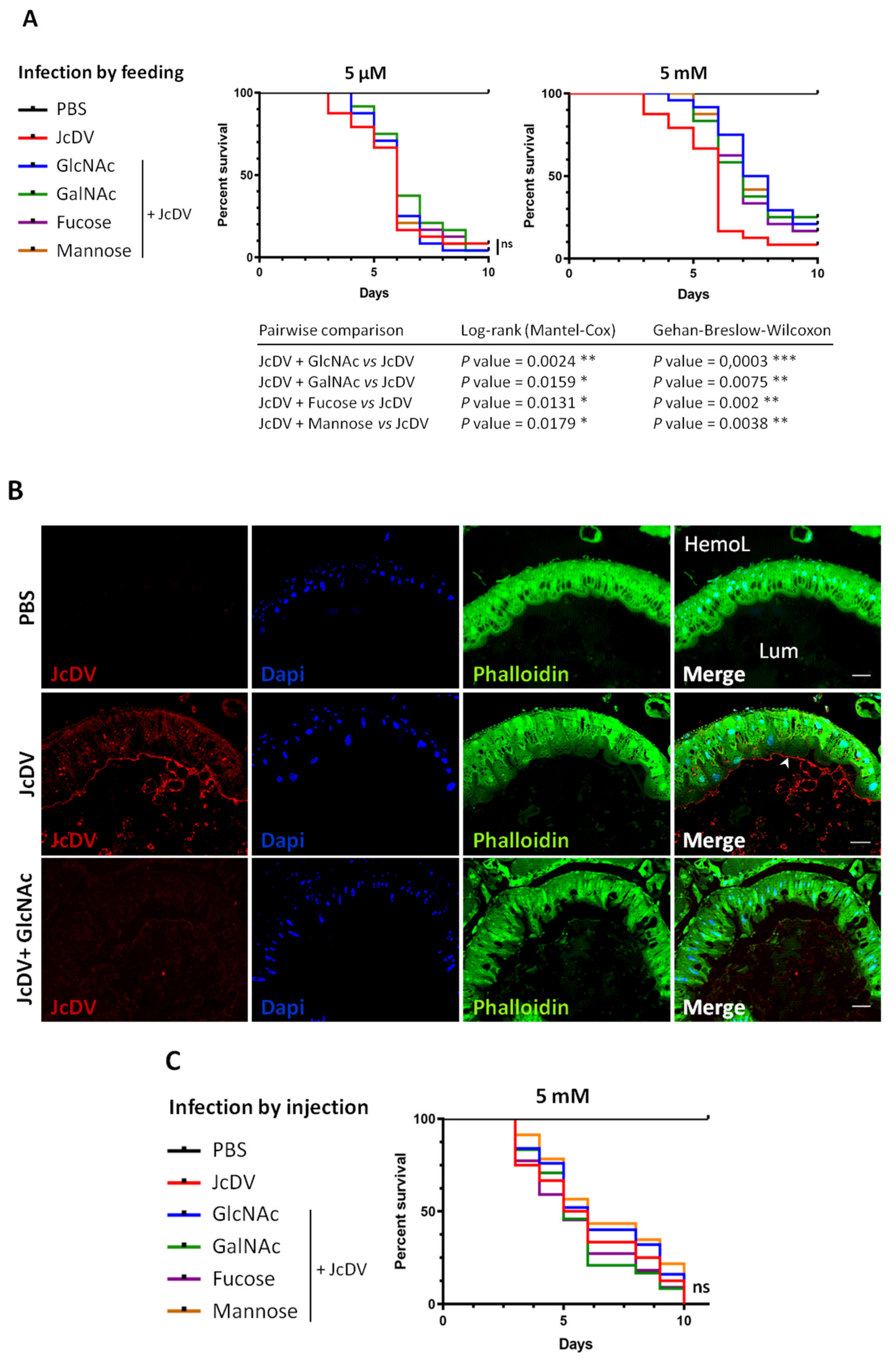
3.2. JcDV Binding to the PM is Required for Oral Infection of S. frugiperda Caterpillars
3.3. JcDV Binds to Both Chitin and Protein Components of the PM
3.4. JcDV Early Pathogenesis Induces Changes in Midgut Metabolism
4. Discussion
4.1. JcDV Recognition and Binding to Glycans
4.2. The Role of PM Glycans in the Species Barrier to Densovirus Infection
4.3. JcDV Early Infection Induces Physiological Changes in the Midgut
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Qiu, J.; Söderlund-Venermo, M.; Young, N.S. Human Parvoviruses. Clin. Microbiol. Rev. 2017, 30, 43–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- France, M.M.; Turner, J.R. The mucosal barrier at a glance. J. Cell Sci. 2017, 130, 307–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stavolone, L.; Lionetti, V. Extracellular Matrix in Plants and Animals: Hooks and Locks for Viruses. Front. Microbiol. 2017, 8, 1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Pendu, J.; Nyström, K.; Ruvoën-Clouet, N. Host-pathogen co-evolution and glycan interactions. Curr. Opin. Virol. 2014, 7, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Agbandje-McKenna, M.; Chiorini, J.A.; Mukha, D.V.; Pintel, D.J.; Qiu, J.; Soderlund-Venermo, M.; Tattersall, P.; Tijssen, P.; Gatherer, D.; et al. The family Parvoviridae. Arch. Virol. 2014, 159, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Mietzsch, M.; Pénzes, J.J.; Agbandje-McKenna, M. Twenty-Five Years of Structural Parvovirology. Viruses 2019, 11, 362. [Google Scholar] [CrossRef] [PubMed]
- Pillay, S.; Meyer, N.L.; Puschnik, A.S.; Davulcu, O.; Diep, J.; Ishikawa, Y.; Jae, L.T.; Wosen, J.E.; Nagamine, C.M.; Chapman, M.S.; et al. An essential receptor for adeno-associated virus infection. Nature 2016, 530, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Goodman, L.B.; Lyi, S.M.; Johnson, N.C.; Cifuente, J.O.; Hafenstein, S.L.; Parrish, C.R. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J. Virol. 2010, 84, 4969–4978. [Google Scholar] [CrossRef]
- Di Pasquale, G.; Chiorini, J.A. AAV transcytosis through barrier epithelia and endothelium. Mol. Ther. 2006, 13, 506–516. [Google Scholar] [CrossRef]
- Walters, R.W.; Pilewski, J.M.; Chiorini, J.A.; Zabner, J. Secreted and Transmembrane Mucins Inhibit Gene Transfer with AAV4 More Efficiently than AAV5. J. Biol. Chem. 2002, 277, 23709–23713. [Google Scholar] [CrossRef] [Green Version]
- Ito, K.; Kidokoro, K.; Shimura, S.; Katsuma, S.; Kadono-Okuda, K. Detailed investigation of the sequential pathological changes in silkworm larvae infected with Bombyx densovirus type 1. J. Invertebr. Pathol. 2013, 112, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gosselin Grenet, A.S.; Castelli, I.; Cermenati, G.; Ravallec, M.; Fiandra, L.; Debaisieux, S.; Multeau, C.; Lautredou, N.; Dupressoir, T.; et al. Densovirus crosses the insect midgut by transcytosis and disturbs the epithelial barrier function. J. Virol. 2013, 87, 12380–12391. [Google Scholar] [CrossRef] [PubMed]
- Mutuel, D.; Ravallec, M.; Chabi, B.; Multeau, C.; Salmon, J.M.; Fournier, P.; Ogliastro, M. Pathogenesis of Junonia coenia densovirus in Spodoptera frugiperda: A route of infection that leads to hypoxia. Virology 2010, 403, 137–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tijssen, P.; Pénzes, J.J.; Yu, Q.; Pham, H.T.; Bergoin, M. Diversity of small, single-stranded DNA viruses of invertebrates and their chaotic evolutionary past. J. Invertebr. Pathol. 2016, 140, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Kidokoro, K.; Katsuma, S.; Sezutsu, H.; Uchino, K.; Kobayashi, I.; Tamura, T.; Yamamoto, K.; Mita, K.; Shimada, T.; et al. A single amino acid substitution in the Bombyx-specific mucin-like membrane protein causes resistance to Bombyx mori densovirus. Sci. Rep. 2018, 8, 7430. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, D.; Erlandson, M.; Gillott, C.; Toprak, U. New insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 2009, 54, 285–302. [Google Scholar] [CrossRef] [PubMed]
- Dias, R.O.; Cardoso, C.; Pimentel, A.C.; Damasceno, T.F.; Ferreira, C.; Terra, W.R. The roles of mucus-forming mucins, peritrophins and peritrophins with mucin domains in the insect midgut. Insect Mol. Biol. 2018, 27, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Terra, W.R. The origin and functions of the insect peritrophic membrane and peritrophic gel. Arch. Insect Biochem. Physiol. 2001, 47, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Kawakita, H.; Miyamoto, K.; Wada, S.; Mitsuhashi, W. Analysis of the ultrastructure and formation pattern of the peritrophic membrane in the cupreous chafer, Anomala cuprea (Coleoptera: Scarabaeidae). Appl. Entomol. Zool. 2016, 51, 133–142. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C. Insect digestive enzymes: Properties, compartmentalization and function. Comp. Biochem. Physiol. Part B Comp. Biochem. 1994, 109, 1–62. [Google Scholar] [CrossRef]
- Chiu, E.; Hijnen, M.; Bunker, R.D.; Boudes, M.; Rajendran, C.; Aizel, K.; Olieric, V.; Schulze-Briese, C.; Mitsuhashi, W.; Young, V.; et al. Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization. Proc. Natl. Acad. Sci. USA 2015, 112, 3973–3978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passarelli, A.L. Barriers to success: How baculoviruses establish efficient systemic infections. Virology 2011, 411, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsuhashi, W.; Miyamoto, K. Disintegration of the peritrophic membrane of silkworm larvae due to spindles of an entomopoxvirus. J. Invertebr. Pathol. 2003, 82, 34–40. [Google Scholar] [CrossRef]
- Mitsuhashi, W.; Shimura, S.; Miyamoto, K.; Sugimoto, T.N. Spatial distribution of orally administered viral fusolin protein in the insect midgut and possible synergism between fusolin and digestive proteases to disrupt the midgut peritrophic matrix. Arch. Virol. 2019, 164, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Koonin, E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA 2017, 114, E2401–E2410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Jousset, F.X.; Giraud, C.; Rolling, F.; Quiot, J.M.; Bergoin, M. A titration procedure of the Junonia coenia densovirus and quantitation of transfection by its cloned genomic DNA in four lepidopteran cell lines. J. Virol. Methods 1996, 57, 47–60. [Google Scholar] [CrossRef]
- Shi, X.; Chamankhah, M.; Visal-Shah, S.; Hemmingsen, S.M.; Erlandson, M.; Braun, L.; Alting-Mees, M.; Khachatourians, G.G.; O’Grady, M.; Hegedus, D.D. Modeling the structure of the type I peritrophic matrix: Characterization of a Mamestra configurata intestinal mucin and a novel peritrophin containing 19 chitin binding domains. Insect Biochem. Mol. Biol. 2004, 34, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Zacharius, R.M.; Zell, T.E.; Morrison, J.H.; Woodlock, J.J. Glycoprotein staining following electrophoresis on acrylamide gels. Anal. Biochem. 1969, 30, 148–152. [Google Scholar] [CrossRef]
- Edmondson, D.G.; Dent, S.Y. Identification of protein interactions by far western analysis. Curr. Protoc. Protein Sci. 2001, 55, 20.6.1–20.6.10. [Google Scholar] [CrossRef]
- Thouvenot, E.; Urbach, S.; Dantec, C.; Poncet, J.; Séveno, M.; Demettre, E.; Jouin, P.; Touchon, J.; Bockaert, J.; Marin, P. Enhanced detection of CNS cell secretome in plasma protein-depleted cerebrospinal fluid. J. Proteome Res. 2008, 7, 4409–4421. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; de Godoy, L.M.F.; Li, G.; Macek, B.; Mortensen, P.; Pesch, R.; Makarov, A.; Lange, O.; Horning, S.; Mann, M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteom. 2005, 4, 2010–2021. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.-M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagné, N.; Darboux, I.; et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.L.; Hogh, A.L.; Emmersen, J. DeepSAGE--digital transcriptomics with high sensitivity, simple experimental protocol and multiplexing of samples. Nucleic Acids Res. 2006, 34, e133. [Google Scholar] [CrossRef] [PubMed]
- De Lorgeril, J.; Zenagui, R.; Rosa, R.D.; Piquemal, D.; Bachere, E. Whole transcriptome profiling of successful immune response to Vibrio infections in the oyster Crassostrea gigas by digital gene expression analysis. PLoS ONE 2011, 6, e23142. [Google Scholar] [CrossRef]
- Velculescu, V.E.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Serial analysis of gene expression. Science 1995, 270, 484–487. [Google Scholar] [CrossRef]
- Legeai, F.; Gimenez, S.; Duvic, B.; Escoubas, J.M.; Gosselin Grenet, A.S.; Blanc, F.; Cousserans, F.; Seninet, I.; Bretaudeau, A.; Mutuel, D.; et al. Establishment and analysis of a reference transcriptome for Spodoptera frugiperda. BMC Genom. 2014, 15, 704. [Google Scholar] [CrossRef]
- Ferreira, C.; Capella, A.N.; Sitnik, R.; Terra, W.R. Properties of the digestive enzymes and the permeability of the peritrophic membrane of Spodoptera frugiperda (Lepidoptera) larvae. Comp. Biochem. Physiol. 1994, 107A, 631–640. [Google Scholar] [CrossRef]
- Herth, W. Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: Evidence for a gap between polymerization and microfibril formation. J. Cell Biol. 1980, 87, 442–450. [Google Scholar] [CrossRef]
- Merzendorfer, H. Chitin synthesis inhibitors: Old molecules and new developments. Insect Sci. 2013, 20, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Granados, R.R. Calcofluor disrupts the midgut defense system in insects. Insect Biochem. Mol. Biol. 2000, 30, 135–143. [Google Scholar] [CrossRef]
- Zhu, K.Y.; Merzendorfer, H.; Zhang, W.; Zhang, J.; Muthukrishnan, S. Biosynthesis, Turnover, and Functions of Chitin in Insects. Annu. Rev. Entomol. 2016, 61, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Liu, K.; Peng, J.; Yang, H.; Hong, H. Optical brightener M2R destroys the peritrophic membrane of Spodoptera exigua (Lepidoptera: Noctuidae) larvae. Pest Manag. Sci. 2007, 63, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Agbandje-McKenna, M.; Llamas-Saiz, A.L.; Wang, F.; Tattersall, P.; Rossmann, M.G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 1998, 6, 1369–1381. [Google Scholar] [CrossRef] [Green Version]
- Tresnan, D.B.; Southard, L.; Weichert, W.; Sgro, J.Y.; Parrish, C.R. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology 1995, 211, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Walski, T.; De Schutter, K.; Van Damme, E.J.M.; Smagghe, G. Diversity and functions of protein glycosylation in insects. Insect Biochem. Mol. Biol. 2017, 83, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Salasc, F.; Mutuel, D.; Debaisieux, S.; Perrin, A.; Dupressoir, T.; Grenet, A.S.; Ogliastro, M. Role of the phosphatidylinositol-3-kinase/Akt/target of rapamycin pathway during ambidensovirus infection of insect cells. J. Gen. Virol. 2016, 97, 233–245. [Google Scholar] [CrossRef]
- Delmas, B.; Gelfi, J.; Sjöström, H.; Noren, O.; Laude, H. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 1993, 342, 293–298. [Google Scholar] [PubMed]
- Li, B.X.; Ge, J.W.; Li, Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007, 365, 166–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linz, L.B.; Liu, S.; Chougule, N.P.; Bonning, B.C. In Vitro Evidence Supports Membrane Alanyl Aminopeptidase N as a Receptor for a Plant Virus in the Pea Aphid Vector. J. Virol. 2015, 89, 11203–11212. [Google Scholar] [CrossRef] [PubMed]
- Tresnan, D.B.; Levis, R.; Holmes, K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996, 70, 8669–8674. [Google Scholar] [PubMed]
- Harper, M.S.; Hopkins, T.L.; Czapla, T.H. Effect of wheat germ agglutinin on formation and structure of the peritrophic membrane in European corn borer (Ostrinia nubilalis) larvae. Tissue Cell 1998, 30, 166–176. [Google Scholar] [CrossRef]
- Martinez, A.-M.; Simon, O.; Williams, T.; Caballero, P. Effect of optical brighteners on the insecticidal activity of a nucleopolyhedrovirus in three instars of Spodoptera frugiperda. Entomol. Exp. Appl. 2003, 109, 139–146. [Google Scholar] [CrossRef]
- Chen, E.; Kolosov, D.; O’Donnell, M.J.; Erlandson, M.A.; McNeil, J.N.; Donly, C. The Effect of Diet on Midgut and Resulting Changes in Infectiousness of AcMNPV Baculovirus in the Cabbage Looper, Trichoplusia ni. Front. Physiol. 2018, 9, 1348. [Google Scholar] [CrossRef] [PubMed]
- Ros, C.; Bayat, N.; Wolfisberg, R.; Almendral, J.M. Protoparvovirus Cell Entry. Viruses 2017, 9, 313. [Google Scholar] [CrossRef]
- López-Bueno, A.; Rubio, M.-P.; Bryant, N.; McKenna, R.; Agbandje-McKenna, M.; Almendral, J.M. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 2006, 80, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.B.; Kohler, D.J.; Ortega, A.; Hoover, E.A.; Grove, D.M.; Holmes, E.C.; Parrish, C.R. Host-Specific Parvovirus Evolution in Nature Is Recapitulated by In Vitro Adaptation to Different Carnivore Species. PLoS Pathog. 2014, 10, e1004475. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.Y.; Halder, S.; Agbandje-McKenna, M. Parvovirus glycan interactions. Curr. Opin. Virol. 2014, 7, 108–118. [Google Scholar] [CrossRef] [Green Version]
- Multeau, C.; Froissart, R.; Perrin, A.; Castelli, I.; Casartelli, M.; Ogliastro, M. Four amino acids of an insect densovirus capsid determine midgut tropism and virulence. J. Virol. 2012, 86, 5937–5941. [Google Scholar] [CrossRef]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, F.H.; Gendrin, M.; Wyer, C.A.S.; Christophides, G.K. Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes. PLoS Pathog. 2017, 13, e1006391. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, M.; Dong, L.; Zhu, H.; Wang, J. PGRP-LD mediates A. stephensi vector competency by regulating homeostasis of microbiota-induced peritrophic matrix synthesis. PLoS Pathog. 2018, 14, e1006899. [Google Scholar] [CrossRef] [PubMed]
- Kuss, S.K.; Best, G.T.; Etheredge, C.A.; Pruijssers, A.J.; Frierson, J.M.; Hooper, L.V.; Dermody, T.S.; Pfeiffer, J.K. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 2011, 334, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.K.; Mainou, B.A. Interactions between Enteric Bacteria and Eukaryotic Viruses Impact the Outcome of Infection. Viruses 2018, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.K.; Yi, H.; Kearns, D.B.; Mainou, B.A. Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog. 2017, 13, e1006768. [Google Scholar] [CrossRef]
- François, S.; Filloux, D.; Roumagnac, P.; Bigot, D.; Gayral, P.; Martin, D.P.; Froissart, R.; Ogliastro, M. Discovery of parvovirus-related sequences in an unexpected broad range of animals. Sci. Rep. 2016, 6, 30880. [Google Scholar] [CrossRef] [Green Version]
- Behdenna, A.; Lembo, T.; Calatayud, O.; Cleaveland, S.; Halliday, J.E.B.; Packer, C.; Lankester, F.; Hampson, K.; Craft, M.E.; Czupryna, A.; et al. Transmission ecology of canine parvovirus in a multi-host, multi-pathogen system. Proc. Biol. Sci. 2019, 286, 20182772. [Google Scholar] [CrossRef] [Green Version]
- Handel, A.; Akin, V.; Pilyugin, S.S.; Zarnitsyna, V.; Antia, R. How sticky should a virus be? The impact of virus binding and release on transmission fitness using influenza as an example. J. R. Soc. Interface 2014, 11, 20131083. [Google Scholar] [CrossRef]
- Kolliopoulou, A.; Taning, C.N.T.; Smagghe, G.; Swevers, L. Viral Delivery of dsRNA for Control of Insect Agricultural Pests and Vectors of Human Disease: Prospects and Challenges. Front. Physiol. 2017, 8, 399. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pigeyre, L.; Schatz, M.; Ravallec, M.; Gasmi, L.; Nègre, N.; Clouet, C.; Seveno, M.; El Koulali, K.; Decourcelle, M.; Guerardel, Y.; et al. Interaction of a Densovirus with Glycans of the Peritrophic Matrix Mediates Oral Infection of the Lepidopteran Pest Spodoptera frugiperda. Viruses 2019, 11, 870. https://doi.org/10.3390/v11090870
Pigeyre L, Schatz M, Ravallec M, Gasmi L, Nègre N, Clouet C, Seveno M, El Koulali K, Decourcelle M, Guerardel Y, et al. Interaction of a Densovirus with Glycans of the Peritrophic Matrix Mediates Oral Infection of the Lepidopteran Pest Spodoptera frugiperda. Viruses. 2019; 11(9):870. https://doi.org/10.3390/v11090870
Chicago/Turabian StylePigeyre, Laetitia, Malvina Schatz, Marc Ravallec, Leila Gasmi, Nicolas Nègre, Cécile Clouet, Martial Seveno, Khadija El Koulali, Mathilde Decourcelle, Yann Guerardel, and et al. 2019. "Interaction of a Densovirus with Glycans of the Peritrophic Matrix Mediates Oral Infection of the Lepidopteran Pest Spodoptera frugiperda" Viruses 11, no. 9: 870. https://doi.org/10.3390/v11090870
APA StylePigeyre, L., Schatz, M., Ravallec, M., Gasmi, L., Nègre, N., Clouet, C., Seveno, M., El Koulali, K., Decourcelle, M., Guerardel, Y., Cot, D., Dupressoir, T., Gosselin-Grenet, A.-S., & Ogliastro, M. (2019). Interaction of a Densovirus with Glycans of the Peritrophic Matrix Mediates Oral Infection of the Lepidopteran Pest Spodoptera frugiperda. Viruses, 11(9), 870. https://doi.org/10.3390/v11090870





