Antibody Responses against Enterovirus Proteases are Potential Markers for an Acute Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Husbandry and Ethics Statement
2.2. Human Samples Ethics Statement
2.3. Preparation of ELISA Antigens
2.4. Protease ELISA
2.5. Capture IgM-ELISA for Human Samples
2.6. Data Processing
3. Results
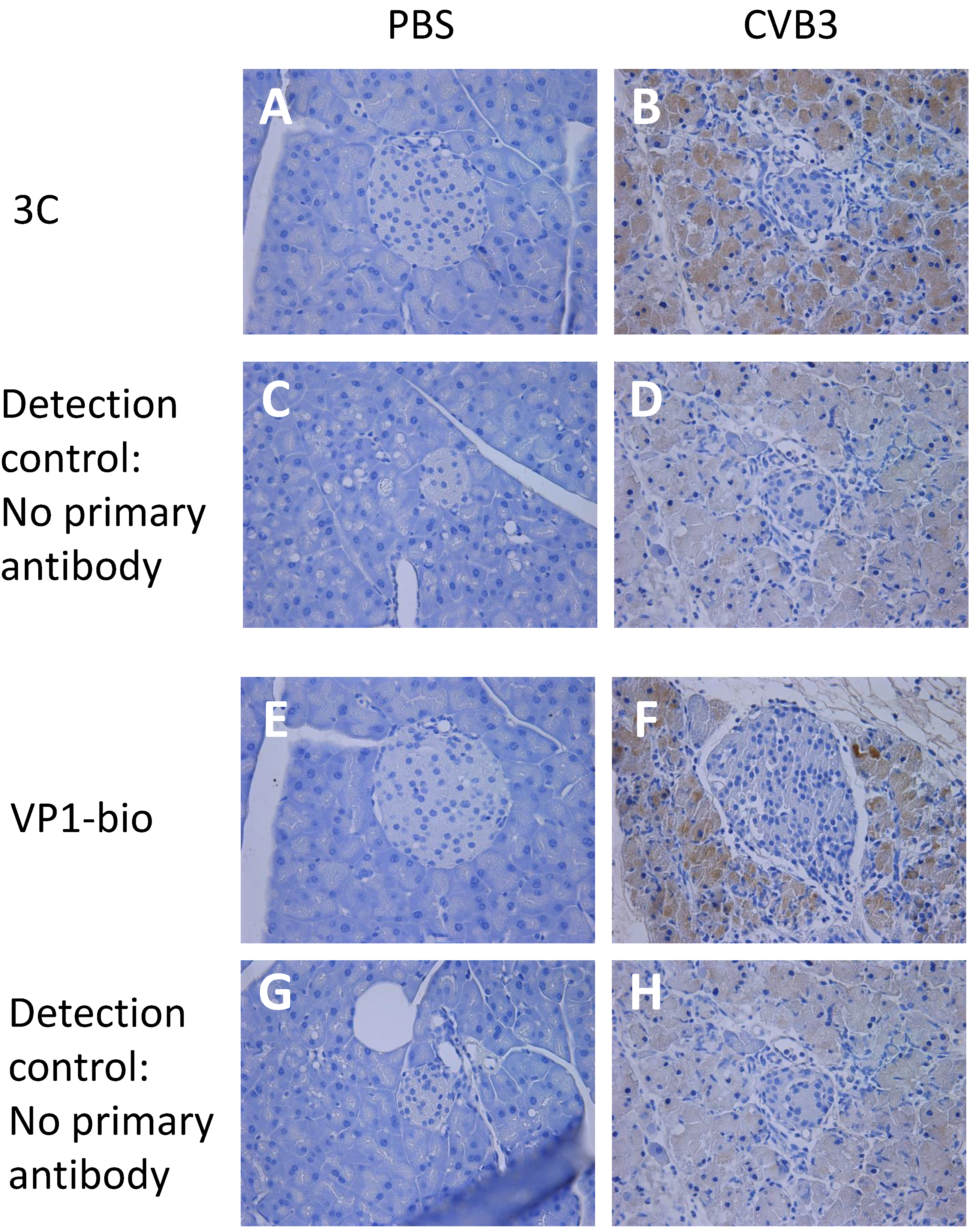
3.1. Enterovirus Proteases Are Expressed in Mice Shortly after CVB3 Infection
3.2. Pooled Mouse and Human Sera Contain Antibodies against Enteroviral Proteases
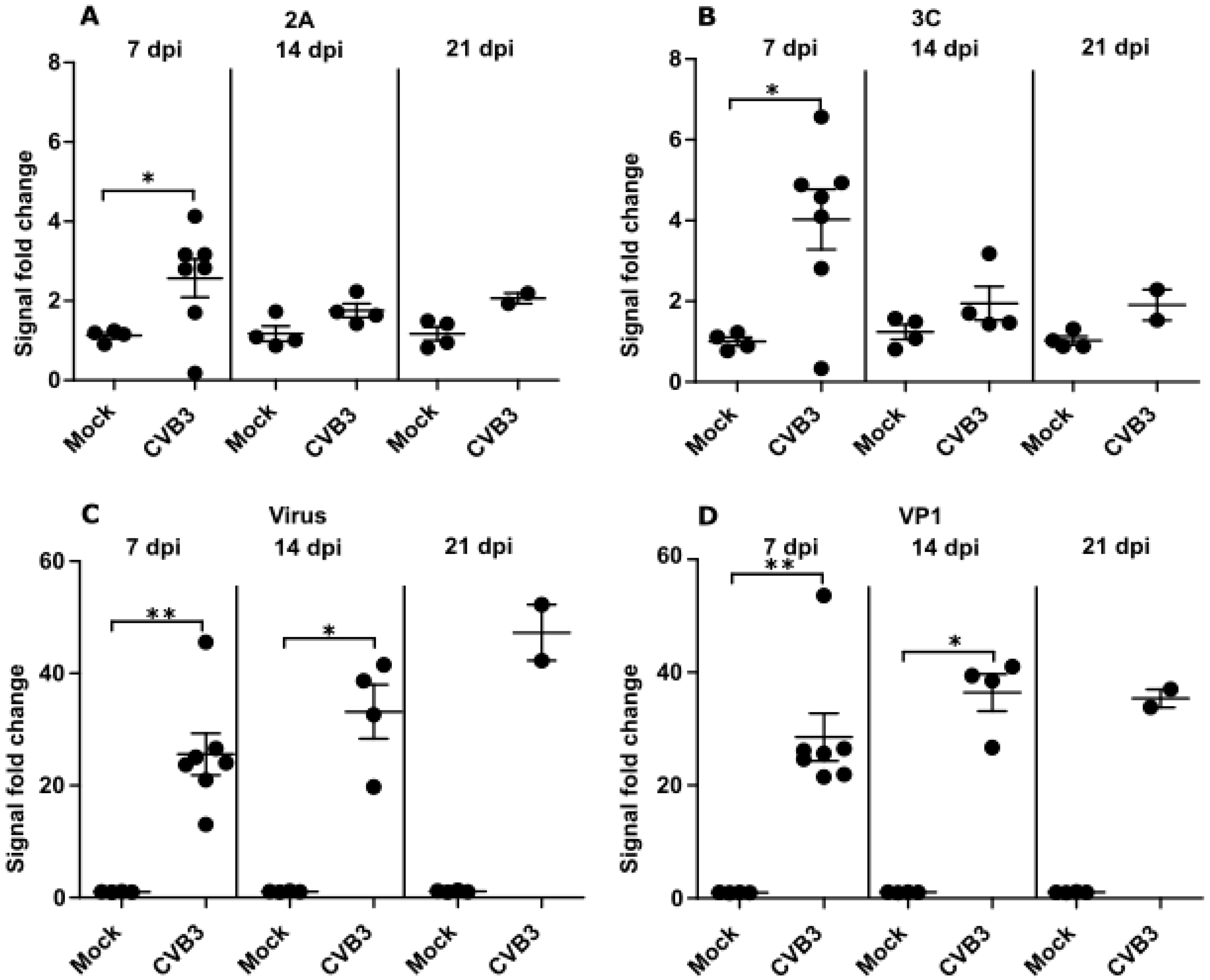
3.3. Time Dependent Antibody Response against Enteroviral Proteases and Structural Proteins Differ in Mice
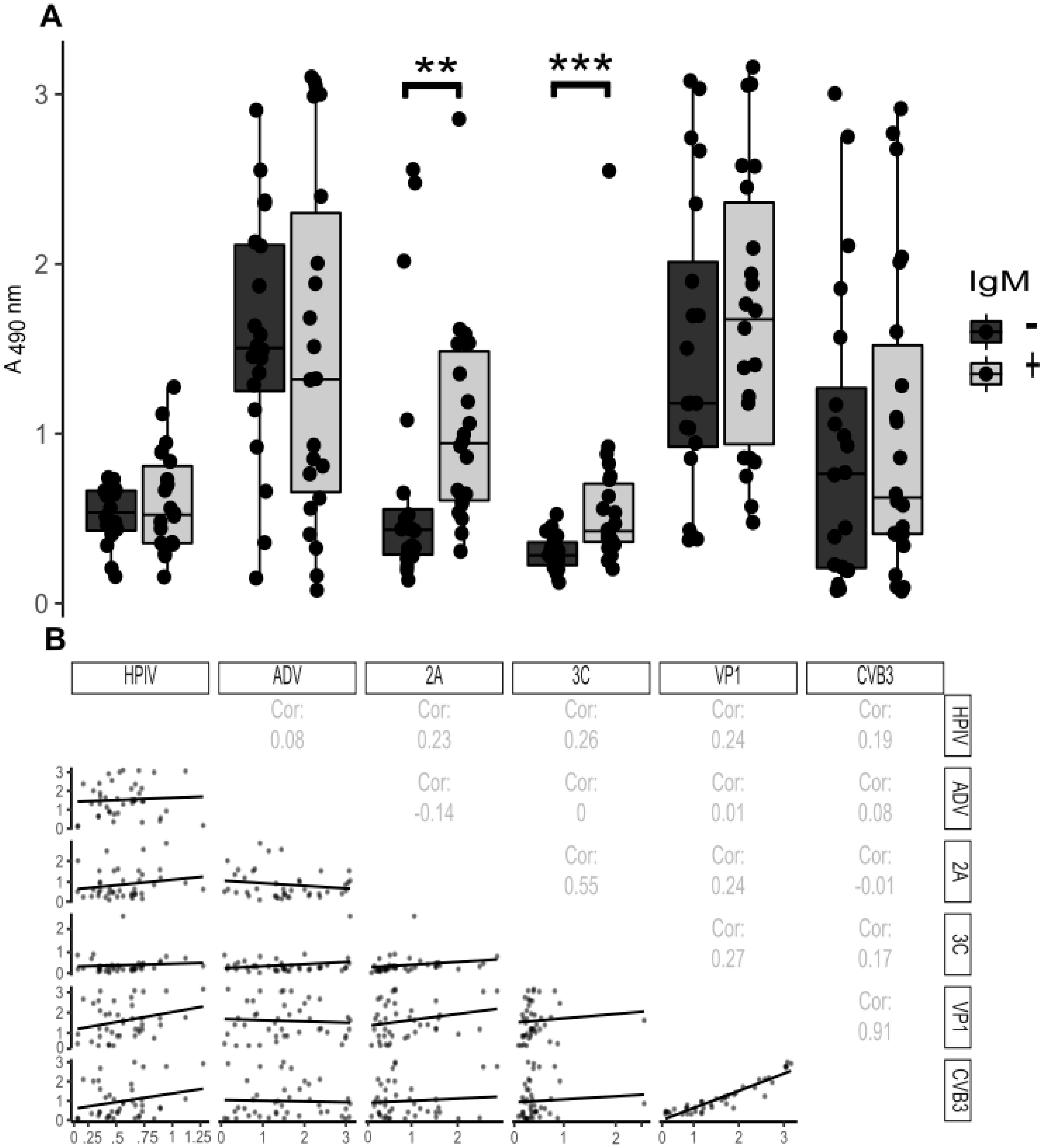
3.4. Antibody Responses towards Viral Proteases during Acute Enterovirus Infection in Humans
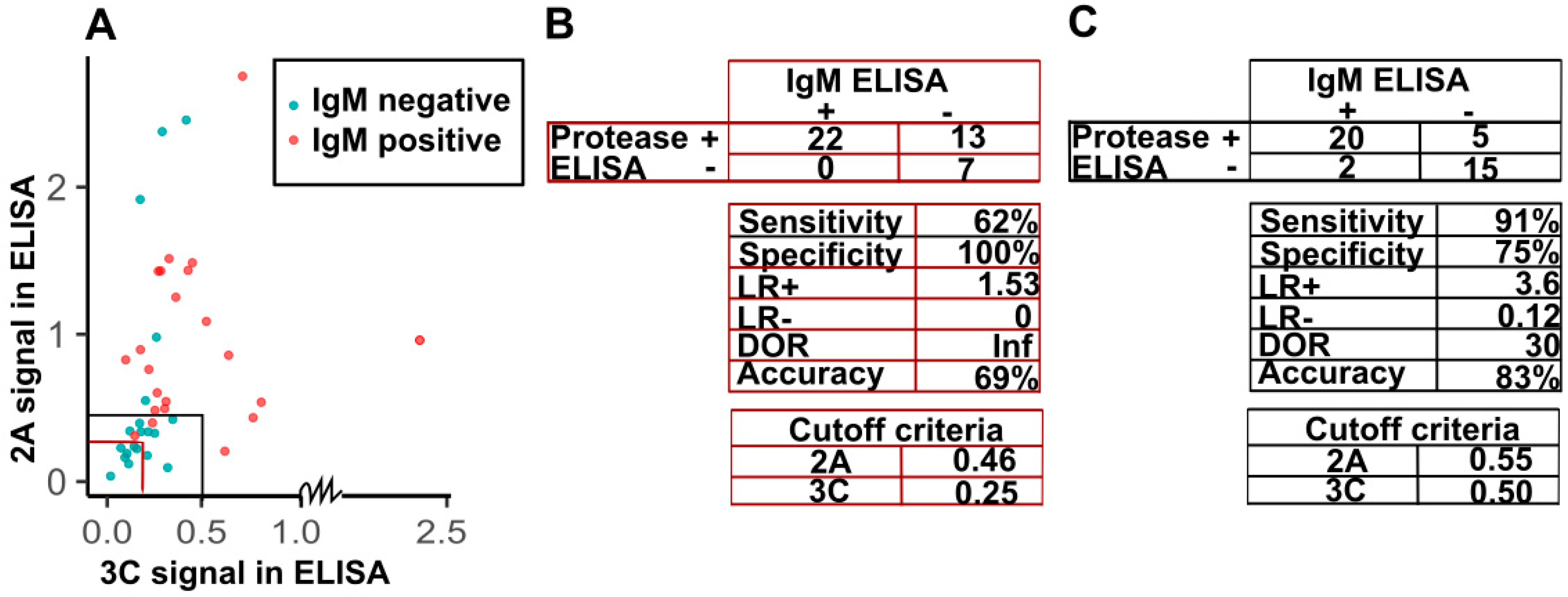
3.5. Sensitivity and Specificity of Detection of IgG against Viral Proteases in the Diagnosis of IgM-Positive Acute Enterovirus Infections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Mouse Infection and Sample Collection
Appendix A.2. Histology
Appendix A.3. Optimization of Protease-ELISA
Appendix A.4. Preparation of ELISA Antigens
Appendix A.5. Virus Culturing
Appendix A.5.1. Culturing Viruses for Infecting Mice
Appendix A.5.2. Culturing Viruses for ELISA
Appendix A.6. Protease ELISA for the Infected Mice
Appendix A.7. Protease ELISA for Human Samples
Appendix A.8. Capture IgM-ELISA for Human Samples
References
- Abzug, M.J.; Michaels, M.G.; Wald, E.; Jacobs, R.F.; Romero, J.R.; Sanchez, P.J.; Wilson, G.; Krogstad, P.; Storch, G.A.; Lawrence, R.; et al. Natl Inst Allergy Infect A Randomized, Double-Blind, Placebo-Controlled Trial of Pleconaril for the Treatment of Neonates with Enterovirus Sepsis. J. Pediatr. Infect. Dis. Soc. 2016, 5, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Green, J.; Casabonne, D.; Newton, R. Coxsackie B virus serology and Type 1 diabetes mellitus: A systematic review of published case-control studies. Diabet. Med. 2004, 21, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Gern, J.E.; Busse, W.W. Association of rhinovirus infections with asthma. Clin. Microbiol. Rev. 1999, 12, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niespodziana, K.; Stenberg-Hammar, K.; Megremis, S.; Cabauatan, C.R.; Napora-Wijata, K.; Vacal, P.C.; Gallerano, D.; Lupinek, C.; Ebner, D.; Schlederer, T.; et al. PreDicta chip-based high resolution diagnosis of rhinovirus-induced wheeze. Nat. Commun. 2018, 9, 2382. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Calvo, T.; von Herrath, M.G. Enterovirus infection and type 1 diabetes: Closing in on a link? Diabetes 2015, 64, 1503–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laitinen, O.H.; Honkanen, H.; Pakkanen, O.; Oikarinen, S.; Hankaniemi, M.M.; Huhtala, H.; Ruokoranta, T.; Lecouturier, V.; Andre, P.; Harju, R.; et al. Coxsackievirus B1 Is Associated with Induction of [beta]-Cell Autoimmunity That Portends Type 1 Diabetes. Diabetes 2014, 63, 446–455. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Song, Z.; Yu, H.; Zhang, X.; Xu, S.; Li, Z.; Li, J.; Xu, H.; Yuan, Z.; Ma, H.; et al. Genome-wide linear B-cell epitopes of enterovirus 71 in a hand, foot and mouth disease (HFMD) population. J. Clin. Virol. 2018, 105, 41–48. [Google Scholar] [CrossRef]
- Liu, D.; Wang, T.; Huang, W.; Chang, L.; Wang, E.; Cheng, S.; Yang, M. Disease burden of enterovirus infection in Taiwan: Implications for vaccination policy. Vaccine 2016, 34, 974–980. [Google Scholar] [CrossRef] [Green Version]
- Arbustini, E.; Porcu, E.; Bellini, O.; Grasso, M.; Pilotto, A.; Dal Bello, B.; Morbini, P.; Diegoli, M.; Gavazzi, A.; Specchia, G.; et al. Enteroviral infection causing fatal myocarditis and subclinical myopathy. Heart 2000, 83, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Freund, M.W.; Kleinveld, G.; Krediet, T.G.; van Loon, A.M.; Verboon-Maciolek, M.A. Prognosis for neonates with enterovirus myocarditis. Arch. Dis. Child. -Fetal Neonatal. Ed. 2010, 95, F206–F212. [Google Scholar] [CrossRef]
- Stene, L.C.; Rewers, M. Immunology in the clinic review series; focus on type 1 diabetes and viruses: The enterovirus link to type 1 diabetes: Critical review of human studies. Clin. Exp. Immunol. 2012, 168, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Laitinen, O.H.; Svedin, E.; Kapell, S.; Nurminen, A.; Hytonen, V.P.; Flodstrom-Tullberg, M. Enteroviral proteases: Structure, host interactions and pathogenicity. Rev. Med. Virol. 2016, 26, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Maher, K.; Pallansch, M.A. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 2004, 78, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitray, M.; Grazioli, S.; Willems, T.; Tshabalala, T.; De Vleeschauwer, A.; Esterhuysen, J.J.; Brocchi, E.; De Clercq, K.; Maree, F.F. Development and validation of a foot-and-mouth disease virus SAT serotype-specific 3ABC assay to differentiate infected from vaccinated animals. J. Virol. Methods 2018, 255, 44–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lubroth, J.; Brown, F. Identification of native foot-and-mouth disease virus non-structural protein 2C as a serological indicator to differentiate infected from vaccinated livestock. Res. Vet. Sci. 1995, 59, 70–78. [Google Scholar] [CrossRef]
- Chen, X.; Qin, B.; Shi, M.; Zhu, L.; Sun, M.; Liu, X.; Zhang, J. Immunoreactivity Analysis of the Nonstructural Proteins of Human Enterovirus 71. Viral Immunol. 2016, 30, 106–110. [Google Scholar] [CrossRef]
- Araujo, A.C.; Astrakhantseva, I.V.; Fields, H.A.; Kamili, S. Distinguishing Acute from Chronic Hepatitis C Virus (HCV) Infection Based on Antibody Reactivities to Specific HCV Structural and Nonstructural Proteins. J. Clin. Microbiol. 2011, 49, 54–57. [Google Scholar] [CrossRef] [Green Version]
- Gronroos, M.; Parajuli, A.; Laitinen, O.H.; Roslund, M.I.; Vari, H.K.; Hyoty, H.; Puhakka, R.; Sinkkonen, A. Short-term direct contact with soil and plant materials leads to an immediate increase in diversity of skin microbiota. Microbiologyopen 2019, 8, e00645. [Google Scholar] [CrossRef]
- Haller, M.J.; Schatz, D.A. The DIPP project: 20 years of discovery in type 1 diabetes. Pediatr. Diabetes 2016, 17, 5–7. [Google Scholar] [CrossRef]
- Saarinen, N.V.V.; Laiho, J.E.; Richardson, S.J.; Zeissler, M.; Stone, V.M.; Marjomaki, V.; Kantoluoto, T.; Horwitz, M.S.; Sioofy-Khojine, A.; Honkimaa, A.; et al. A novel rat CVB1-VP1 monoclonal antibody 3A6 detects a broad range of enteroviruses. Sci. Rep. 2018, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Laitinen, O.H.; Svedin, E.; Kapell, S.; Hankaniemi, M.M.; Larsson, P.G.; Domsgen, E.; Stone, V.M.; Maatta, J.A.E.; Hyoty, H.; Hytonen, V.P.; et al. New Coxsackievirus 2Apro and 3Cpro protease antibodies for virus detection and discovery of pathogenic mechanisms. J. Virol. Methods 2018, 255, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Hankaniemi, M.M.; Laitinen, O.H.; Stone, V.M.; Sioofy-Khojine, A.; Maatta, J.A.E.; Larsson, P.G.; Marjomaki, V.; Hyoty, H.; Flodstrom-Tullberg, M.; Hytonen, V.P. Optimized production and purification of Coxsackievirus B1 vaccine and its preclinical evaluation in a mouse model. Vaccine 2017, 35, 3718–3725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viskari, H.; Knip, M.; Tauriainen, S.; Huhtala, H.; Veijola, R.; Ilonen, J.; Simell, O.; Surcel, H.; Hyoty, H. Maternal enterovirus infection as a risk factor for type 1 diabetes in the exposed offspring. Diabetes Care 2012, 35, 1328–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorries, R.; ter Meulen, V. Specificity of IgM antibodies in acute human coxsackievirus B infections, analysed by indirect solid phase enzyme immunoassay and immunoblot technique. J. Gen. Virol. 1983, 64, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.J.; Kula, T.; Xu, Q.; Li, M.Z.; Vernon, S.D.; Ndung’u, T.; Ruxrungtham, K.; Sanchez, J.; Brander, C.; Chung, R.T. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015, 348, aaa0698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pou, C.; Nkulikiyimfura, D.; Henckel, E.; Olin, A.; Lakshmikanth, T.; Mikes, J.; Wang, J.; Chen, Y.; Bernhardsson, A.K.; Gustafsson, A.; et al. The repertoire of maternal anti-viral antibodies in human newborns. Nat. Med. 2019, 25, 591–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuelson, A.; Forsgren, M.; Johansson, B.; Wahren, B.; Sallberg, M. Molecular basis for serological cross-reactivity between enteroviruses. Clin. Diagn. Lab. Immunol. 1994, 1, 336–341. [Google Scholar] [CrossRef]
- Niespodziana, K.; Napora, K.; Cabauatan, C.; Focke-Tejkl, M.; Keller, W.; Niederberger, V.; Tsolia, M.; Christodoulou, I.; Papadopoulos, N.G.; Valenta, R. Misdirected antibody responses against an N-terminal epitope on human rhinovirus VP1 as explanation for recurrent RV infections. FASEB J. 2012, 26, 1001–1008. [Google Scholar] [CrossRef]
- Hyoty, H.; Leon, F.; Knip, M. Developing a vaccine for type 1 diabetes by targeting coxsackievirus B. Expert Rev. Vaccines 2018, 17, 1071–1083. [Google Scholar] [CrossRef]
- Hansson, S.F.; Korsgren, S.; Ponten, F.; Korsgren, O. Enteroviruses and the pathogenesis of type 1 diabetes revisited: Cross-reactivity of enterovirus capsid protein (VP1) antibodies with human mitochondrial proteins. J. Pathol. 2013, 229, 719–728. [Google Scholar] [CrossRef]
- Laitinen, O.H.; Airenne, K.J.; Hytonen, V.P.; Peltomaa, E.; Mahonen, A.J.; Wirth, T.; Lind, M.M.; Makela, K.A.; Toivanen, P.I.; Schenkwein, D.; et al. A multipurpose vector system for the screening of libraries in bacteria, insect and mammalian cells and expression in vivo. Nucleic Acids Res. 2005, 33, e42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heikura, T.; Nieminen, T.; Roschier, M.M.; Karvinen, H.; Kaikkonen, M.U.; Mahonen, A.J.; Lesch, H.P.; Rissanen, T.T.; Laitinen, O.H.; Airenne, K.J.; et al. Baculovirus-mediated vascular endothelial growth factor-D(DeltaNDeltaC) gene transfer induces angiogenesis in rabbit skeletal muscle. J. Gene Med. 2012, 14, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Hankaniemi, M.M.; Stone, V.M.; Sioofy-Khojine, A.; Heinimaki, S.; Marjomaki, V.; Hyoty, H.; Blazevic, V.; Laitinen, O.H.; Flodstrom-Tullberg, M.; Hytonen, V.P. A comparative study of the effect of UV and formalin inactivation on the stability and immunogenicity of a Coxsackievirus B1 vaccine. Vaccine 2019, 37, 5962–5971. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saarinen, N.V.V.; Stone, V.M.; Hankaniemi, M.M.; Mazur, M.A.; Vuorinen, T.; Flodström-Tullberg, M.; Hyöty, H.; Hytönen, V.P.; Laitinen, O.H. Antibody Responses against Enterovirus Proteases are Potential Markers for an Acute Infection. Viruses 2020, 12, 78. https://doi.org/10.3390/v12010078
Saarinen NVV, Stone VM, Hankaniemi MM, Mazur MA, Vuorinen T, Flodström-Tullberg M, Hyöty H, Hytönen VP, Laitinen OH. Antibody Responses against Enterovirus Proteases are Potential Markers for an Acute Infection. Viruses. 2020; 12(1):78. https://doi.org/10.3390/v12010078
Chicago/Turabian StyleSaarinen, Niila V. V., Virginia M. Stone, Minna M. Hankaniemi, Magdalena A. Mazur, Tytti Vuorinen, Malin Flodström-Tullberg, Heikki Hyöty, Vesa P. Hytönen, and Olli H. Laitinen. 2020. "Antibody Responses against Enterovirus Proteases are Potential Markers for an Acute Infection" Viruses 12, no. 1: 78. https://doi.org/10.3390/v12010078
APA StyleSaarinen, N. V. V., Stone, V. M., Hankaniemi, M. M., Mazur, M. A., Vuorinen, T., Flodström-Tullberg, M., Hyöty, H., Hytönen, V. P., & Laitinen, O. H. (2020). Antibody Responses against Enterovirus Proteases are Potential Markers for an Acute Infection. Viruses, 12(1), 78. https://doi.org/10.3390/v12010078







