Host Range and Coding Potential of Eukaryotic Giant Viruses
Abstract
:1. Introduction
2. Giant Viruses Infect Every Major Eukaryotic Lineage
2.1. The Founding Members of NCLDVs
2.2. The Age of Discovery for Giant Viruses Infecting Microbial Eukaryotes
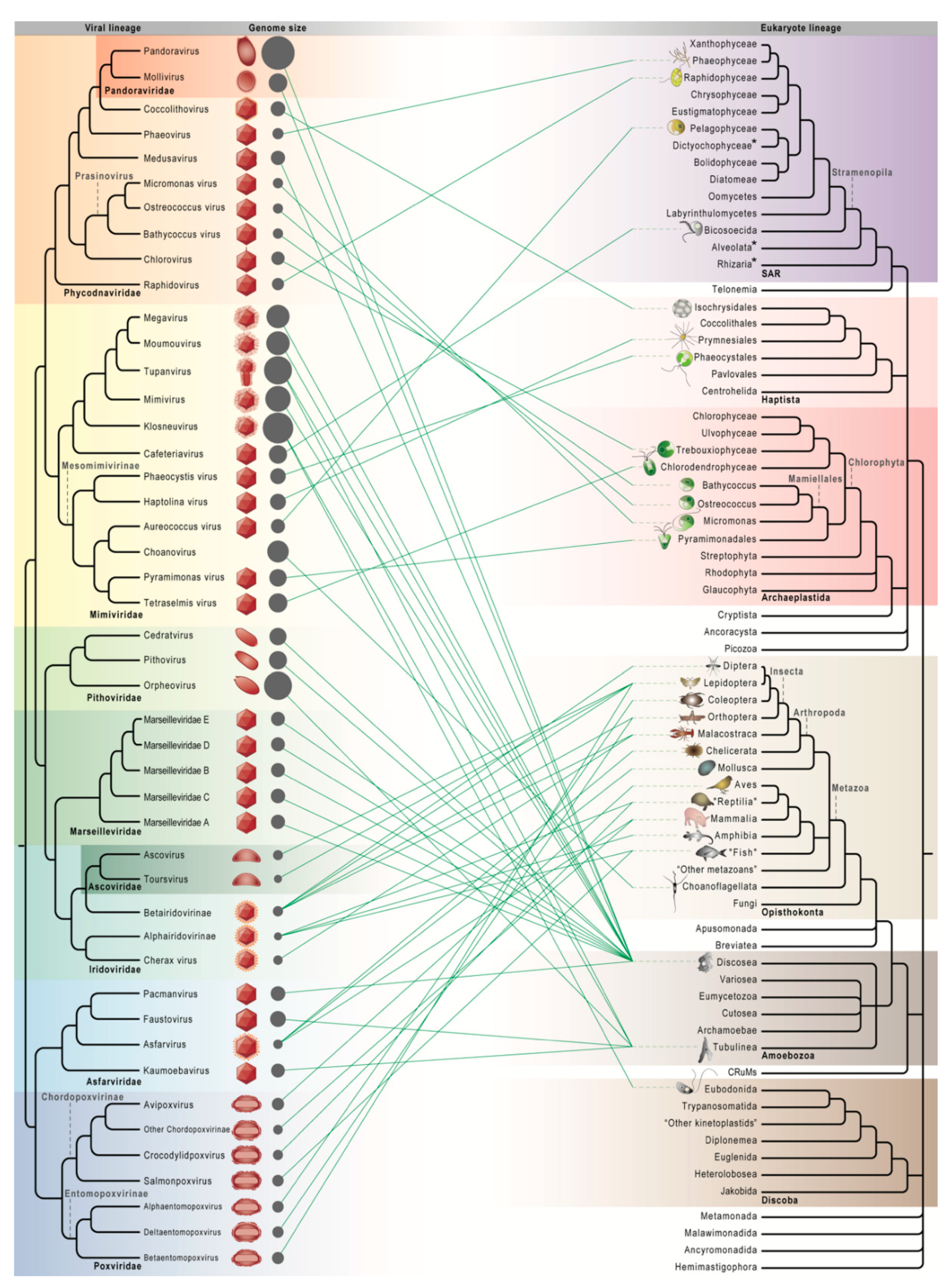
2.3. Increasingly Non-Algal Phycodnaviridae and Increasingly Non-Amoebal Mimiviridae
2.4. Undiscovered Virus–Host Relationships
3. Variation and Evolution of Host Range
4. Functional Potential of Virus-Encoded Proteins
4.1. Information Storage and Flow
4.2. Energy Metabolism
4.3. Synthesis of Biomolecules
4.4. Membrane Transport and Sensing
5. Evolution of Genome Content
5.1. Expansive Evolution
5.2. Reductive Evolution
5.3. Generalist Viruses and Genome Evolution
5.4. Origin of Giant Viruses and Their Families
6. Future Perspective
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iyer, L.M.; Aravind, L.; Koonin, E.V. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001, 75, 11720–11734. [Google Scholar] [CrossRef] [Green Version]
- Iyer, L.M.; Balaji, S.; Koonin, E.V.; Aravind, L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006, 117, 156–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, e00061-19. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses Executive Committee. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Philippe, N.; Legendre, M.; Doutre, G.; Couté, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: Amoeba Viruses with Genomes Up to 2.5 Mb Reaching That of Parasitic Eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, K.; Miyazaki, N.; Song, C.; Maia, F.R.N.C.; Reddy, H.K.N.; Abergel, C.; Claverie, J.M.; Hajdu, J.; Svenda, M.; Murata, K. Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high-voltage electron cryo-tomography and energy-filtered electron cryo-microscopy. Sci. Rep. 2017, 7, 13291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Etten, J.L.; Meints, R.H. Giant viruses infecting algae. Annu. Rev. Microbiol. 1999, 53, 447–494. [Google Scholar] [CrossRef] [Green Version]
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; De Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.M.; Raoult, D. A giant virus in amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef]
- Fischer, M.G.; Allen, M.J.; Wilson, W.H.; Suttle, C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef] [Green Version]
- Yutin, N.; Koonin, E.V. Proteorhodopsin genes in giant viruses. Biol. Direct 2012, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, S.W.; Coy, S.R.; Gann, E.R.; Moniruzzaman, M.; Stough, J.M.A. Standing on the shoulders of giant viruses: Five lessons learned about large viruses infecting small eukaryotes and the opportunities they create. PLoS Pathog. 2016, 12, e1005752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claverie, J.-M.; Abergel, C. Giant viruses: The difficult breaking of multiple epistemological barriers. Stud. Hist. Philos. Biol. Biomed. Sci. 2016, 59, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V.; Yutin, N. Multiple evolutionary origins of giant viruses. F1000Research 2018, 7, 1840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Needham, D.M.; Yoshizawa, S.; Hosaka, T.; Poirier, C.; Choi, C.J.; Hehenberger, E.; Irwin, N.A.T.T.; Wilken, S.; Yung, C.-M.M.; Bachy, C.; et al. A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc. Natl. Acad. Sci. USA 2019, 116, 20574–20583. [Google Scholar] [CrossRef] [Green Version]
- Brandes, N.; Linial, M. Giant Viruses—Big Surprises. Viruses 2019, 11, 404. [Google Scholar] [CrossRef] [Green Version]
- Rosenwasser, S.; Ziv, C.; van Creveld, S.G.; Vardi, A. Virocell metabolism: Metabolic innovations during host–virus interactions in the ocean. Trends Microbiol. 2016, 24, 821–832. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Martinez-Gutierrez, C.A.; Weinheimer, A.R.; Aylward, F.O. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat. Commun. 2020, 11, 1710. [Google Scholar] [CrossRef] [Green Version]
- Chinchar, V.G.; Waltzek, T.B.; Subramaniam, K. Ranaviruses and other members of the family Iridoviridae: Their place in the virosphere. Virology 2017, 511, 259–271. [Google Scholar] [CrossRef]
- Van Etten, J.L.; Agarkova, I.V.; Dunigan, D.D. Chloroviruses. Viruses 2020, 12, 20. [Google Scholar] [CrossRef] [Green Version]
- Mayer, J.A.; Taylor, F.J.R. A virus which lyses the marine nanoflagellate Micromonas pusilla. Nature 1979, 281, 299–301. [Google Scholar] [CrossRef]
- Bratbak, G.; Egge, J.K.; Heldal, M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 1993, 93, 39–48. [Google Scholar] [CrossRef]
- Garza, D.R.; Suttle, C.A. Large double-stranded DNA viruses which cause the lysis of a marine heterotrophic nanoflagellate (Bodo sp.) occur in natural marine viral communities. Aquat. Microb. Ecol. 1995, 9, 203–210. [Google Scholar] [CrossRef]
- Nagasaki, K.; Yamaguchi, M. Isolation of a virus infectious to the harmful bloom causing microalga Heterosigma akashiwo (Raphidophyceae). Aquat. Microb. Ecol. 1997, 13, 135–140. [Google Scholar] [CrossRef]
- Wilson, W.H. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 2005, 309, 1090–1092. [Google Scholar] [CrossRef] [PubMed]
- Moreau, H.; Piganeau, G.; Desdevises, Y.; Cooke, R.; Derelle, E.; Grimsley, N. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J. Virol. 2010, 84, 12555–12563. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, F.; Ueki, S. Evolution and phylogeny of large DNA viruses, Mimiviridae and Phycodnaviridae including mewly characterized Heterosigma akashiwo Virus. Front. Microbiol. 2016, 7, 1942. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [Green Version]
- Marciano-Cabral, F.; Cabral, G. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 2003, 16, 273–307. [Google Scholar] [CrossRef] [Green Version]
- Boyer, M.; Yutin, N.; Pagnier, I.; Barrassi, L.; Fournous, G.; Espinosa, L.; Robert, C.; Azza, S.; Sun, S.; Rossmann, M.G.; et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA 2009, 106, 21848–21853. [Google Scholar] [CrossRef] [Green Version]
- Arslan, D.; Legendre, M.; Seltzer, V.; Abergel, C.; Claverie, J.M. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. USA 2011, 108, 17486–17491. [Google Scholar] [CrossRef] [Green Version]
- Yoosuf, N.; Yutin, N.; Colson, P.; Shabalina, S.A.; Pagnier, I.; Robert, C.; Azza, S.; Klose, T.; Wong, J.; Rossmann, M.G.; et al. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the Megavirus lineage. Genome Biol. Evol. 2012, 4, 1324–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legendre, M.; Bartoli, J.; Shmakova, L.; Jeudy, S.; Labadie, K.; Adrait, A.; Lescot, M.; Poirot, O.; Bertaux, L.; Bruley, C.; et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 4274–4279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-depth study of Mollivirus sibericum, a new 30,000-yold giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreani, J.; Aherfi, S.; Bou Khalil, J.; Di Pinto, F.; Bitam, I.; Raoult, D.; Colson, P.; La Scola, B. Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses 2016, 8, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreani, J.; Khalil, J.Y.B.; Sevvana, M.; Benamar, S.; Di Pinto, F.; Bitam, I.; Colson, P.; Klose, T.; Rossmann, M.G.; Raoult, D.; et al. Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfarviridae and faustoviruses. J. Virol. 2017, 91, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Abrahão, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, G.; Blanc-Mathieu, R.; Song, C.; Kayama, Y.; Mochizuki, T.; Murata, K.; Ogata, H.; Takemurad, M.; Blanc-Mathieu, R.; Song, C.; et al. Medusavirus, a novel large DNA virus discovered from hot spring water. J. Virol. 2019, 93, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Reteno, D.G.; Benamar, S.; Khalil, J.B.; Andreani, J.; Armstrong, N.; Klose, T.; Rossmann, M.; Colson, P.; Raoult, D.; La Scola, B. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J. Virol. 2015, 89, 6585–6594. [Google Scholar] [CrossRef] [Green Version]
- Bajrai, L.H.; Benamar, S.; Azhar, E.I.; Robert, C.; Levasseur, A.; Raoult, D.; La Scola, B. Kaumoebavirus, a new virus that clusters with Faustoviruses and Asfarviridae. Viruses 2016, 8, 278. [Google Scholar] [CrossRef] [Green Version]
- Andreani, J.; Khalil, J.Y.B.; Baptiste, E.; Hasni, I.; Michelle, C.; Raoult, D.; Levasseur, A.; La Scola, B. Orpheovirus IHUMI-LCC2: A new virus among the giant viruses. Front. Microbiol. 2018, 8, 2643. [Google Scholar] [CrossRef] [Green Version]
- Bajrai, L.H.; Mougari, S.; Andreani, J.; Baptiste, E.; Delerce, J.; Raoult, D.; Azhar, E.I.; La Scola, B.; Levasseur, A. Isolation of Yasminevirus, the first member of Klosneuvirinae isolated in coculture with Vermamoeba vermiformis, demonstrates an extended arsenal of translational apparatus components. J. Virol. 2019, 94, e01534-19. [Google Scholar] [CrossRef] [PubMed]
- Needham, D.M.; Poirier, C.; Hehenberger, E.; Jiménez, V.; Swalwell, J.E.; Santoro, A.E.; Worden, A.Z. Targeted metagenomic recovery of four divergent viruses reveals shared and distinctive characteristics of giant viruses of marine eukaryotes. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, F.; Alteio, L.; Goudeau, D.; Ryan, E.M.; Yu, F.B.; Malmstrom, R.R.; Blanchard, J.; Woyke, T. Hidden diversity of soil giant viruses. Nat. Commun. 2018, 9, 4881. [Google Scholar] [CrossRef] [PubMed]
- Bellec, L.; Clerissi, C.; Edern, R.; Foulon, E.; Simon, N.; Grimsley, N.; Desdevises, Y. Cophylogenetic interactions between marine viruses and eukaryotic picophytoplankton. BMC Evol. Biol. 2014, 14, 59. [Google Scholar] [CrossRef] [Green Version]
- Derelle, R.; López-García, P.; Timpano, H.; Moreira, D. A phylogenomic framework to study the diversity and evolution of stramenopiles (=heterokonts). Mol. Biol. Evol. 2016, 33, 2890–2898. [Google Scholar] [CrossRef] [Green Version]
- Ševčíková, T.; Yurchenko, T.; Fawley, K.P.; Amaral, R.; Strnad, H.; Santos, L.M.A.; Fawley, M.W.; Eliáš, M. Plastid genomes and proteins illuminate the evolution of eustigmatophyte algae and their bacterial endosymbionts. Genome Biol. Evol. 2019, 11, 362–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fučíková, K.; Leliaert, F.; Cooper, E.D.; Škaloud, P.; D’Hondt, S.; De Clerck, O.; Gurgel, C.F.D.; Lewis, L.A.; Lewis, P.O.; Lopez-Bautista, J.M.; et al. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front. Ecol. Evol. 2014, 2, 63. [Google Scholar]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The new tree of eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Kang, S.; Tice, A.K.; Spiegel, F.W.; Silberman, J.D.; Pánek, T.; Čepička, I.; Kostka, M.; Kosakyan, A.; Alcântara, D.M.C.; Roger, A.J.; et al. Between a pod and a hard test: The deep evolution of amoebae. Mol. Biol. Evol. 2017, 34, 2258–2270. [Google Scholar] [CrossRef] [Green Version]
- Kukovetz, K.; Hertel, B.; Schvarcz, C.R.; Saponaro, A.; Manthey, M.; Burk, U.; Greiner, T.; Steward, G.F.; Van Etten, J.L.; Moroni, A.; et al. A functional K+ channel from Tetraselmis virus 1, a member of the Mimiviridae. Viruses 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Schvarcz, C.R. Cultivation and Characterization of Viruses Infecting Eukaryotic Phytoplankton from the Tropical North Pacific Ocean. Ph.D. Thesis, University of Hawai’i at Manoa, Honolulu, HI, USA, 2018. [Google Scholar]
- Ogata, H.; Toyoda, K.; Tomaru, Y.; Nakayama, N.; Shirai, Y.; Claverie, J.M.; Nagasaki, K. Remarkable sequence similarity between the dinoflagellate-infecting marine girus and the terrestrial pathogen African swine fever virus. Virol. J. 2009, 6, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzakhanyan, Y.; Gershon, P.D. Multisubunit DNA-dependent RNA polymerases from vaccinia virus and other nucleocytoplasmic large-DNA viruses: Impressions from the age of structure. Microbiol. Mol. Biol. Rev. 2017, 81, e00010-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abergel, C.; Legendre, M.; Claverie, J.M. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol. Rev. 2015, 39, 779–796. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Fabre, E.; Poirot, O.; Jeudy, S.; Lartigue, A.; Alempic, J.-M.M.; Beucher, L.; Philippe, N.N.; Bertaux, L.; Christo-Foroux, E.; et al. Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 2018, 9, 2285. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; La Scola, B.; Levasseur, A.; Caetano-Anollés, G.; Raoult, D. Mimivirus: Leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 2017, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Blanca, L.; Christo-Foroux, E.; Rigou, S.; Legendre, M. Comparative Analysis of the Circular and Highly Asymmetrical Marseilleviridae Genomes. Viruses 2020, 12. [Google Scholar] [CrossRef]
- Fabre, E.; Jeudy, S.; Santini, S.; Legendre, M.; Trauchessec, M.; Couté, Y.; Claverie, J.M.; Abergel, C. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Federici, B.A.; Vlak, J.M.; Hamm, J.J. Comparative study of virion structure, protein composition and genomic DNA of three ascovirus isolates. J. Gen. Virol. 1990, 71, 1661–1668. [Google Scholar] [CrossRef]
- Bideshi, D.K.; Demattei, M.-V.; Rouleux-Bonnin, F.; Stasiak, K.; Tan, Y.; Bigot, S.; Bigot, Y.; Federici, B.A. Genomic Sequence of Spodoptera frugiperda Ascovirus 1a, an Enveloped, Double-Stranded DNA Insect Virus That Manipulates Apoptosis for Viral Reproduction. J. Virol. 2006, 80, 11791–11805. [Google Scholar] [CrossRef] [Green Version]
- Majji, S.; Thodima, V.; Sample, R.; Whitley, D.; Deng, Y.; Mao, J.; Chinchar, V.G. Transcriptome analysis of Frog virus 3, the type species of the genus Ranavirus, family Iridoviridae. Virology 2009, 391, 293–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M. ICTV virus taxonomy profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Galindo, I.; Alonso, C. African swine fever virus: A review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condit, R.C.; Moussatche, N.; Traktman, P. In A Nutshell: Structure and Assembly of the Vaccinia Virion. Adv. Virus Res. 2006, 65, 31–124. [Google Scholar]
- Deeg, C.M.; Chow, C.E.T.; Suttle, C.A. The kinetoplastid-infecting Bodo saltans virus (Bsv), a window into the most abundant giant viruses in the sea. Elife 2018, 7, e33014. [Google Scholar] [CrossRef]
- Sandaa, R.A.; Heldal, M.; Castberg, T.; Thyrhaug, R.; Bratbak, G. Isolation and characterization of two viruses with large genome size infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae). Virology 2001, 290, 272–280. [Google Scholar] [CrossRef] [Green Version]
- Brussaard, C.P.D.; Short, S.M.; Frederickson, C.M.; Suttle, C.A. Isolation and phylogenetic analysis of novel viruses infecting the phytoplankton Phaeocystis globosa (Prymnesiophyceae). Appl. Environ. Microbiol. 2004, 70, 3700–3705. [Google Scholar] [CrossRef] [Green Version]
- Rowe, J.M.; Dunlap, J.R.; Gobler, C.J.; Anderson, O.R.; Gastrich, M.D.; Wilhelm, S.W. Isolation of a non-phage-like lytic virus infecting Aureococcus anophagefferens. J. Phycol. 2008, 44, 71–76. [Google Scholar] [CrossRef]
- Wilson, W.H.; Gilg, I.C.; Moniruzzaman, M.; Field, E.K.; Koren, S.; LeCleir, G.R.; Martínez Martínez, J.; Poulton, N.J.; Swan, B.K.; Stepanauskas, R.; et al. Genomic exploration of individual giant ocean viruses. ISME J. 2017, 11, 1736–1745. [Google Scholar] [CrossRef]
- Schulz, F.; Yutin, N.; Ivanova, N.N.; Ortega, D.R.; Lee, T.K.; Vierheilig, J.; Daims, H.; Horn, M.; Wagner, M.; Jensen, G.J.; et al. Giant viruses with an expanded complement of translation system components. Science 2017, 356, 82–85. [Google Scholar] [CrossRef] [Green Version]
- Yau, S.; Lauro, F.M.; DeMaere, M.Z.; Brown, M.V.; Thomas, T.; Raftery, M.J.; Andrews-Pfannkoch, C.; Lewis, M.; Hoffman, J.M.; Gibson, J.A.; et al. Virophage control of antarctic algal host-virus dynamics. Proc. Natl. Acad. Sci. USA 2011, 108, 6163–6168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schvarcz, C.R.; Steward, G.F. A giant virus infecting green algae encodes key fermentation genes. Virology 2018, 518, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Colson, P.; Raoult, D.; Koonin, E.V. Mimiviridae: Clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol. J. 2013, 10, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallot-Lavallée, L.; Blanc, G. A glimpse of nucleo-cytoplasmic large DNA virus biodiversity through the eukaryotic genomics window. Viruses 2017, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Gallot-Lavallée, L.; Blanc, G.; Claverie, J.-M. Comparative genomics of Chrysochromulina ericina virus and other microalga-infecting large DNA viruses highlights their intricate evolutionary relationship with the established Mimiviridae family. J. Virol. 2017, 91, e00230-17. [Google Scholar] [CrossRef] [Green Version]
- Blanc-Mathieu, R.; Dahle, H.; Hofgaard, A.; Brandt, D.; Ogata, H.; Sandaa, R.-A. The genome of a persistent giant algal virus encodes an unprecedented number of genes involved in energy metabolism. bioRxiv 2020. [Google Scholar] [CrossRef]
- Matsuyama, T.; Takano, T.; Nishiki, I.; Fujiwara, A.; Kiryu, I.; Inada, M.; Sakai, T.; Terashima, S.; Matsuura, Y.; Isowa, K.; et al. A novel Asfarvirus-like virus identified as a potential cause of mass mortality of abalone. Sci. Rep. 2020, 10, 4620. [Google Scholar] [CrossRef]
- Rozenberg, A.; Oppermann, J.; Wietek, J.; Fernandez Lahore, R.G.; Sandaa, R.-A.; Bratbak, G.; Hegemann, P.; Beja, O. Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses. Curr. Biol. 2020, 30. in press. [Google Scholar]
- Keeling, P.J.; Burki, F. Progress towards the Tree of Eukaryotes. Curr. Biol. 2019, 29, R808–R817. [Google Scholar] [CrossRef]
- Schulz, F.; Roux, S.; Paez-Espino, D.; Jungbluth, S.; Walsh, D.A.; Denef, V.J.; McMahon, K.D.; Konstantinidis, K.T.; Eloe-Fadrosh, E.A.; Kyrpides, N.C.; et al. Giant virus diversity and host interactions through global metagenomics. Nature 2020, 578, 432–436. [Google Scholar] [CrossRef]
- Maumus, F.; Epert, A.; Nogué, F.; Blanc, G. Plant genomes enclose footprints of past infections by giant virus relatives. Nat. Commun. 2014, 5, 4268. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, M. Plasmodesmata: Channels for viruses on the move. Methods Mol. Biol. 2014, 1217, 25–52. [Google Scholar]
- Dornas, F.P.; Khalil, J.Y.B.; Pagnier, I.; Raoult, D.; Abrahão, J.; La Scola, B. Isolation of new Brazilian giant viruses from environmental samples using a panel of protozoa. Front. Microbiol. 2015, 6, 1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, E.; Oosterhof, M.; Sandaa, R.A.; Larsen, A.; Pagarete, A. Emerging interaction patterns in the Emiliania huxleyi-EhV system. Viruses 2017, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Ku, C.; Hu, J.-M. Phylogenetic and cophylogenetic analyses of the leaf-nodule symbiosis in Ardisia Subgenus Crispardisia (Myrsinaceae): Evidence from nuclear and chloroplast markers and bacterial rrn operons. Int. J. Plant Sci. 2014, 175, 92–109. [Google Scholar] [CrossRef]
- Monier, A.; Chambouvet, A.; Milner, D.S.; Attah, V.; Terrado, R.; Lovejoy, C.; Moreau, H.; Santoro, A.E.; Derelle, É.; Richards, T.A. Host-derived viral transporter protein for nitrogen uptake in infected marine phytoplankton. Proc. Natl. Acad. Sci. USA 2017, 114, E7489–E7498. [Google Scholar] [CrossRef] [Green Version]
- Guglielmini, J.; Woo, A.C.; Krupovic, M.; Forterre, P.; Gaia, M. Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes. Proc. Natl. Acad. Sci. USA 2019, 116, 19585–19592. [Google Scholar] [CrossRef] [Green Version]
- DeAngelis, P.L.; Jing, W.; Graves, M.V.; Burbank, D.E.; Van Etten, J.L. Hyaluronan synthase of chlorella virus PBCV-1. Science 1997, 278, 1800–1803. [Google Scholar] [CrossRef] [Green Version]
- Tonetti, M.; Zanardi, D.; Gurnon, J.R.; Fruscione, F.; Armirotti, A.; Damonte, G.; Sturla, L.; De Flora, A.; Van Etten, J.L. Paramecium bursaria chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. J. Biol. Chem. 2003, 278, 21559–21565. [Google Scholar] [CrossRef] [Green Version]
- Chothi, M.P.; Duncan, G.A.; Armirotti, A.; Abergel, C.; Gurnon, J.R.; Van Etten, J.L.; Bernardi, C.; Damonte, G.; Tonetti, M. Identification of an L-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J. Virol. 2010, 84, 8829–8838. [Google Scholar] [CrossRef] [Green Version]
- Piacente, F.; Marin, M.; Molinaro, A.; De Castro, C.; Seltzer, V.; Salis, A.; Damonte, G.; Bernardi, C.; Claverie, J.M.; Abergel, C.; et al. Giant DNA virus Mimivirus encodes pathway for biosynthesis of unusual sugar 4-amino-4,6-dideoxy-D-glucose (Viosamine). J. Biol. Chem. 2012, 287, 3009–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graves, M.V.; Bernadt, C.T.; Cerny, R.; Van Etten, J.L. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 2001, 285, 332–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hülsmeier, A.J.; Hennet, T. O-Linked glycosylation in Acanthamoeba polyphaga mimivirus. Glycobiology 2014, 24, 703–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, M.; Myllyharju, J.; Tu, H.; Hellman, M.; Kivirikko, K.I. Evidence for 4-hydroxyproline in viral proteins. Characterization of a viral prolyl 4-hydroxylase and its peptide substrates. J. Biol. Chem. 1999, 274, 22131–22134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, I.; Redrejo-Rodríguez, M.; Rodríguez, J.M.; Alejo, A.; Salas, J.; Salas, M.L. African swine fever virus pB119L protein is a flavin adenine dinucleotide-linked sulfhydryl oxidase. J. Virol. 2006, 80, 3157–3166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakim, M.; Ezerina, D.; Alon, A.; Vonshak, O.; Fass, D. Exploring ORFan domains in giant viruses: Structure of mimivirus sulfhydryl oxidase R596. PLoS ONE 2012, 7, e50649. [Google Scholar] [CrossRef] [PubMed]
- Thai, V.; Renesto, P.; Fowler, C.A.; Brown, D.J.; Davis, T.; Gu, W.; Pollock, D.D.; Kern, D.; Raoult, D.; Eisenmesser, E.Z. Structural, biochemical, and in vivo characterization of the first virally encoded cyclophilin from the mimivirus. J. Mol. Biol. 2008, 378, 71–86. [Google Scholar] [CrossRef]
- Redrejo-Rodríguez, M.; Salas, M.L. Repair of base damage and genome maintenance in the nucleo-cytoplasmic large DNA viruses. Virus Res. 2014, 179, 12–25. [Google Scholar] [CrossRef]
- Yutin, N.; Koonin, E.V. Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: A case of hidden complexity. Biol. Direct 2009, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Aherfi, S.; Brahim Belhaouari, D.; Pinault, L.; Decloquement, P.; Abrahao, J.; Colson, P.; Levasseur, A.; Lamb, D.C.; Chabriere, E.; Raoult, D.; et al. Tricarboxylic acid cycle and proton gradient in Pandoravirus massiliensis: Is it still a virus? bioRxiv 2020. [Google Scholar] [CrossRef]
- Zabelskii, D.; Alekseev, A.; Kovalev, K.; Oliveira, A.-S.; Balandin, T.; Soloviov, D.; Bratanov, D.; Volkov, D.; Vaganova, S.; Astashkin, R.; et al. Viral channelrhodopsins: Calcium-dependent Na+/K+ selective light-gated channels. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Baumann, S.; Sander, A.; Gurnon, J.R.; Yanai-Balser, G.M.; Van Etten, J.L.; Piotrowski, M. Chlorella viruses contain genes encoding a complete polyamine biosynthetic pathway. Virology 2007, 360, 209–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeudy, S.; Lartigue, A.; Claverie, J.-M.; Abergel, C. Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase. J. Virol. 2009, 83, 7142–7150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landstein, D.; Graves, M.V.; Burbank, D.E.; Deangelis, P.; Van Etten, J.L. Chlorella virus PBCV-1 encodes functional glutamine: Fructose-6- phosphate amidotransferase and UDP-glucose dehydrogenase enzymes. Virology 1998, 250, 388–396. [Google Scholar] [CrossRef]
- Piacente, F.; Bernardi, C.; Marin, M.; Blanc, G.; Abergel, C.; Tonetti, M.G. Characterization of a UDP-N-acetylglucosamine biosynthetic pathway encoded by the giant DNA virus mimivirus. Glycobiology 2014, 24, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Piacente, F.; De Castro, C.; Jeudy, S.; Molinaro, A.; Salis, A.; Damonte, G.; Bernardi, C.; Abergel, C.; Tonetti, M.G. Giant virus Megavirus chilensis encodes the biosynthetic pathway for uncommon acetamido sugars. J. Biol. Chem. 2014, 289, 24428–24439. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Adams, B.; Gurnon, J.R.; Ye, Y.; Van Etten, J.L. Characterization of two chitinase genes and one chitosanase gene encoded by Chlorella virus PBCV-1. Virology 1999, 263, 376–387. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Gurnon, J.R.; Adams, B.J.; Graves, M.V.; Van Etten, J.L. Characterization of a beta-1,3-glucanase encoded by chlorella virus PBCV-1. Virology 2000, 276, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Suda, K.; Tanji, Y.; Hori, K.; Unno, H. Evidence for a novel chlorella virus-encoded alginate lyase. FEMS Microbiol. Lett. 1999, 180, 45–53. [Google Scholar] [CrossRef]
- Sugimoto, I.; Hiramatsu, S.; Murakami, D.; Fujie, M.; Usami, S.; Yamada, T. Algal-lytic activities encoded by Chlorella virus CVK2. Virology 2000, 277, 119–126. [Google Scholar] [CrossRef]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J.; et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010, 22, 2943–2955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klose, T.; Herbst, D.A.; Zhu, H.; Max, J.P.; Kenttämaa, H.I.; Rossmann, M.G. A mimivirus enzyme that participates in viral entry. Structure 2015, 23, 1058–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vardi, A.; Van Mooy, B.A.S.; Fredricks, H.F.; Popendorf, K.J.; Ossolinski, J.E.; Haramaty, L.; Bidle, K.D. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science 2009, 326, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Monier, A.; Pagarete, A.; De Vargas, C.; Allen, M.J.; Read, B.; Claverie, J.M.; Ogata, H. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Res. 2009, 19, 1441–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plugge, B.; Gazzarrini, S.; Nelson, M.; Cerana, R.; Van Etten, J.L.; Derst, C.; DiFrancesco, D.; Moroni, A.; Thiel, G. A potassium channel protein encoded by Chlorella virus PBCV-1. Science 2000, 287, 1641–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cock, J.M.; Sterck, L.; Rouzé, P.; Scornet, D.; Allen, A.E.; Amoutzias, G.; Anthouard, V.; Artiguenave, F.; Aury, J.M.; Badger, J.H.; et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010, 465, 617–621. [Google Scholar] [CrossRef] [Green Version]
- Thiel, G.; Moroni, A.; Blanc, G.; Van Etten, J.L. Potassium ion channels: Could they have evolved from viruses? Plant Physiol. 2013, 162, 1215–1224. [Google Scholar] [CrossRef]
- Siotto, F.; Martin, C.; Rauh, O.; Van Etten, J.L.; Schroeder, I.; Moroni, A.; Thiel, G. Viruses infecting marine picoplancton encode functional potassium ion channels. Virology 2014, 466–467, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Gazzarrini, S.; Kang, M.; Epimashko, S.; Van Etten, J.L.; Dainty, J.; Thiel, G.; Moroni, A. Chlorella virus MT325 encodes water and potassium channels that interact synergistically. Proc. Natl. Acad. Sci. USA 2006, 103, 5355–5360. [Google Scholar] [CrossRef] [Green Version]
- Greiner, T.; Moroni, A.; Van Etten, J.L.; Thiel, G. Genes for membrane transport proteins: Not so rare in viruses. Viruses 2018, 10, 456. [Google Scholar] [CrossRef]
- Monier, A.; Welsh, R.M.; Gentemann, C.; Weinstock, G.; Sodergren, E.; Armbrust, E.V.; Eisen, J.A.; Worden, A.Z. Phosphate transporters in marine phytoplankton and their viruses: Cross-domain commonalities in viral-host gene exchanges. Environ. Microbiol. 2012, 14, 162–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greiner, T.; Ramos, J.; Alvarez, M.C.; Gurnon, J.R.; Kang, M.; Van Etten, J.L.; Moroni, A.; Thiel, G. Functional HAK/KUP/KT-like potassium transporter encoded by chlorella viruses. Plant J. 2011, 68, 977–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonza, M.C.; Martin, H.; Kang, M.; Lewis, G.; Greiner, T.; Giacometti, S.; Van Etten, J.L.; De Michelis, M.I.; Thiel, G.; Moroni, A. A functional calcium-transporting ATPase encoded by chlorella viruses. J. Gen. Virol. 2010, 91, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.N.; Greenleaf, W.B.; Ostrov, D.A.; Moyer, R.W. Amsacta moorei entomopoxvirus expresses an active superoxide dismutase. J. Virol. 2004, 78, 10265–10275. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.; Duncan, G.A.; Kuszynski, C.; Oyler, G.; Zheng, J.; Becker, D.F.; Van Etten, J.L. Chlorovirus PBCV-1 encodes an active copper-zinc superoxide dismutase. J. Virol. 2014, 88, 12541–12550. [Google Scholar] [CrossRef] [Green Version]
- Lartigue, A.; Burlat, B.; Coutard, B.; Chaspoul, F.; Claverie, J.-M.; Abergel, C. The megavirus chilensis Cu,Zn-superoxide dismutase: The first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme. J. Virol. 2015, 89, 824–832. [Google Scholar] [CrossRef] [Green Version]
- Legendre, M.; Audic, S.; Poirot, O.; Hingamp, P.; Seltzer, V.; Byrne, D.; Lartigue, A.; Lescot, M.; Bernadac, A.; Poulain, J.; et al. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010, 20, 664–674. [Google Scholar] [CrossRef] [Green Version]
- Ku, C.; Sheyn, U.; Sebé-Pedrós, A.; Ben-Dor, S.; Schatz, D.; Tanay, A.; Rosenwasser, S.; Vardi, A. A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Sci. Adv. 2020, 6, eaba4137. [Google Scholar] [CrossRef]
- Müller, M.; Mentel, M.; van Hellemond, J.J.; Henze, K.; Woehle, C.; Gould, S.B.; Yu, R.-Y.; van der Giezen, M.; Tielens, A.G.M.; Martin, W.F. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 2012, 76, 444–495. [Google Scholar] [CrossRef] [Green Version]
- Van Etten, J.; Agarkova, I.; Dunigan, D.; Tonetti, M.; De Castro, C.; Duncan, G.; Van Etten, J.L.; Agarkova, I.; Dunigan, D.D.; Tonetti, M.; et al. Chloroviruses Have a Sweet Tooth. Viruses 2017, 9, 88. [Google Scholar] [CrossRef]
- Rosenwasser, S.; Mausz, M.A.; Schatz, D.; Sheyn, U.; Malitsky, S.; Aharoni, A.; Weinstock, E.; Tzfadia, O.; Ben-Dor, S.; Feldmesser, E.; et al. Rewiring Host Lipid Metabolism by Large Viruses Determines the Fate of Emiliania huxleyi, a Bloom-Forming Alga in the Ocean. Plant Cell 2014, 26, 2689–2707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thamatrakoln, K.; Talmy, D.; Haramaty, L.; Maniscalco, C.; Latham, J.; Knowles, B.; Natale, F.; Coolen, M.J.L.; Follows, M.J.; Bidle, K.D. Light regulation of coccolithophore host-virus interactions. New Phytol. 2019, 221, 1289–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pushkarev, A.; Inoue, K.; Larom, S.; Flores-uribe, J.; Singh, M.; Konno, M.; Tomida, S.; Philosof, A.; Sharon, I.; Yutin, N.; et al. A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 2018, 558, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Santini, S.; Jeudy, S.; Bartoli, J.; Poirot, O.; Lescot, M.; Abergel, C.; Barbe, V.; Wommack, K.E.; Noordeloos, A.A.M.; Brussaard, C.P.D.; et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. USA 2013, 110, 10800–10805. [Google Scholar] [CrossRef] [Green Version]
- Filée, J.; Pouget, N.; Chandler, M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses. BMC Evol. Biol. 2008, 8, 320. [Google Scholar] [CrossRef] [Green Version]
- Christo-Foroux, E.; Alempic, J.; Lartigue, A.; Santini, S.; Labadie, K.; Legendre, M.; Abergel, C.; Claverie, J. Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses. J. Virol. 2020, 94, 1–16. [Google Scholar] [CrossRef]
- Doutre, G.; Philippe, N.; Abergel, C.; Claverie, J.-M. Genome Analysis of the First Marseilleviridae Representative from Australia Indicates that Most of Its Genes Contribute to Virus Fitness. J. Virol. 2014, 88, 14340–14349. [Google Scholar] [CrossRef] [Green Version]
- Claverie, J.M. Fundamental difficulties prevent the reconstruction of the deep phylogeny of viruses. Viruses 2020, 12, 1130. [Google Scholar] [CrossRef]
- Boyer, M.; Azza, S.; Barrassi, L.; Klose, T.; Campocasso, A.; Pagnier, I.; Fournous, G.; Borg, A.; Robert, C.; Zhang, X.; et al. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. USA 2011, 108, 10296–10301. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.F.; Steward, G.F. Host traits drive viral life histories across phytoplankton viruses. Am. Nat. 2018, 191, 566–581. [Google Scholar] [CrossRef]
- Weynberg, K.D.; Allen, M.J.; Wilson, W.H. Marine prasinoviruses and their tiny plankton hosts: A review. Viruses 2017, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Wolf, Y.I.; Koonin, E.V. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology 2014, 466–467, 38–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betts, H.C.; Puttick, M.N.; Clark, J.W.; Williams, T.A.; Donoghue, P.C.J.; Pisani, D. Integrated genomic and fossil evidence illuminates life’s early evolution and eukaryote origin. Nat. Ecol. Evol. 2018, 2, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Imachi, H.; Nobu, M.K.; Nakahara, N.; Morono, Y.; Ogawara, M.; Takaki, Y.; Takano, Y.; Uematsu, K.; Ikuta, T.; Ito, M.; et al. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 2020, 577, 519–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brueckner, J.; Martin, W.F. Bacterial genes outnumber archaeal genes in eukaryotic genomes. Genome Biol. Evol. 2020, 12, 282–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, C.; Nelson-Sathi, S.; Roettger, M.; Garg, S.; Hazkani-Covo, E.; Martin, W.F. Endosymbiotic gene transfer from prokaryotic pangenomes: Inherited chimerism in eukaryotes. Proc. Natl. Acad. Sci. USA 2015, 112, 10139–10146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, C.; Sun, T. Did giant and large dsDNA viruses originate before their eukaryotic hosts? Proc. Natl. Acad. Sci. USA 2020, 117, 2747–2748. [Google Scholar] [CrossRef]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Popa, O.; Dagan, T. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 2011, 14, 615–623. [Google Scholar] [CrossRef]
- McInerney, J.O.; McNally, A.; O’Connell, M.J. Why prokaryotes have pangenomes. Nat. Microbiol. 2017, 2, 17040. [Google Scholar] [CrossRef]
- Nagies, F.S.P.; Brueckner, J.; Tria, F.D.K.; Martin, W.F. A spectrum of verticality across genes. PLoS Genet. 2020, 16, e1009200. [Google Scholar] [CrossRef] [PubMed]
| Family | Virus (Lineage) | Shape | Size (nm) | Genome Size (kb) | DNA | tRNAs | RNA Polymerase Subunits [54] | Replication Cycle | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Pandoraviridae (Phycodnaviridae) | Pandoravirus salinus (Pandoravirus) | Ovoid with a pore | 1200 × 500 | 2770 | Linear | 3 | 4 | Nuclear and cytoplasmic | [5,55,56] |
| Phycodnaviridae | Paramecium bursaria Chlorella virus 1 (Chlorovirus) | Icosahedral with a spike | 170 | 331 | Linear | 11 | 0 | Nuclear and cytoplasmic | [7,19] |
| Mimiviridae | Acanthamoeba polyphaga mimivirus (Mimivirus) | Icosahedral (with fibers) | 390 (630) | 1181 | Linear | 6 | 9 | Cytoplasmic | [55,57] |
| Pithoviridae | Pithovirus sibericum (Pithovirus) | Ovoid with a capped pore | 1500 × 800 | 610 | Circular | 0 | 4 | Cytoplasmic | [6,32,55] |
| Marseilleviridae | Marseillevirus marseillevirus (Marseilleviridae A) | Icosahedral | 250 | 368 | Circular | 0 | 3 | Cytoplasmic, involving the nucleus | [29,58,59] |
| Ascoviridae (Iridoviridae) | Spodoptera frugiperda ascovirus 1a (Ascovirus) | Bacilliform or allantoid | 400 × 130 | 157 | Circular | 0 | 4 | Nuclear and cytoplasmic | [60,61] |
| Iridoviridae | Frog virus 3 (Alphairidovirinae) | Icosahedral | 175 | 106 | Linear | 0 | 2 | Nuclear and cytoplasmic | [18,62] |
| Asfarviridae | African swine fever virus BA71V (Asfarvirus) | Icosahedral | 200 | 170 | Linear | 0 | 7 | Nuclear and cytoplasmic | [63,64] |
| Poxviridae | Vaccinia virus (other Chordopoxvirinae) | Brick-shaped | 310 × 240 | 195 | Linear | 0 | 9 | Cytoplasmic | [65] |
| COG Category | Function | Distribution | Putative LGT Source | Reference |
|---|---|---|---|---|
| Cellular Processes and Signaling | ||||
| [M] Cell wall/membrane/envelope biogenesis | Hyaluronan synthesis | Chlorovirus | - | [89] |
| Fucose synthesis | Chlorovirus | - | [90] | |
| L-rhamnose synthesis | Chlorovirus; Mimivirus | Trebouxiophyceae; eukaryotes | [91] | |
| 3-deoxy-D-manno-octulosonate synthesis | Cafeteriavirus | Phagocytosed bacteria in the Cafeteria host | [9] | |
| 4-Amino-4,6-dideoxy-D-glucose (Viosamine) synthesis | Mimivirus | - | [92] | |
| [O] Posttranslational modification, protein turnover, chaperones | Protein glycosylase | Chlorovirus; Mimivirus | - | [93,94] |
| Prolyl 4-hydroxylase | Chlorovirus | - | [95] | |
| Sulfhydryl oxidase | Asfarvirus; Mimivirus | - | [96,97] | |
| Isomerization of peptide bonds (Cyclophilin) | Mimivirus | - | [98] | |
| Information Storage and Processing | ||||
| [J] Translation, ribosomal structure, and biogenesis | Aminoacyl tRNA synthetase | Mimiviridae except Mesomimivirinae; Pandoravirus; Orpheovirus | - | [13] |
| Translation factors | Mimiviridae; Chlorovirus; Pandoraviridae; Marseilleviridae; Asfarviridae; Alphairidovirinae; Pithoviridae; Alphaentomopoxvirus | - | [13] | |
| [K] Transcription | DNA-dependent RNA polymerase (DDRP) subunits | Most NCLDVs except many phycodnavirids | - | [37,54] |
| Transcription elongation factors (TFIIS) | Most NCLDVs | - | [54] | |
| General transcription factors (TBP-like) | Some phycodnavirids and Mesomimivirinae | - | [54] | |
| General transcription factors (TFIIB-like) | Asfarviridae; Mimiviridae; Marseilleviridae; Pithovirus; Prasinovirus | - | [54] | |
| [L] Replication, recombination, and repair | DNA Glycosylase | Marseilleviridae; most mimivirids; Poxviridae | - | [99] |
| NAD/ATP dependent DNA ligase | Most NCLDV families | - | [100] | |
| Metabolism | ||||
| [C] Energy production and conversion | Tricarboxylic acid (TCA) cycle | Mimiviridae MAGs; Prymnesium virus; Pandoravirus | - | [17,77,101] |
| Cellular fermentation | Tetraselmis virus | Tetraselmis or related chlorophytes | [73] | |
| [C] Energy production and conversion; [T] Signal transduction mechanisms | Rhodopsin | Phaeocystis virus; Choanovirus; metagenomic contigs | - | [10,14,79,102] |
| [E] Amino acid transport and metabolism | Polyamine synthesis | Chlorovirus | - | [103] |
| Amino acid synthesis | Prasinovirus | Chlorophyta or bacteria | [25] | |
| [F] Nucleotide transport and metabolism | Nucleoside-diphosphate kinase | Mimivirus | - | [104] |
| [G] Carbohydrate transport and metabolism | UDP-N-acetylglucosamine synthesis | Chlorovirus; Mimiviridae | Bacteria; - | [105,106] |
| UDP-2-acetamido-2,6-dideoxy-hexose synthesis | Megavirus | - | [107] | |
| Cell wall (polysaccharide) degradation | Chlorovirus; Mimivirus | Non-Chloroplastida sources; - | [108,109,110,111,112,113] | |
| [I] Lipid transport and metabolism | Sphingolipid synthesis | Coccolithovirus | Isochrysidales | [24,114,115] |
| [P] Inorganic ion transport and metabolism | Potassium ion channel | Chlorovirus; Phaeovirus; Prasinovirus; many Mesomimivirinae viruses | - | [51,116,117,118,119] |
| Aquaporin | Chlorovirus | - | [120] | |
| Sodium/phosphate symporter | Coccolithovirus; Prasinovirus | Isochrysidales; Mamiellales | [121,122] | |
| Potassium ion transporter | Chlorovirus | Trebouxiophyceae | [121,123] | |
| Ammonium transporter | Ostreococcus virus; Haptolina virus | Ostreococcus; - | [87,121] | |
| Calcium-transporting ATPase | Chlorovirus | - | [124] | |
| Cu/Zn superoxide dismutase | Betaentomopoxvirus; Chlorovirus; Megavirus | - | [125,126,127] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.-W.; Yang, C.-L.; Kao, T.-T.; Wang, T.-H.; Lai, M.-W.; Ku, C. Host Range and Coding Potential of Eukaryotic Giant Viruses. Viruses 2020, 12, 1337. https://doi.org/10.3390/v12111337
Sun T-W, Yang C-L, Kao T-T, Wang T-H, Lai M-W, Ku C. Host Range and Coding Potential of Eukaryotic Giant Viruses. Viruses. 2020; 12(11):1337. https://doi.org/10.3390/v12111337
Chicago/Turabian StyleSun, Tsu-Wang, Chia-Ling Yang, Tzu-Tong Kao, Tzu-Haw Wang, Ming-Wei Lai, and Chuan Ku. 2020. "Host Range and Coding Potential of Eukaryotic Giant Viruses" Viruses 12, no. 11: 1337. https://doi.org/10.3390/v12111337
APA StyleSun, T.-W., Yang, C.-L., Kao, T.-T., Wang, T.-H., Lai, M.-W., & Ku, C. (2020). Host Range and Coding Potential of Eukaryotic Giant Viruses. Viruses, 12(11), 1337. https://doi.org/10.3390/v12111337






