A Cell-Based ELISA to Improve the Serological Analysis of Anti-SARS-CoV-2 IgG
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects Recruitment
2.2. Ethics Approval
2.3. Cell-ELISA
2.4. Serology
2.5. Statistical Analysis
3. Results
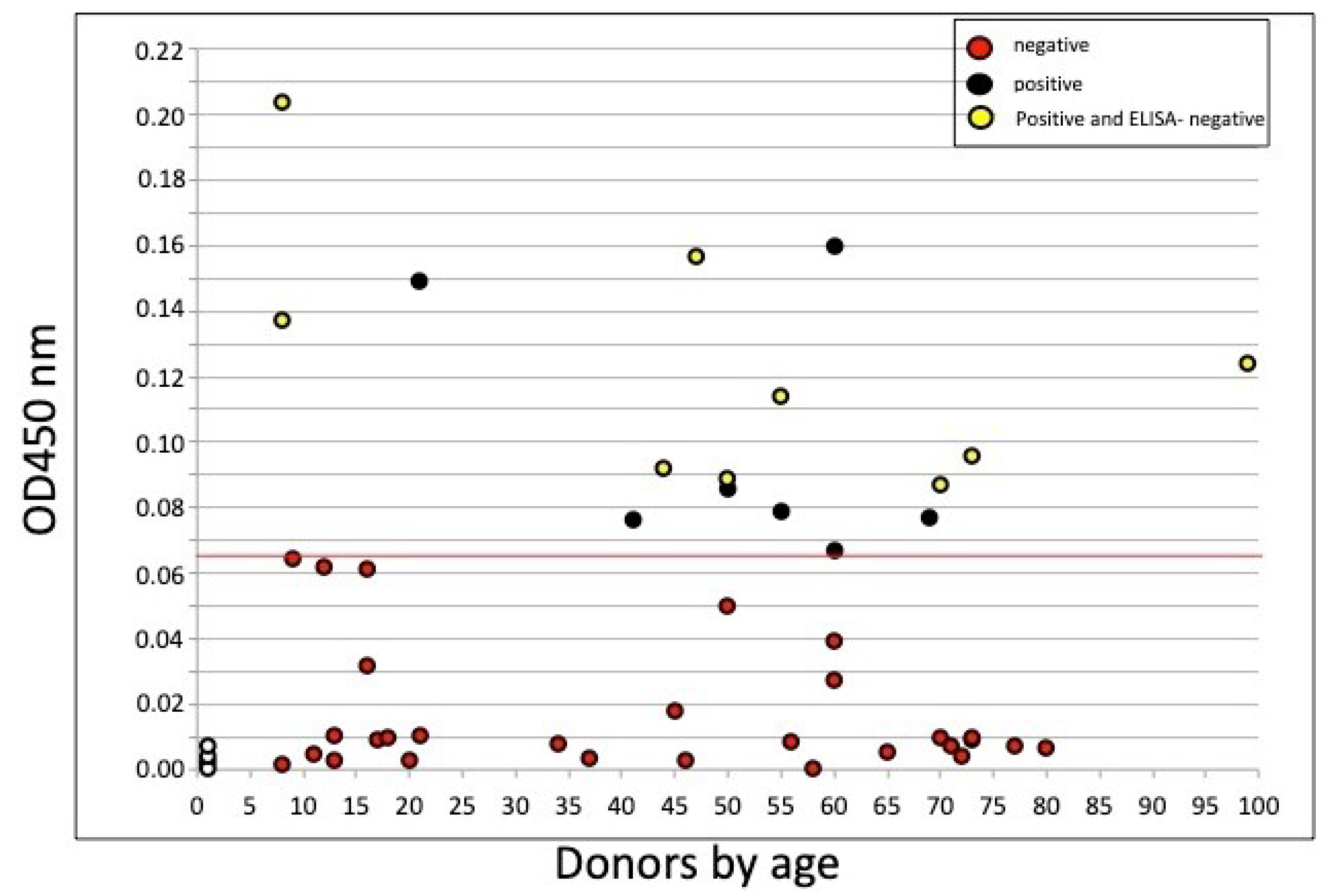
3.1. Cell-ELISA
3.2. Combined ELISA/Cell-ELISA
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Buisman, A.; De Rond, C.; Öztürk, K.; Hulscher, H.T.; Van Binnendijk, R. Long-term presence of memory B-cells specific for different vaccine components. Vaccine 2009, 28, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.B.; Koelsch, K.A. B-Cell ELISPOT: For the Identification of Antigen-Specific Antibody-Secreting Cells. Adv. Struct. Saf. Stud. 2015, 1312, 419–426. [Google Scholar] [CrossRef]
- Jahnmatz, P.; Sundling, C.; Makower, B.; Sondén, K.; Färnert, A.; Ahlborg, N.; Zuber, B. Multiplex analysis of antigen-specific memory B cells in humans using reversed B-cell FluoroSpot. J. Immunol. Methods 2020, 478, 112715. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.; Jilg, W. Hepatitis B surface antigen-specific T and B cell memory in individuals who had lost protective antibodies after hepatitis B vaccination. Vaccine 2006, 24, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Halliley, J.L.; Kyu, S.; Kobie, J.J.; Walsh, E.E.; Falsey, A.R.; Randall, T.D.; Treanor, J.; Feng, C.; Sanz, I.; Lee, F.E.-H. Peak frequencies of circulating human influenza-specific antibody secreting cells correlate with serum antibody response after immunization. Vaccine 2010, 28, 3582–3587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meloni, S.; Scapigliati, G. Evaluation of immunoglobulins produced in vitro by head-kidney leucocytes of sea bass Dicentrarchus labrax by immunoenzymatic assay. Fish Shellfish Immunol. 2000, 10, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Beavis, K.G.; Matushek, S.M.; Abeleda, A.P.F.; Bethel, C.; Hunt, C.; Gillen, S.; Moran, A.; Tesic, V. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020, 129, 104468. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Genet. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rongqing, Z.; Li, M.; Song, H.; Chen, J.; Ren, W.; Feng, Y.; Gao, G.F.; Song, J.; Peng, Y.; Su, B.; et al. Early Detection of Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies as a Serologic Marker of Infection in Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tong, X.; Wang, J.; Huang, W.; Yin, S.; Huang, R.; Yang, H.; Chen, Y.; Huang, A.; Liu, Y.; et al. High SARS-CoV-2 antibody prevalence among healthcare workers exposed to COVID-19 patients. J. Infect. 2020, 81, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Tu, D.; Shu, J.; Wu, X.; Li, H.; Xia, Z.; Zhang, Y.; Fang, Y.; Shen, S.; Guan, W.; Wang, H.; et al. Immunological detection of serum antibodies in pediatric medical workers exposed to varying levels of SARS-CoV-2. J. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ndungu, F.M.; Olotu, A.; Mwacharo, J.; Nyonda, M.; Apfeld, J.; Mramba, L.K.; Fegan, G.W.; Bejon, P.; Marsh, K. Memory B cells are a more reliable archive for historical antimalarial responses than plasma antibodies in no-longer exposed children. Proc. Natl. Acad. Sci. USA 2012, 109, 8247–8252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latner, D.R.; McGrew, M.; Williams, N.; Lowe, L.; Werman, R.; Warnock, E.; Gallagher, K.; Doyle, P.; Smole, S.; Lett, S.; et al. Enzyme-Linked Immunospot Assay Detection of Mumps-Specific Antibody-Secreting B Cells as an Alternative Method of Laboratory Diagnosis. Clin. Vaccine Immunol. 2010, 18, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nat. Cell Biol. 2020, 584, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.-W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nat. Cell Biol. 2020, 584, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Mankarious, S.; Lee, M.; Fischer, S.; Pyun, K.H.; Ochs, H.D.; Oxelius, V.A.; Wedgwood, R.J. The half-lives of IgG subclasses and specific antibodies in patients with primary immunodeficiency who are receiving intravenously administered immunoglobulin. J. Lab. Clin. Med. 1988, 112, 634–640. [Google Scholar] [PubMed]
- Blink, E.J.; Light, A.; Kallies, A.; Nutt, S.L.; Hodgkin, P.D.; Tarlinton, D.M. Early appearance of germinal center–derived memory B cells and plasma cells in blood after primary immunization. J. Exp. Med. 2005, 201, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef] [PubMed]

| All Subjects | Age Group | |||
|---|---|---|---|---|
| <18 | 18–55 | >55 | ||
| Total n | 45 | 11 | 16 | 18 |
| Gender (male, n) | 25 | 6 | 9 | 10 |
| Asymptomatic (n) | 24 | 6 | 7 | 11 |
| Mild symptoms (n) | 19 | 5 | 9 | 5 |
| Severe symptoms * (n) | 2 | - | - | 2 |
| Rt-PCR (tested, n) | 5 | - | 2 | 3 |
| Rt-PCR (positive, n) | 5 | 2 | 3 | |
| Serology IgG (tested, n) | 45 | 11 | 16 | 18 |
| Serology IgG $ (positive, n) | 12 | 0 | 7 | 4 |
| Cell-ELISA (tested, n) | 45 | 11 | 16 | 18 |
| Cell-ELISA(positive, n) | 16 | 2 | 8 | 6 |
| Sample/Age | Day 0 | Day 35–60 | |||
|---|---|---|---|---|---|
| ELISA 1 * | ELISA 2 * | Cell-ELISA ° | IgG ^ (ng/well) | ELISA2 vs. Cell-ELISA | |
| #4, 60 | 4.3 | 1.8 | 0.16 | 35 | +/+ |
| #5, 60 | 5.5 | 3.3 | 0.066 | 7 | +/+ |
| #6, 55 | 3.6 | 1 | 0.079 | 9 | −/+ |
| #7, 50 | 4.2 | 2.1 | 0.085 | 10 | ++ |
| #13, 55 | 3.5 | 0.43 | 0.114 | 16 | −/+ |
| #14, 41 | 0.19 | 0.16 | 0.076 | 8.5 | −/+ |
| #16, 8 | 0.18 | 0.17 | 0.203 | 50 | −/+ |
| #24, 24 | 3.64 | 1.51 | 0.092 | 11 | +/+ |
| #25, 69 | 4.77 | 4.21 | 0.077 | 8.5 | +/+ |
| #26, 50 | 3.01 | 0.88 | 0.088 | 10 | −/+ |
| #31, 21 | 4.96 | 1.02 | 0.149 | 32 | −/+ |
| #33, 8 | 0.16 | 0.20 | 0.137 | 30 | −/+ |
| #36, 99 | 0.23 | 0.23 | 0.124 | 18 | −/+ |
| #37, 73 | 1.21 | 0.74 | 0.096 | 11.5 | −/+ |
| #38, 70 | 1.05 | 0.26 | 0.087 | 10 | −/+ |
| #50, 47 | 0.33 | 0.18 | 0.157 | 34 | −/+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarletti, G.; Tiberi, M.; De Molfetta, V.; Bossù, M.; Toppi, E.; Bossù, P.; Scapigliati, G. A Cell-Based ELISA to Improve the Serological Analysis of Anti-SARS-CoV-2 IgG. Viruses 2020, 12, 1274. https://doi.org/10.3390/v12111274
Zarletti G, Tiberi M, De Molfetta V, Bossù M, Toppi E, Bossù P, Scapigliati G. A Cell-Based ELISA to Improve the Serological Analysis of Anti-SARS-CoV-2 IgG. Viruses. 2020; 12(11):1274. https://doi.org/10.3390/v12111274
Chicago/Turabian StyleZarletti, Gianpaolo, Massimo Tiberi, Veronica De Molfetta, Maurizio Bossù, Elisa Toppi, Paola Bossù, and Giuseppe Scapigliati. 2020. "A Cell-Based ELISA to Improve the Serological Analysis of Anti-SARS-CoV-2 IgG" Viruses 12, no. 11: 1274. https://doi.org/10.3390/v12111274
APA StyleZarletti, G., Tiberi, M., De Molfetta, V., Bossù, M., Toppi, E., Bossù, P., & Scapigliati, G. (2020). A Cell-Based ELISA to Improve the Serological Analysis of Anti-SARS-CoV-2 IgG. Viruses, 12(11), 1274. https://doi.org/10.3390/v12111274






