(Non-)Sense of Milk Testing in Small Ruminant Lentivirus Control Programs in Goats. Comparative Analysis of Antibody Detection and Molecular Diagnosis in Blood and Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Origin
2.2. Processing of Blood Samples
2.3. Processing of Milk Samples
2.4. Serological Analysis by ELISA
2.5. Evaluation of the Analytical Sensitivity of ELISA Tests
2.6. DNA Extraction and qPCR Analysis
2.7. Statistical Analysis
3. Results
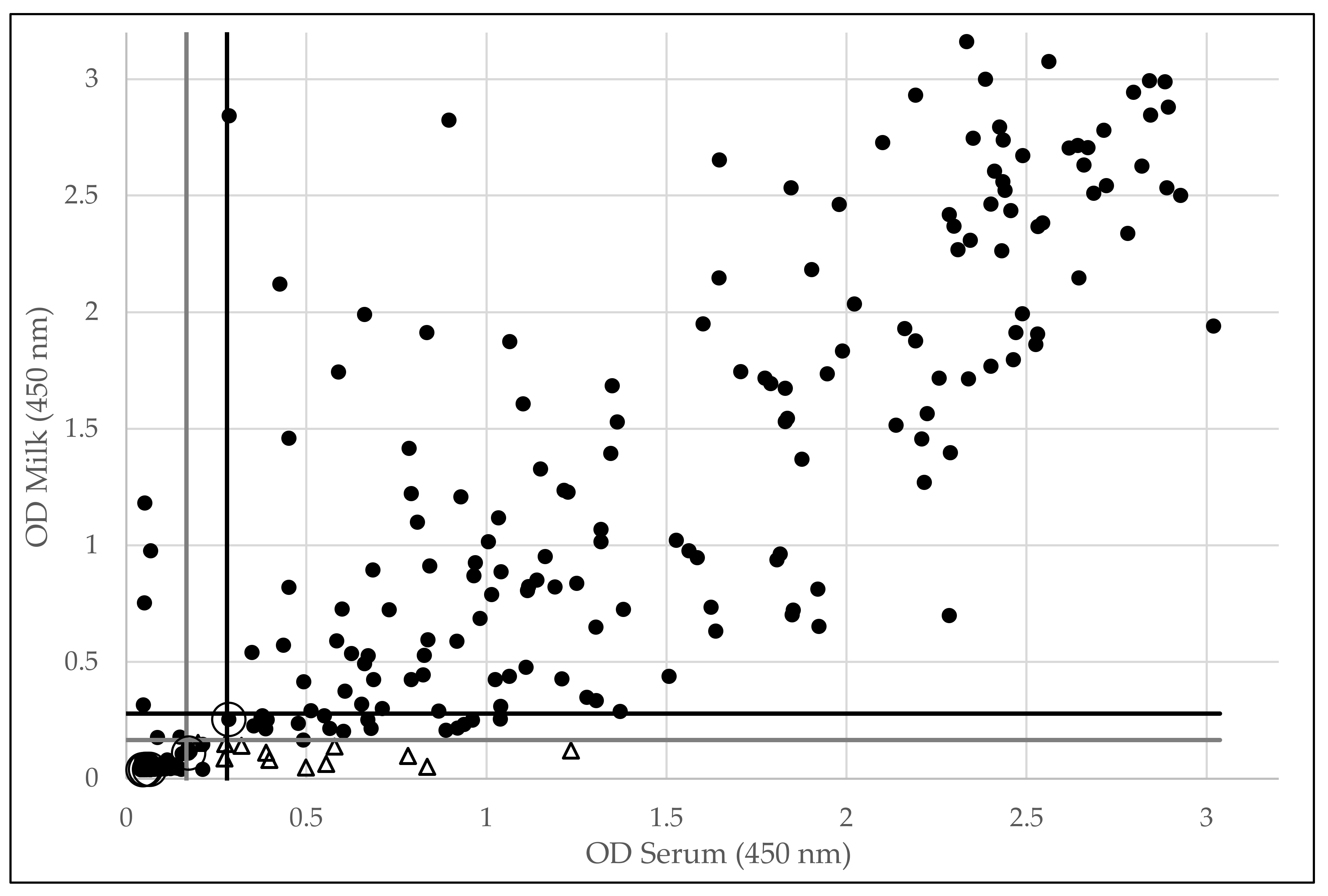
3.1. SRLV-Specific Antibody Detection in Serum and Milk via ELISA
3.2. Molecular SRLV Detection in PBMCs and Milk Cell Pellets by qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Herrmann-Hoesing, L.M. Diagnostic assays used to control small ruminant lentiviruses. J. Vet. Diagn. Invest. 2010, 22, 843–855. [Google Scholar] [CrossRef] [Green Version]
- Minguijón, E.; Reina, R.; Pérez, M.; Polledo, L.; Villoria, M.; Ramírez, H.; Leginagoikoaa, I.; Badiola, J.J.; García-Marín, J.F.; de Andrés, D.; et al. Small ruminant lentivirus infections and diseases. J. Vet. Microbiol. 2015, 181, 75–89. [Google Scholar] [CrossRef]
- De Andrés, D.; Klein, D.; Watt, N.J.; Berriatua, E.; Torsteinsdottir, S.; Blacklaws, B.A.; Harkiss, G.D. Diagnostic tests for small ruminant lentiviruses. J. Vet. Microbiol. 2005, 107, 49–62. [Google Scholar] [CrossRef]
- Blacklaws, B.A.; Berriatua, E.; Torsteinsdottir, S.; Watt, N.J.; De Andres, D.; Klein, D.; Harkiss, G.D. Transmission of small ruminant lentiviruses. J. Vet. Microbiol. 2004, 101, 199–208. [Google Scholar] [CrossRef]
- Blacklaws, B.A. Small ruminant lentiviruses: Immunopathogenesis of visna-maedi and caprine arthritis and encephalitis virus. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, H.; Reina, R.; Amorena, B.; de Andrés, D.; Martínez, H.A. Small ruminant Lentiviruses: Genetic variability, tropism and diagnosis. Viruses 2013, 5, 1175–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stonos, N.; Wootton, S.K.; Karrow, N. Immunogenetics of small ruminant lentiviral infections. Viruses 2014, 6, 3311–3333. [Google Scholar] [CrossRef] [PubMed]
- Zahno, M.L.; Bertoni, G. An immunodominant region of the envelope glycoprotein of small ruminant lentiviruses may function as decoy antigen. Viruses 2018, 10, 231. [Google Scholar] [CrossRef] [Green Version]
- Grego, E.; Reina, R.; Lanfredini, S.; Tursi, M.; Favole, A.; Profiti, M.; Lungu, M.M.; Perona, G.; Gay, L.; Stella, M.C.; et al. Viral load, tissue distribution and histopathological lesions in goats naturally and experimentally infected with the Small Ruminant Lentivirus Genotype E (subtype E1 Roccaverano strain). Res. Vet. Sci. 2018, 118, 107–114. [Google Scholar] [CrossRef]
- Pérez, M.; Biescas, E.; de Andrés, X.; Leginagoikoa, I.; Salazar, E.; Berriatua, E.; Reina, R.; Bolea, R.; de Andrés, D.; Juste, R.A.; et al. Visna/maedi virus serology in sheep: Survey, risk factors and implementation of a successful control programme in Aragón (Spain). Vet. J. 2010, 186, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Tavella, A.; Bettini, A.; Ceol, M.; Zambotto, P.; Stifter, E.; Kusstatscher, N.; Lombardi, O.; Nardeli, S.; Beato, M.S.; Capello, K.; et al. Achievements of an eradication programme against caprine arthritis encephalitis virus in South Tyrol, Italy. Vet. Rec. 2017, 182, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keen, J.E.; Hungerford, L.L.; Littledike, E.T.; Wittum, T.E.; Kwang, J. Effect of ewe ovine lentivirus infection on ewe and lamb productivity. Prev. Vet. Med. 1997, 30, 155–169. [Google Scholar] [CrossRef]
- Reina, R.; Berriatua, E.; Luján, L.; Juste, R.; Sánchez, A.; de Andrés, D.; Amorena, B. Prevention strategies against small ruminant lentiviruses: An update. Vet. J. 2009, 182, 31–37. [Google Scholar] [CrossRef] [PubMed]
- De Andrés, X.; Ramírez, H.; Bertolotti, L.; San Román, B.; Glaria, I.; Crespo, H.; Jauregui, P.; Minguijon, E.; Juste, R.; Leginagoikoa, I.; et al. An insight into a combination of ELISA strategies to diagnose small ruminant lentivirus infections. Vet. Immunol. Immunopathol. 2013, 152, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Michiels, R.; Van Mael, E.; Quinet, C.; Adjadj, N.R.; Cay, A.B.; De Regge, N. Comparative analysis of different serological and molecular tests for the detection of small ruminant lentiviruses (Srlvs) in Belgian sheep and goats. Viruses 2018, 10, 696. [Google Scholar] [CrossRef] [Green Version]
- Cortez-Moreira, M.; Oelemann, W.M.R.; Lilenbaum, W. Comparison Of Serological Methods For The Diagnostic Of Caprine Arthritis-Encephalitis (CAE) in Rio de Janeiro, Brazil. Braz. J. Microbiol. 2005, 36, 48–50. [Google Scholar] [CrossRef] [Green Version]
- Panneum, S.; Rukkwamsuk, T. Diagnosis of Caprine Arthritis Encephalitis Virus infection in dairy goats by ELISA, PCR and Viral Culture. Pol. J. Vet. Sci. 2017, 20, 347–353. [Google Scholar] [CrossRef] [Green Version]
- De Pablo-Maiso, L.; Doménech, A.; Echeverría, I.; Gómez-Arrebola, C.; de Andrés, D.; Rosati, S.; Gómez-Lucia, E.; Reina, R. Prospects in Innate Immune Responses as Potential Control Strategies against Non-Primate Lentiviruses. Viruses 2018, 10, 435. [Google Scholar] [CrossRef] [Green Version]
- Barquero, N.; Arjona, A.; Domenech, A.; Toural, C.; de Las Heras, A.; Fernández-Garayzabal, J.F.; Ruiz-Santa Quiteria, J.A.; Gomez-Lucia, E. Diagnostic performance of PCR and ELISA on blood and milk samples and serological survey for small ruminant lentiviruses in central Spain. Vet. Record. 2011, 168, 20. [Google Scholar] [CrossRef]
- Barquero, N.; Domenech, A.; Arjona, A.; Fernández-Garayzabal, J.F.; Ruiz-Santa-Quiteria, J.A.; Gomez-Lucia, E. Comparison of two PCR and one ELISA techniques for the detection of small ruminant lentiviruses (SRLVs) in milk of sheep and goats. Res. Vet. Sci. 2013, 94, 817–819. [Google Scholar] [CrossRef]
- Brinkhof, J.M.A.; Houwers, D.J.; Moll, L.; Dercksen, D.; van Maanen, C. Diagnostic performance of ELISA and PCR in identifying SRLV-infected sheep and goats using serum, plasma and milk samples and in early detection of infection in dairy flocks through bulk milk testing. Vet. Microbiol. 2010, 142, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Larson, B.L.; Heary, H.L.; Devery, J.E. Immunoglobulin Production and Transport by the Mammary Gland. J. Dairy Sci. 1980, 63, 665–671. [Google Scholar] [CrossRef]
- McManaman, J.L.; Neville, M.C. Mammary physiology and milk secretion. Adv. Drug Deliv. Rev. 2003, 55, 629–641. [Google Scholar] [CrossRef]
- Pisoni, G.; Moroni, P.; Turin, L.; Bertoni, G. Compartmentalization of small ruminant lentivirus between blood and colostrum in infected goats. Virology 2007, 369, 119–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolea, R.; Monleon, E.; Carrasco, L.; Vargas, A.; de Andrès, D.; Amorena, B.; Badiola, J.J.; Lujan, L. Maedi-visna virus infection of ovine mammary epithelial cells. Vet. Res. 2006, 37, 655–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Kampen, C.; Mallard, B.A.; Wilkie, B.N. Adhesion molecules and lymphocyte subsets in milk and blood of periparturient Holstein cows. Vet. Immunol. Immunopathol. 1999, 69, 23–32. [Google Scholar] [CrossRef]
- Extramiana, A.B.; González, L.; Cortabarría, N.; García, M.; Juste, R.A. Evaluation of a PCR technique for the detection of Maedi-Visna proviral DNA in blood, milk and tissue samples of naturally infected sheep. Small Rumin. Res. 2002, 44, 109–118. [Google Scholar] [CrossRef]
- Keen, J.; Kwang, J.; Littledike, E.T.; Hungerford, L.L. Ovine lentivirus antibody detection in serum, colostrum and milk using a recombinant transmembrane protein ELISA. Vet. Immunol. Immunopathol. 1996, 51, 253–275. [Google Scholar] [CrossRef]
- Mazzei, M.; Carrozza, M.L.; Bandecchi, P.; Mazzanti, G.; Mannelli, A.; Tolari, F. Evaluation of an ELISA to detect antibodies to maedi-visna virus in individual and pooled samples of milk from sheep. Vet. Rec. 2005, 157, 552–555. [Google Scholar] [CrossRef]
- Plaza, M.; Sánchez, A.; Corrales, J.C.; De la Fe, C.; Contreras, A. Caprine arthritis encephalitis virus diagnosed by ELISA in lactating goats using milk samples. Small Rumin. Res. 2009, 81, 189–192. [Google Scholar] [CrossRef]
- Reddy, P.G.; Sapp, W.J.; Heneine, W. Detection of caprine arthritis-encephalitis virus by polymerase chain reaction. J. Clin. Microbiol. 1993, 31, 3042–3043. [Google Scholar] [PubMed]
- Michiels, R.; Van Mael, E.; Quinet, C.; Welby, S.; Cay, A.B.; De Regge, N. Seroprevalence and risk factors related to small ruminant lentivirus infections in Belgian sheep and goats. Prev. Vet. Med. 2018, 151, 13–20. [Google Scholar] [CrossRef] [PubMed]
- De Regge, N.; Cay, B. Development, validation and evaluation of added diagnostic value of a q(RT)-PCR for the detection of genotype A strains of small ruminant lentiviruses. J. Virol. Methods 2013, 194, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Harp, J.A.; Waters, T.E.; Goff, J.P. Lymphocyte subsets and adhesion molecule expression in milk and blood of periparturient dairy cattle. Vet. Immunol. Immunopathol. 2004, 102, 9–17. [Google Scholar] [CrossRef]
- Motha, J.M.X.; Ralston, J.C. Evaluation of ELISA for detection of antibodies to CAEV in milk. Vet. Microbiol. 1994, 38, 359–367. [Google Scholar] [CrossRef]
- Park, Y.H.; Fox, L.K.; Hamilton, M.J.; Davis, W.C. Bovine Mononuclear Leukocyte Subpopulations in Peripheral Blood and Mammary Gland Secretions During Lactation. J. Dairy Sci. 1992, 75, 998–1006. [Google Scholar] [CrossRef]
- Winnicka, A.; Klucinski, W.; Hoser, G.; Sikora, J.; Kawiak, J. Flow Cytometry Analysis of Milk and Peripheral Blood Cells from Goats during Lactation*. J. Vet. Med. A 1999, 46, 459–464. [Google Scholar] [CrossRef]
- Dulin, A.M.; Paape, M.J.; Schultze, W.D.; Weinland, B.T. Effect of Parity, Stage of Lactation, and Intramammary Infection on Concentration of Somatic Cells and Cytoplasmic Particles in Goat Milk. J. Dairy Sci. 1983, 66, 2426–2433. [Google Scholar] [CrossRef]
- Manlongat, N.; Yang, T.J.; Hinckley, L.S.; Bendel, R.B.; Krider, H.M. Physiologic-chemoattractant-induced migration of polymorphonuclear leukocytes in milk. Clin. Diagn. Lab. Immunol. 1998, 5, 375–381. [Google Scholar]
- Klintevall, K.; Näslund, K.; Svedlund, G.; Hajdu, L.; Linde, N.; Klingeborn, B. Evaluation of an indirect ELISA for the detection of antibodies to bovine leukaemia virus in milk and serum. J. Virol. Methods 1991, 33, 319–333. [Google Scholar] [CrossRef]
- Grego, E.; Profiti, M.; Giammarioli, M.; Giannino, L.; Rutili, D.; Woodall, C.; Rosati, S. Genetic heterogeneity of small ruminant lentiviruses involves immunodominant epitope of capsid antigen and affects sensitivity of single-strain-based immunoassay. Clin. Diagn. Lab. Immunol. 2002, 9, 828–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardinaux, L.; Zahno, M.L.; Deubelbeiss, M.; Zanoni, R.; Vogt, H.R.; Bertoni, G. Virological and phylogenetic characterization of attenuated small ruminant lentivirus isolates eluding efficient serological detection. Vet. Microbiol. 2013, 162, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Brinkhof, J.; Van Maanen, C. Evaluation of five enzyme-linked immunosorbent assays and an agar gel immunodiffusion test for detection of antibodies to small ruminant lentiviruses. Clin. Vaccine Immunol. 2007, 14, 1210–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lechat, E.; Milhau, N.; Brun, P.; Bellaton, C.; Greenland, T.; Mornex, J.F.; Le Jan, C. Goat endothelial cells may be infected in vitro by transmigration of caprine arthritis-encephalitis virus-infected leucocytes. Vet. Immunol. Immunopathol. 2005, 104, 257–263. [Google Scholar] [CrossRef]
- Milhau, N.; Renson, P.; Dreesen, I.; Greenland, T.; Bellaton, C.; Guiguen, F.; Mornex, J.F.; Le Jan, C. Viral expression and leukocyte adhesion after in vitro infection of goat mammary gland cells with caprine arthritis-encephalitis virus. Vet. Immunol. Immunopathol. 2005, 103, 93–99. [Google Scholar] [CrossRef]
- Ismail, H.I.; Hashimoto, Y.; Kon, Y.; Okada, K.; Davis, W.C.; Iwanaga, T. Lymphocyte subpopulations in the mammary gland of the goat. Vet. Immunol. Immunopathol. 1996, 52, 201–212. [Google Scholar] [CrossRef]
- Ravazzolo, A.P.; Nenci, C.; Vogt, H.R.; Waldvogel, A.; Obexer-Ruff, G.; Peterhans, E.; Bertoni, G. Viral load, organ distribution, histopathological lesions, and cytokine mRNA expression in goats infected with a molecular clone of the caprine arthritis encephalitis virus. Virology 2006, 350, 116–127. [Google Scholar] [CrossRef]
- Akers, R.M.; Nickerson, S.C. Mastitis and its impact on structure and function in the ruminant mammary gland. J. Mammary Gland Biol. Neoplasia 2011, 16, 275–289. [Google Scholar] [CrossRef]
- Santos, B.P.; Souza, F.N.; Blagitz, M.G.; Batista, C.F.; Bertagnon, H.G.; Diniz, S.A.; Silva, M.X.; Haddad, J.P.A.; Della Libera, A.M.M.P. Blood and milk polymorphonuclear leukocyte and monocyte/macrophage functions in naturally caprine arthritis encephalitis virus infection in dairy goats. Vet. Immunol. Immunopathol. 2017, 188, 21–26. [Google Scholar] [CrossRef]
- Clements, J.E.; Zink, M.C. Molecular biology and pathogenesis of animal lentivirus infections. Clin. Microbiol. Rev. 1996, 9, 100–117. [Google Scholar] [CrossRef]
- Gayo, E.; Polledo, L.; Magalde, A.; Balseiro, A.; García Iglesias, M.J.; Pérez Martínez, C.; Preziuso, S.; Rossi, G.; García Marín, J.F. Characterization of minimal lesions related to the presence of visna/maedi virus in the mammary gland and milk of dairy sheep. BMC Vet. Res. 2019, 15, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mselli-lakhal, L.; Fornazero, C.; Du, J.; Favier, C.; Durand, J.; Grezel, D.; Balleydier, S.; Mornex, J.F.; Chebloune, Y. Goat Milk Epithelial Cells Are Highly Permissive to CAEV Infection in Vitro. Virology 1999, 259, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mselli-Lakhal, L.; Guiguen, F.; Fornazero, C.; Favier, C.; Durand, J.; Grezel, D.; Moussa, A.; Mornex, J.F.; Chebloune, Y. Immortalized goat milk epithelial cell lines replicate CAEV at high level. Vet. Res. 2001, 32, 429–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutinaud, M.; Jammes, H. Potential uses of milk epithelial cells: A review. Reprod. Nutr. Dev. 2002, 42, 133–147. [Google Scholar] [CrossRef]
- Schwertfeger, K.L.; Rosen, J.M.; Cohen, D.A. Mammary Gland Macrophages: Pleiotropic Functions in Mammary Development. J. Mammary Gland Biol. Neoplasia 2006, 11, 229–238. [Google Scholar] [CrossRef]
- Zink, M.C.; Yager, J.A.; Myers, J.D. Pathogenesis of caprine arthritis encephalitis virus: Cellular localization of viral transcripts in tissues of infected goats. Am. J. Pathol. 1990, 136, 843–854. [Google Scholar]
- De la Concha-Bermejillo, A. Maedi-Visna and Ovine Progressive Pneumonia. Vet. Clin. North. Am. Food Anim. Pract. 1997, 13, 13–34. [Google Scholar] [CrossRef]



| Elitest® | ||||||
| Infection status in milk | ||||||
| Pos | Neg | Total | Relative sensitivity | Relative specificity | ||
| Infection status in serum | Pos | 168 | 12 | 180 | 93% | 97% |
| Neg | 4 | 137 | 141 | |||
| Total | 172 | 149 | 321 | |||
| IDscreen® | ||||||
| Infection status in milk | ||||||
| Pos | Neg | Total | Relative sensitivity | Relative specificity | ||
| Infection status in serum | Pos | 194 | 7 | 201 | 97% | 100% |
| Neg | 0 | 120 | 120 | |||
| Total | 194 | 127 | 321 | |||
| PBMCs | Milk Cell Pellets | ||||
|---|---|---|---|---|---|
| Pos | Neg | Total | Relative Sensitivity | Relative Specificity | |
| Pos | 134 | 31 | 165 | 81% | 88% |
| Neg | 18 | 138 | 156 | ||
| Total | 152 | 169 | 321 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adjadj, N.R.; Vicca, J.; Michiels, R.; De Regge, N. (Non-)Sense of Milk Testing in Small Ruminant Lentivirus Control Programs in Goats. Comparative Analysis of Antibody Detection and Molecular Diagnosis in Blood and Milk. Viruses 2020, 12, 3. https://doi.org/10.3390/v12010003
Adjadj NR, Vicca J, Michiels R, De Regge N. (Non-)Sense of Milk Testing in Small Ruminant Lentivirus Control Programs in Goats. Comparative Analysis of Antibody Detection and Molecular Diagnosis in Blood and Milk. Viruses. 2020; 12(1):3. https://doi.org/10.3390/v12010003
Chicago/Turabian StyleAdjadj, Nadjah Radia, Jo Vicca, Rodolphe Michiels, and Nick De Regge. 2020. "(Non-)Sense of Milk Testing in Small Ruminant Lentivirus Control Programs in Goats. Comparative Analysis of Antibody Detection and Molecular Diagnosis in Blood and Milk" Viruses 12, no. 1: 3. https://doi.org/10.3390/v12010003




