Characterization and Diversity of 243 Complete Human Papillomavirus Genomes in Cervical Swabs Using Next Generation Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. Anyplex II HPV28
2.3. Next Generation Sequencing
2.3.1. Library Preparation
2.3.2. Bioinformatic Analysis
Full Genome Assembly and Annotation
HPV Detection
2.4. Phylogenetic Analysis and Lineage Classification
2.5. Statistical Analysis
2.6. Data Access
2.7. Ethical Approval
3. Results
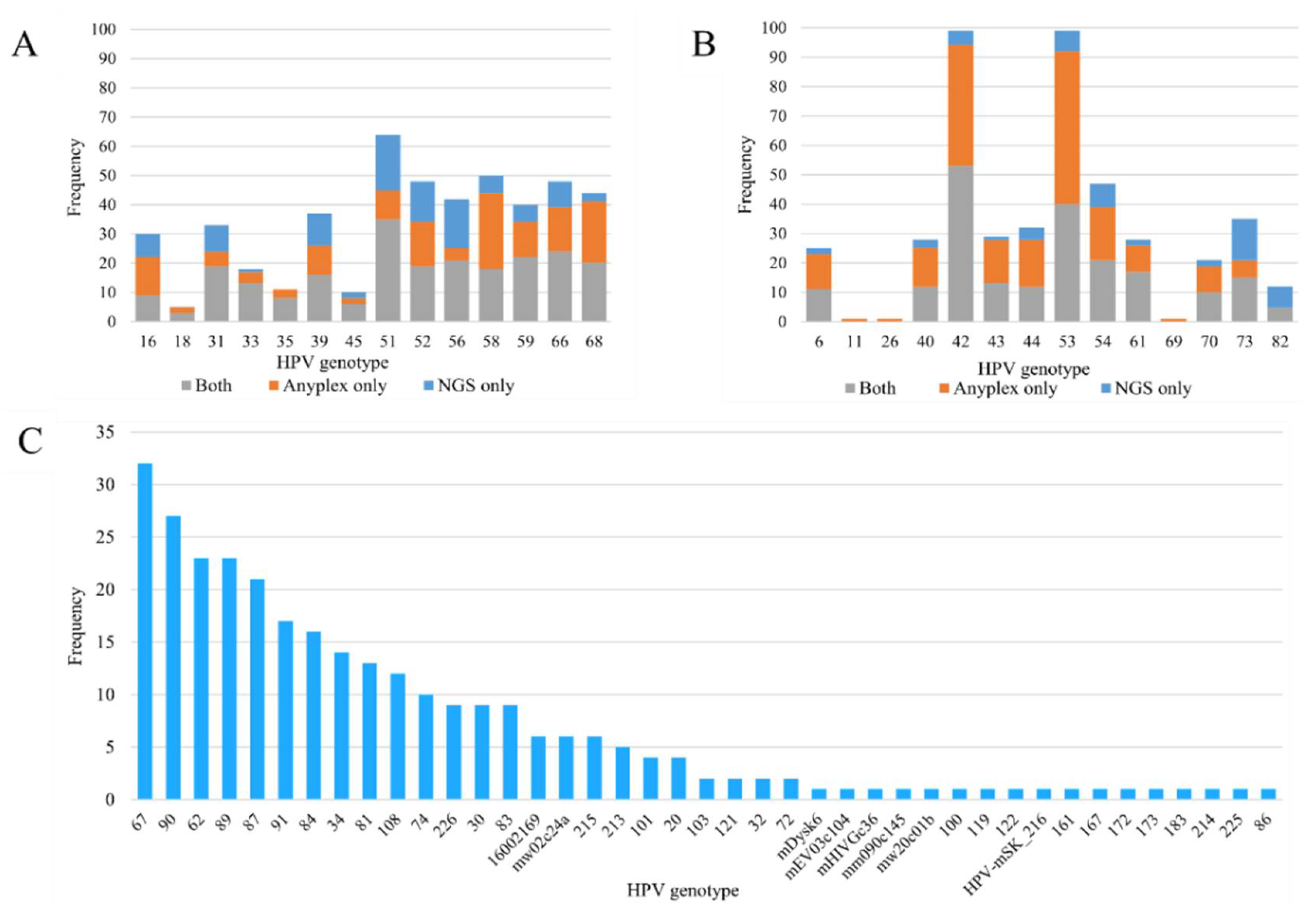
3.1. NGS and Anyplex II HPV 28
3.2. HPV Variant Distribution and Lineage Classification
3.2.1. High-Risk HPV
3.2.2. Low-Risk HPV
3.2.3. Gamma HPV
4. Discussion
Supplementary Materials
 ) indicates novel lineage/sublineage. The phylogenetic trees are shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number. Figure S3A–H. Phylogenetic trees and pairwise distance comparisons of high-risk HPV complete genome variants. Phylogenetic trees were inferred from a global alignment of the complete genome nucleotide sequences of the variants from following high risk HPV genotypes using RAxML: (A) HPV16, (B) HPV31, (C) HPV33, (D) HPV35, (E) HPV39, (F) HPV51, (G) HPV56, (H) HPV58, (I) 68. The phylogenetic trees are shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number. Figure S4A–H. Phylogenetic trees and pairwise distance comparisons of low-risk complete genome variants. Phylogenetic trees were inferred from a global alignment of the complete genome nucleotide sequences of the variants from following low risk HPV genotypes using RAxML: (A) HPV6, (B) HPV30, (C) HPV44, (D) HPV53, (E) HPV70, (F) HPV82, (G) HPV87, (H) HPV89. Phylogenetic trees are shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.
) indicates novel lineage/sublineage. The phylogenetic trees are shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number. Figure S3A–H. Phylogenetic trees and pairwise distance comparisons of high-risk HPV complete genome variants. Phylogenetic trees were inferred from a global alignment of the complete genome nucleotide sequences of the variants from following high risk HPV genotypes using RAxML: (A) HPV16, (B) HPV31, (C) HPV33, (D) HPV35, (E) HPV39, (F) HPV51, (G) HPV56, (H) HPV58, (I) 68. The phylogenetic trees are shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number. Figure S4A–H. Phylogenetic trees and pairwise distance comparisons of low-risk complete genome variants. Phylogenetic trees were inferred from a global alignment of the complete genome nucleotide sequences of the variants from following low risk HPV genotypes using RAxML: (A) HPV6, (B) HPV30, (C) HPV44, (D) HPV53, (E) HPV70, (F) HPV82, (G) HPV87, (H) HPV89. Phylogenetic trees are shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Einstein, M.H.; Schiller, J.T.; Viscidi, R.P.; Strickler, H.D.; Coursaget, P.; Tan, T.; Halsey, N.; Jenkins, D. Clinician’s guide to human papillomavirus immunology: Knowns and unknowns. Lancet Infect. Dis. 2009, 9, 347–356. [Google Scholar] [CrossRef]
- de Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Chan, P.K.; Zhang, C.; Park, J.S.; Smith-McCune, K.K.; Palefsky, J.M.; Giovannelli, L.; Coutlee, F.; Hibbitts, S.; Konno, R.; Settheetham-Ishida, W.; et al. Geographical distribution and oncogenic risk association of human papillomavirus type 58 E6 and E7 sequence variations. Int. J. Cancer 2013, 132, 2528–2536. [Google Scholar] [CrossRef]
- Chen, Z.; Schiffman, M.; Herrero, R.; Desalle, R.; Anastos, K.; Segondy, M.; Sahasrabuddhe, V.V.; Gravitt, P.E.; Hsing, A.W.; Burk, R.D. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS ONE 2011, 6, e20183. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; Ghissassi, F.E.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part B: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- Arbyn, M.; Tommasino, M.; Depuydt, C.; Dillner, J. Are 20 human papillomavirus types causing cervical cancer? J. Pathol. 2014, 234, 431–435. [Google Scholar] [CrossRef]
- Xi, L.F.; Kiviat, N.B.; Hildesheim, A.; Galloway, D.A.; Wheeler, C.M.; Ho, J.; Koutsky, L.A. Human papillomavirus type 16 and 18 variants: Race-related distribution and persistence. J. Natl. Cancer Inst. 2006, 98, 1045–1052. [Google Scholar] [CrossRef] [Green Version]
- Xi, L.F.; Schiffman, M.; Koutsky, L.A.; Hulbert, A.; Lee, S.K.; Defilippis, V.; Shen, Z.; Kiviat, N.B. Association of human papillomavirus type 31 variants with risk of cervical intraepithelial neoplasia grades 2–3. Int. J. Cancer 2012, 131, 2300–2307. [Google Scholar] [CrossRef]
- Mirabello, L.; Yeager, M.; Cullen, M.; Boland, J.F.; Chen, Z.; Wentzensen, N.; Zhang, X.; Yu, K.; Yang, Q.; Mitchell, J.; et al. HPV16 Sublineage Associations With Histology-Specific Cancer Risk Using HPV Whole-Genome Sequences in 3200 Women. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [Green Version]
- Godínez, J.M.; Heideman, D.A.M.; Gheit, T.; Alemany, L.; Snijders, P.J.F.; Tommasino, M.; Meijer, C.J.L.M.; de Sanjosé, S.; Bosch, F.X.; Bravo, I.G. Differential presence of Papillomavirus variants in cervical cancer: An analysis for HPV33, HPV45 and HPV58. Infect. Genet. Evol. 2013, 13, 96–104. [Google Scholar] [CrossRef]
- Nicolás-Párraga, S.; Gandini, C.; Pimenoff, V.N.; Alemany, L.; de Sanjosé, S.; Bosch, F.X.; Bravo, I.G.; The RIS HPV TT and HPV VVAP study groups. HPV16 variants distribution in invasive cancers of the cervix, vulva, vagina, penis, and anus. Cancer Med. 2016, 5, 2909–2919. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.F.; Schiffman, M.; Koutsky, L.A.; Hughes, J.P.; Winer, R.L.; Mao, C.; Hulbert, A.; Lee, S.K.; Shen, Z.; Kiviat, N.B. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Schiffman, M.; Herrero, R.; DeSalle, R.; Anastos, K.; Segondy, M.; Sahasrabuddhe, V.V.; Gravitt, P.E.; Hsing, A.W.; Chan, P.K.S.; et al. Classification and evolution of human papillomavirus genome variants: Alpha-5 (HPV26, 51, 69, 82), Alpha-6 (HPV30, 53, 56, 66), Alpha-11 (HPV34, 73), Alpha-13 (HPV54) and Alpha-3 (HPV61). Virology 2018, 516, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.D.; Alves, B.M.; Prellwitz, I.M.; Furtado, C.; Meyrelles, A.R.; Machado, E.S.; Seuanez, H.N.; Soares, M.A.; Soares, E.A. Identification of novel human papillomavirus lineages and sublineages in HIV/HPV-coinfected pregnant women by next-generation sequencing. Virology 2016, 493, 202–208. [Google Scholar] [CrossRef]
- Jelen, M.M.; Chen, Z.; Kocjan, B.J.; Hošnjak, L.; Burt, F.J.; Chan, P.K.S.; Chouhy, D.; Combrinck, C.E.; Estrade, C.; Fiander, A.; et al. Global Genomic Diversity of Human Papillomavirus 11 Based on 433 Isolates and 78 Complete Genome Sequences. J. Virol. 2016, 90, 5503–5513. [Google Scholar] [CrossRef] [Green Version]
- Poljak, M.; Valenčak, A.O.; Domjanič, G.G.; Xu, L.; Arbyn, M. Commercially available molecular tests for human papillomaviruses: A global overview. Clin. Microbiol. Infect. 2020. [Google Scholar] [CrossRef]
- Ostrbenk, A.; Xu, L.; Arbyn, M.; Poljak, M. Clinical and Analytical Evaluation of the Anyplex II HPV HR Detection Assay within the VALGENT-3 Framework. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [Green Version]
- Hesselink, A.T.; Sahli, R.; Berkhof, J.; Snijders, P.J.; van der Salm, M.L.; Agard, D.; Bleeker, M.C.; Heideman, D.A. Clinical validation of Anyplex II HPV HR Detection according to the guidelines for HPV test requirements for cervical cancer screening. J. Clin. Virol. 2016, 76, 36–39. [Google Scholar] [CrossRef] [Green Version]
- Latsuzbaia, A.; Tapp, J.; Nguyen, T.; Fischer, M.; Arbyn, M.; Weyers, S.; Mossong, J. Analytical performance evaluation of Anyplex II HPV28 and Euroarray HPV for genotyping of cervical samples. Diagn. Microbiol. Infect. Dis. 2016, 85, 318–322. [Google Scholar] [CrossRef]
- Latsuzbaia, A.; Arbyn, M.; Tapp, J.; Fischer, M.; Weyers, S.; Pesch, P.; Mossong, J. Effectiveness of bivalent and quadrivalent human papillomavirus vaccination in Luxembourg. Cancer Epidemiol. 2019, 63, 101593. [Google Scholar] [CrossRef]
- Arroyo, L.S.; Smelov, V.; Bzhalava, D.; Eklund, C.; Hultin, E.; Dillner, J. Next generation sequencing for human papillomavirus genotyping. J. Clin. Virol. 2013, 58, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Muhr, L.S.A.; Bzhalava, D.; Lagheden, C.; Eklund, C.; Johansson, H.; Forslund, O.; Dillner, J.; Hultin, E. Does human papillomavirus-negative condylomata exist? Virology 2015, 485, 283–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galati, L.; Brancaccio, R.N.; Robitaille, A.; Cuenin, C.; Luzi, F.; Fiorucci, G.; Chiantore, M.V.; Marascio, N.; Matera, G.; Liberto, M.C.; et al. Detection of human papillomaviruses in paired healthy skin and actinic keratosis by next generation sequencing. Papillomavirus Res. 2020, 9, 100196. [Google Scholar] [CrossRef] [PubMed]
- Murahwa, A.T.; Meiring, T.L.; Mbulawa, Z.Z.A.; Williamson, A.-L. Complete Genome Sequences of Four Novel Human Gammapapillomavirus Types, HPV-219, HPV-220, HPV-221, and HPV-222, Isolated from Penile Skin Swabs from South African Men. Genome Announc. 2018, 6, e00584-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocjan, B.J.; Steyer, A.; Sagadin, M.; Hosnjak, L.; Poljak, M. Novel human papillomavirus type 174 from a cutaneous squamous cell carcinoma. Genome Announc. 2013, 1, e00445-13. [Google Scholar] [CrossRef] [Green Version]
- Latsuzbaia, A.; Arbyn, M.; Dutta, S.; Fischer, M.; Gheit, T.; Tapp, J.; Tommasino, M.; Weyers, S.; Mossong, J. Complete Genome Sequence of a Novel Human Gammapapillomavirus Isolated from a Cervical Swab in Luxembourg. Genome Announc. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Van Doorslaer, K.; Li, Z.; Xirasagar, S.; Maes, P.; Kaminsky, D.; Liou, D.; Sun, Q.; Kaur, R.; Huyen, Y.; McBride, A.A. The Papillomavirus Episteme: A major update to the papillomavirus sequence database. Nucleic Acids Res. 2017, 45, D499–D506. [Google Scholar] [CrossRef]
- Mühr, L.S.A.; Eklund, C.; Dillner, J. Towards quality and order in human papillomavirus research. Virology 2018, 519, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.R.; Cai, Y.C.; Giesler, T.L.; Farchaus, J.W.; Sundaram, S.T.; Ortiz-Rivera, M.; Hosta, L.P.; Hewitt, P.L.; Mamone, J.A.; Palaniappan, C.; et al. TempliPhi, phi29 DNA polymerase based rolling circle amplification of templates for DNA sequencing. Biotechniques 2002, 32, 44–47. [Google Scholar] [CrossRef] [Green Version]
- Rector, A.; Tachezy, R.; Van Ranst, M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004, 78, 4993–4998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murahwa, A.T.; Meiring, T.L.; Mbulawa, Z.Z.A.; Williamson, A.L. Discovery, characterisation and genomic variation of six novel Gammapapillomavirus types from penile swabs in South Africa. Papillomavirus Res. 2019, 7, 102–111. [Google Scholar] [CrossRef]
- Pastrana, D.V.; Peretti, A.; Welch, N.L.; Borgogna, C.; Olivero, C.; Badolato, R.; Notarangelo, L.D.; Gariglio, M.; FitzGerald, P.C.; McIntosh, C.E.; et al. Metagenomic Discovery of 83 New Human Papillomavirus Types in Patients with Immunodeficiency. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brancaccio, R.N.; Robitaille, A.; Dutta, S.; Rollison, D.E.; Fischer, N.; Grundhoff, A.; Tommasino, M.; Gheit, T. Complete Genome Sequence of a Novel Human Gammapapillomavirus Isolated from Skin. Genome Announc. 2017, 5, e00833-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latsuzbaia, A.; Hebette, G.; Fischer, M.; Arbyn, M.; Weyers, S.; Vielh, P.; Schmitt, F.; Mossong, J. Introduction of liquid-based cytology and human papillomavirus testing in cervical cancer screening in Luxembourg. Diagn. Cytopathol. 2017, 45, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Brestovac, B.; Wong, M.E.; Costantino, P.S.; Groth, D. A rapid DNA extraction method suitable for human papillomavirus detection. J. Med. Virol. 2014, 86, 653–657. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Lee, B.; Lee, K.N.; Kim, Y.; Oh, E.J. Clinical Validation of Anyplex II HPV HR Detection Test for Cervical Cancer Screening in Korea. Arch. Pathol. Lab. Med. 2016, 140, 276–280. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 15 November 2018).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Shean, R.C.; Makhsous, N.; Stoddard, G.D.; Lin, M.J.; Greninger, A.L. VAPiD: A lightweight cross-platform viral annotation pipeline and identification tool to facilitate virus genome submissions to NCBI GenBank. BMC Bioinform. 2019, 20, 48. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef] [Green Version]
- Arbyn, M.; Depuydt, C.; Benoy, I.; Bogers, J.; Cuschieri, K.; Schmitt, M.; Pawlita, M.; Geraets, D.; Heard, I.; Gheit, T.; et al. VALGENT: A protocol for clinical validation of human papillomavirus assays. J. Clin. Virol. 2016, 76, S14–S21. [Google Scholar] [CrossRef] [PubMed]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Baasland, I.; Romundstad, P.R.; Eide, M.L.; Jonassen, C.M. Clinical performance of Anyplex II HPV28 by human papillomavirus type and viral load in a referral population. PLoS ONE 2019, 14, e0210997. [Google Scholar] [CrossRef] [Green Version]
- Latsuzbaia, A.; Arbyn, M.; Weyers, S.; Mossong, J. Human papillomavirus vaccination coverage in Luxembourg—Implications of lowering and restricting target age groups. Vaccine 2018, 36, 2411–2416. [Google Scholar] [CrossRef]
- Park, Y.; Lee, E.; Choi, J.; Jeong, S.; Kim, H.S. Comparison of the Abbott RealTime High-Risk Human Papillomavirus (HPV), Roche Cobas HPV, and Hybrid Capture 2 assays to direct sequencing and genotyping of HPV DNA. J. Clin. Microbiol. 2012, 50, 2359–2365. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Unger, E.R.; Batra, D.; Sheth, M.; Steinau, M.; Jasinski, J.; Jones, J.; Rajeevan, M.S. Universal Human Papillomavirus Typing Assay: Whole-Genome Sequencing following Target Enrichment. J. Clin. Microbiol. 2017, 55, 811–823. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Unger, E.R.; Rajeevan, M.S. Universal human papillomavirus typing by whole genome sequencing following target enrichment: Evaluation of assay reproducibility and limit of detection. BMC Genom. 2019, 20, 231. [Google Scholar] [CrossRef]
- Meiring, T.L.; Salimo, A.T.; Coetzee, B.; Maree, H.J.; Moodley, J.; Hitzeroth, I.I.; Freeborough, M.J.; Rybicki, E.P.; Williamson, A.L. Next-generation sequencing of cervical DNA detects human papillomavirus types not detected by commercial kits. Virol. J. 2012, 9, 164. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, Y.; Ni, T.; Xie, X.; Zhu, J.; Zheng, Z.-M. Genome sequencing accuracy by RCA-seq versus long PCR template cloning and sequencing in identification of human papillomavirus type 58. Cell Biosci. 2014, 4, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Schiffman, M.; Herrero, R.; Desalle, R.; Burk, R.D. Human papillomavirus (HPV) types 101 and 103 isolated from cervicovaginal cells lack an E6 open reading frame (ORF) and are related to gamma-papillomaviruses. Virology 2007, 360, 447–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Doorslaer, K.; McBride, A.A. Molecular archeological evidence in support of the repeated loss of a papillomavirus gene. Sci. Rep. 2016, 6, 33028. [Google Scholar] [CrossRef] [PubMed]
- Nobre, R.J.; Herraez-Hernandez, E.; Fei, J.W.; Langbein, L.; Kaden, S.; Grone, H.J.; de Villiers, E.M. E7 oncoprotein of novel human papillomavirus type 108 lacking the E6 gene induces dysplasia in organotypic keratinocyte cultures. J. Virol. 2009, 83, 2907–2916. [Google Scholar] [CrossRef] [Green Version]
- Ameur, A.; Meiring, T.L.; Bunikis, I.; Haggqvist, S.; Lindau, C.; Lindberg, J.H.; Gustavsson, I.; Mbulawa, Z.Z.; Williamson, A.L.; Gyllensten, U. Comprehensive profiling of the vaginal microbiome in HIV positive women using massive parallel semiconductor sequencing. Sci. Rep. 2014, 4, 4398. [Google Scholar] [CrossRef]

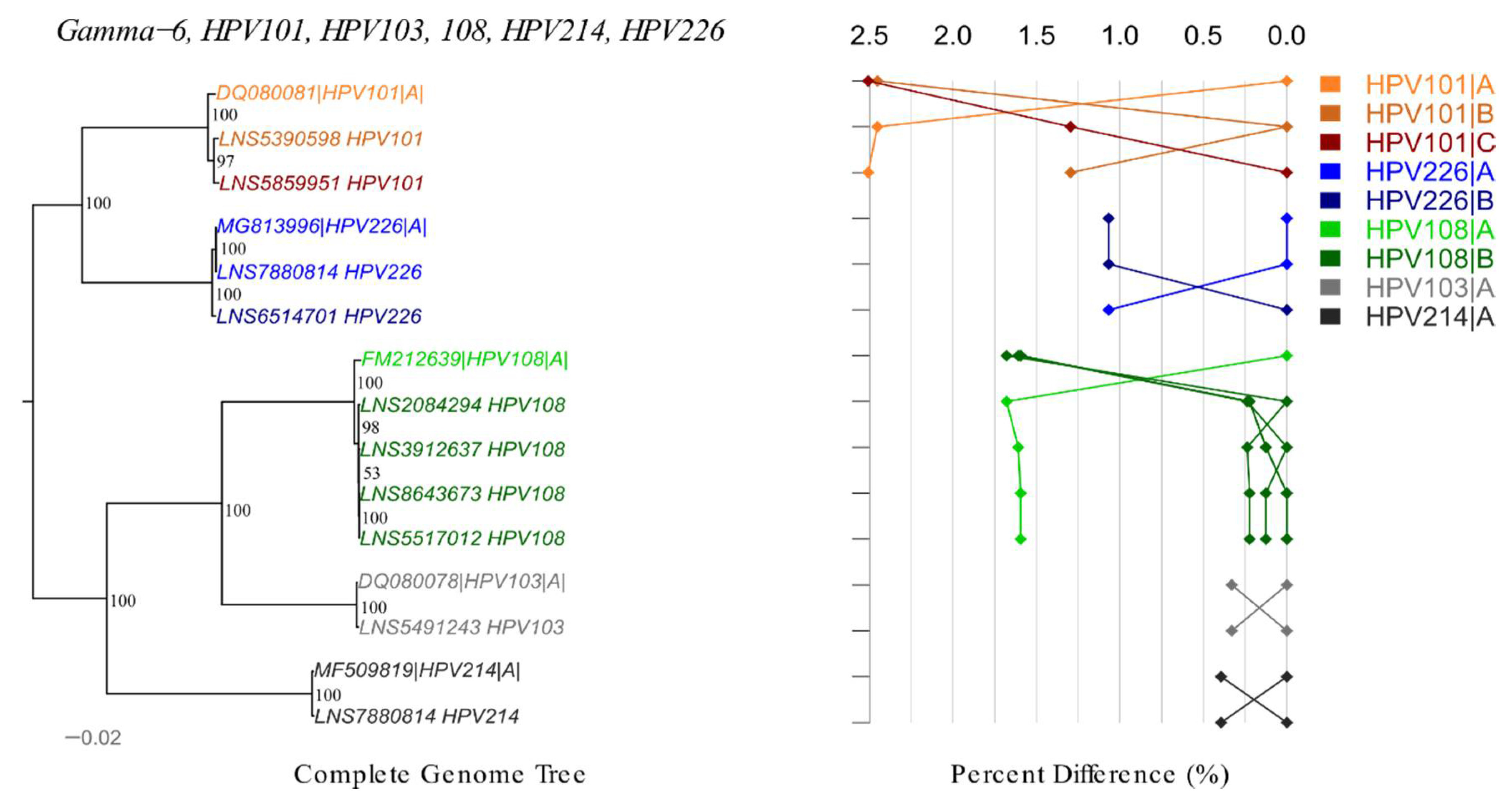
 ) indicates novel lineage/sublineage. The phylogenetic topology is shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.
) indicates novel lineage/sublineage. The phylogenetic topology is shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.
 ) indicates novel lineage/sublineage. The phylogenetic topology is shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.
) indicates novel lineage/sublineage. The phylogenetic topology is shown on the left, pairwise differences for each isolate are shown on the right. Values for each isolate are connected by lines of different colors to distinguish each lineage and sublineage. Genomes from our collection are indicated with LNS identification number.

| Number of Genotypes Detected | Number of Reads per Genotype | Number of Reads per Sample | |||||
|---|---|---|---|---|---|---|---|
| Anyplex Viral Load | Anyplex | Concordant by NGS (%) | p Value | Mean | p Value | Mean | p Value |
| Low (+) | 248 | 40 (16.1) | <0.001 | 1017 | <0.001 | 905 | <0.001 |
| Medium (++) | 344 | 223 (64.8) | 1028 | 2390 | |||
| High (+++) | 186 | 178 (95.7) | 9994 | 18,162 | |||
| HPV Types 1 | A+/NGS+ 2 | A+/NGS− 3 | A-/NGS+ 4 | A-/NGS− 5 | % Agreement | Kappa (se) | Interp-Retation 6 |
|---|---|---|---|---|---|---|---|
| 28 HPV types 7 | 258 | 108 | 32 | 331 | 80.8 | 0.616 (0.036) | G |
| 28 HPV types (++/+++) 8 | 236 | 46 | 54 | 393 | 86.3 | 0.712 (0.037) | G |
| 14 HPV types 9 | 179 | 71 | 40 | 439 | 84.8 | 0.652 (0.036) | G |
| 14 HPV types (++/+++) 10 | 167 | 52 | 42 | 468 | 87.1 | 0.689 (0.037) | G |
| Species | HPV Type | Genomes Sequenced | No. of Genomes on NCBI 1 | No. of Genomes Used 2 | Genome Size 3 | GC Content (%) | Existing Lineage/Sublineage Sequenced | Novel Lineage/Sublineage Proposed |
|---|---|---|---|---|---|---|---|---|
| α-9 | HPV16 | 2 | 566 | 10 | 7906–7907 | 36 | A1 | - |
| α-9 | HPV31 | 11 | 29 | 7 | 7878–7920 | 37 | A1, B2, C2–C3 | - |
| α-9 | HPV33 | 2 | 29 | 5 | 7833–7911 | 36 | A1–A2 | - |
| α-9 | HPV35 | 2 | 30 | 2 | 7879–7880 | 36 | A1 | - |
| α-7 | HPV39 | 11 | 22 | 3 | 7817–7885 | 40 | A1–A2 | - |
| α-7 | HPV45 | 3 | 23 | 5 | 7849–7866 | 39 | A1, B2 | C |
| α-5 | HPV51 | 22 | 31 | 6 | 7811–7815 | 39 | A1–A3, B1 | - |
| α-9 | HPV52 | 7 | 93 | 7 | 7933–7962 | 38 | A1–A2, | E |
| α-6 | HPV56 | 12 | 13 | 3 | 7790–7866 | 37 | A1–A2, B | - |
| α-9 | HPV58 | 8 | 149 | 8 | 7823–7825 | 37–38 | A2, B1–B2 | - |
| α-7 | HPV59 | 18 | 14 | 4 | 7898–7913 | 38 | A1, B1 | B2 |
| α-6 | HPV66 | 15 | 16 | 3 | 7816–7824 | 38 | A, B2 | C |
| α-7 | HPV68 | 5 | 31 | 10 | 7814–7835 | 39–40 | A1–A2, B, F2 | - |
| Species | HPV Type | Genomes Sequenced | No. of Genomes on NCBI 1 | No. of Genomes Used 2 | Genome Size 3 | GC Content (%) | Existing Lineage/Sublineage Sequenced | Novel Lineage/Sublineage Proposed |
|---|---|---|---|---|---|---|---|---|
| α-10 | HPV6 | 4 | 214 | 4 | 8020–8032 | 40 | B1, B3 | – |
| α-6 | HPV30 | 2 | 17 | 6 | 7843–7881 | 40 | A2–A3 | – |
| α-11 | HPV34 | 4 | 17 | 5 | 7668–7788 | 37–38 | A2, C2 | D |
| α-8 | HPV40 | 3 | 4 | 4 | 7905–7909 | 43 | – | A2–A4 |
| α-1 | HPV42 | 21 | 8 | 8 | 7901–7920 | 39 | A1, A2 | A3, B, C |
| α-8 | HPV43 | 2 | 2 | 2 | 7986–8007 | 40 | – | B1–B2 |
| α-10 | HPV44 | 3 | 6 | 6 | 7822–7837 | 40–41 | A, B | – |
| α-6 | HPV53 | 21 | 29 | 7 | 7859–7864 | 40 | A, C, D1–D2 | – |
| α-13 | HPV54 | 6 | 12 | 5 | 7760–7776 | 41 | A2 | D |
| α-3 | HPV61 | 1 | 12 | 4 | 7989 | 46 | – | A3 |
| α-3 | HPV62 | 3 | 4 | 4 | 8092–8092 | 45–46 | A1 | A2 |
| α-9 | HPV67 | 12 | 12 | 3 | 7806–7809 | 38 | B1 | B2 |
| α-11 | HPV73 | 7 | 16 | 3 | 7694–7716 | 36 | A2, B | C |
| α-7 | HPV70 | 2 | 10 | 2 | 7905–7911 | 40 | A | – |
| α-10 | HPV74 | 3 | 1 | 1 | 7893–7902 | 40–41 | – | B, C |
| α-5 | HPV82 | 2 | 22 | 10 | 7868–7874 | 40 | A3, B1 | – |
| α-3 | HPV84 | 2 | 1 | 1 | 7956–7974 | 46 | – | B1–B2 |
| α-3 | HPV87 | 4 | 4 | 4 | 7998–8001 | 45 | A1 | – |
| α-3 | HPV89 | 1 | 4 | 4 | 8072 | 45 | A2 | – |
| α-14 | HPV90 | 7 | 3 | 8016–8032 | 46 | A1 | A3, B | |
| α-8 | HPV91 | 4 | 1 | 1 | 7959–7959 | 40 | – | A2 |
| γ-6 | HPV101 | 2 | 1 | 1 | 7258–7259 | 43 | – | B, C |
| γ-6 | HPV103 | 1 | 1 | 1 | 7263 | 41 | – | – |
| γ-6 | HPV108 | 4 | 1 | 1 | 7158–7158 | 42 | – | B |
| γ-6 | HPV214 | 1 | 1 | 1 | 7357 | 41 | – | – |
| γ-6 | HPV226 | 2 | 1 | 1 | 7313–7315 | 42 | B | |
| γ-13 | HPV213 | 1 | - | 7096 | 39 | – | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latsuzbaia, A.; Wienecke-Baldacchino, A.; Tapp, J.; Arbyn, M.; Karabegović, I.; Chen, Z.; Fischer, M.; Mühlschlegel, F.; Weyers, S.; Pesch, P.; et al. Characterization and Diversity of 243 Complete Human Papillomavirus Genomes in Cervical Swabs Using Next Generation Sequencing. Viruses 2020, 12, 1437. https://doi.org/10.3390/v12121437
Latsuzbaia A, Wienecke-Baldacchino A, Tapp J, Arbyn M, Karabegović I, Chen Z, Fischer M, Mühlschlegel F, Weyers S, Pesch P, et al. Characterization and Diversity of 243 Complete Human Papillomavirus Genomes in Cervical Swabs Using Next Generation Sequencing. Viruses. 2020; 12(12):1437. https://doi.org/10.3390/v12121437
Chicago/Turabian StyleLatsuzbaia, Ardashel, Anke Wienecke-Baldacchino, Jessica Tapp, Marc Arbyn, Irma Karabegović, Zigui Chen, Marc Fischer, Friedrich Mühlschlegel, Steven Weyers, Pascale Pesch, and et al. 2020. "Characterization and Diversity of 243 Complete Human Papillomavirus Genomes in Cervical Swabs Using Next Generation Sequencing" Viruses 12, no. 12: 1437. https://doi.org/10.3390/v12121437
APA StyleLatsuzbaia, A., Wienecke-Baldacchino, A., Tapp, J., Arbyn, M., Karabegović, I., Chen, Z., Fischer, M., Mühlschlegel, F., Weyers, S., Pesch, P., & Mossong, J. (2020). Characterization and Diversity of 243 Complete Human Papillomavirus Genomes in Cervical Swabs Using Next Generation Sequencing. Viruses, 12(12), 1437. https://doi.org/10.3390/v12121437






