Influenza A H1 and H3 Transmembrane Domains Interact Differently with Each Other and with Surrounding Membrane Lipids
Abstract
:1. Introduction
2. Materials and Methods
3. Results
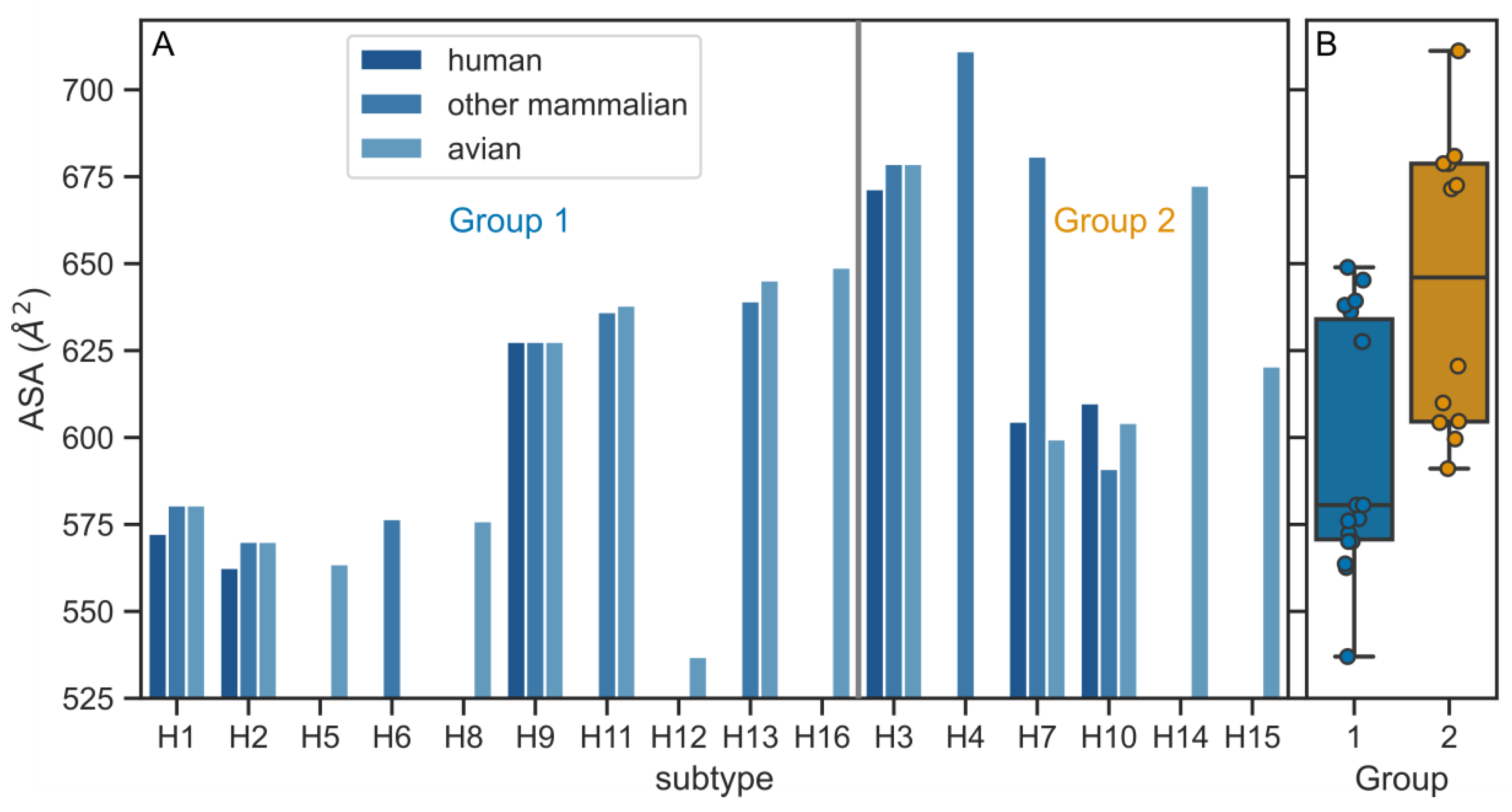
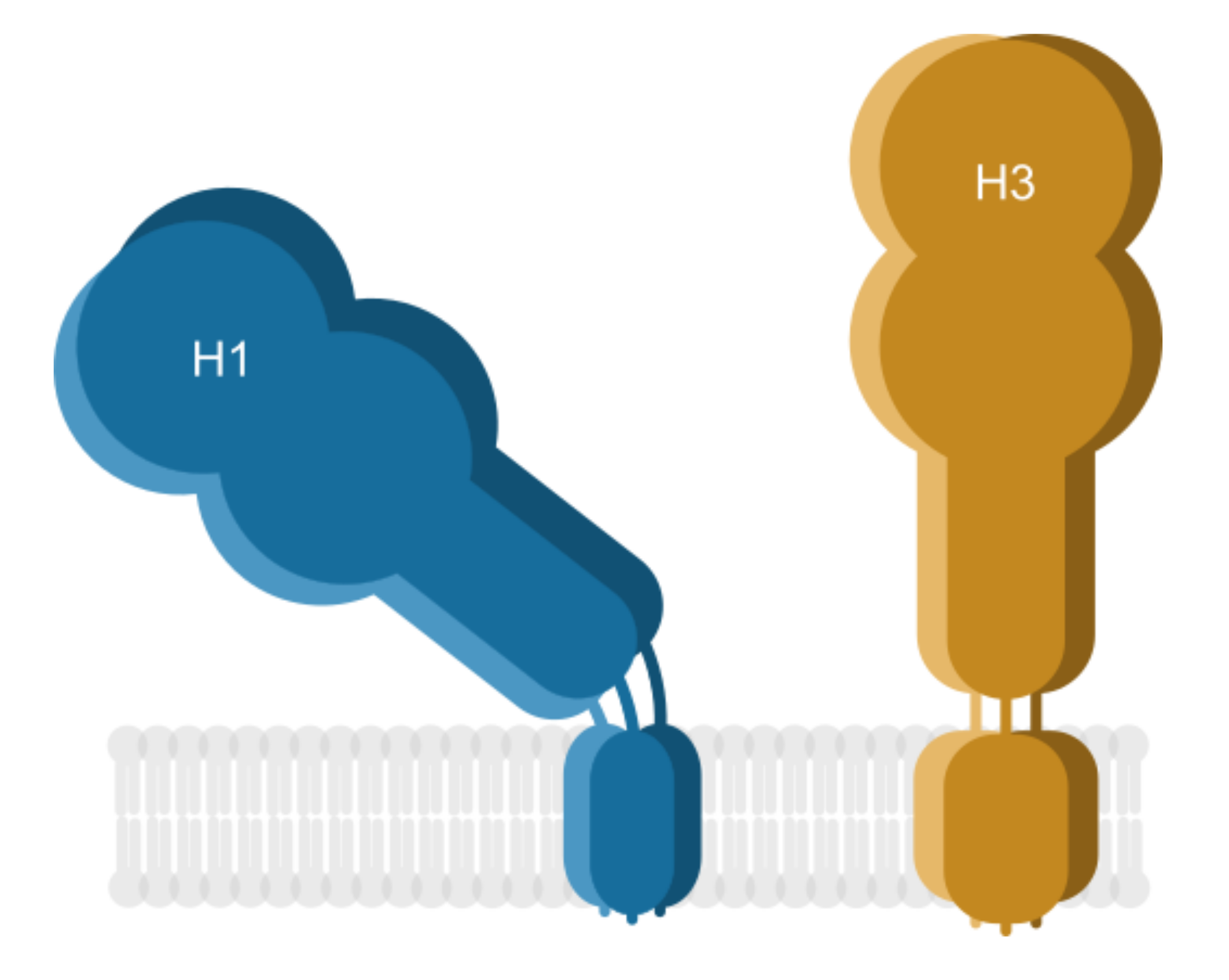
3.1. HA TMDs Differ between H1 and H3 Subtypes
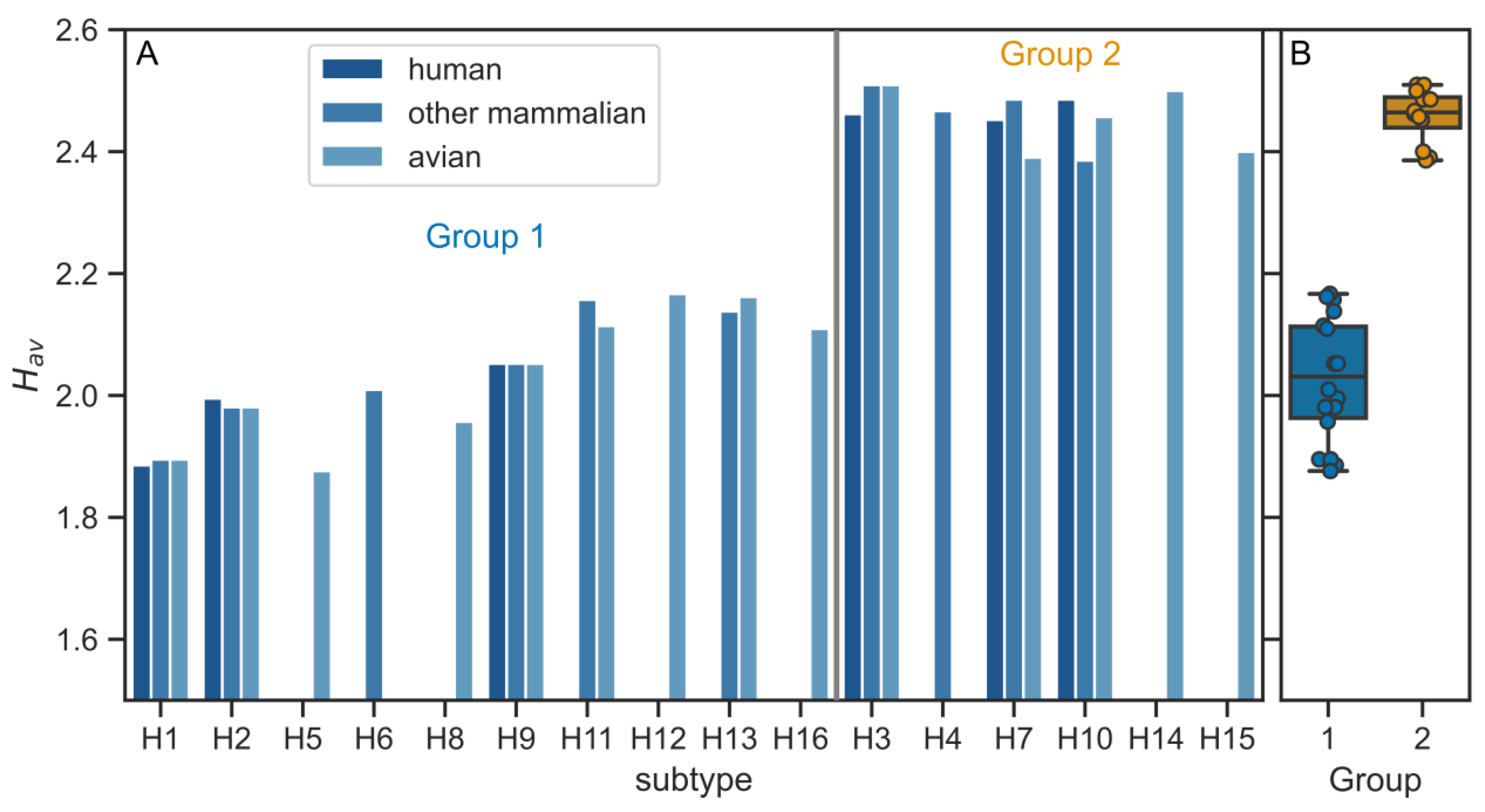
3.2. Physico-Chemical Parameters of HA TMDs Differ between Phylogenetic Groups
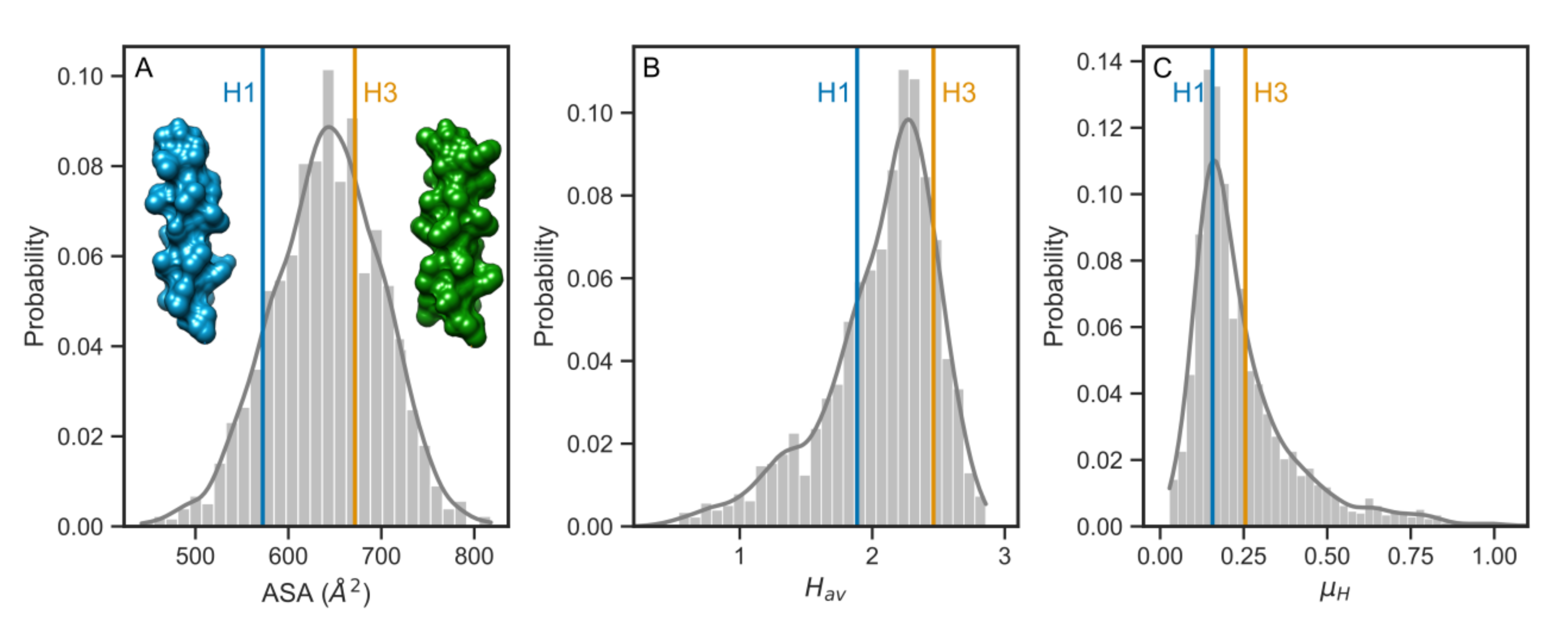
3.3. Comparison with the Human Single-Pass TMDs
3.4. The Pre-TMD Region Also Differs between H1 and H3 Subtypes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Drake, J.W. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA 1993, 90, 4171–4175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heaton, N.S.; Sachs, D.; Chen, C.J.; Hai, R.; Palese, P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 20248–20253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.R.; Liu, Y.M.; Tseng, Y.C.; Ma, C. Better influenza vaccines: An industry perspective. J. Biomed. Sci. 2020, 27, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Copeland, C.S.; Doms, R.W.; Bolzau, E.M.; Webster, R.G.; Helenius, A. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J. Cell Biol. 1986, 103, 1179–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvier, N.; Palese, P. The Biology of Influenza Viruses. Vaccine 2008, 26, D49–D53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.; Friesen, R.H.E.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope: Implications for universal prevention and therapy. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, S.; Holmes, E.C.; Pybus, O.G. The genomic rate of molecular adaptation of the human influenza A virus. Mol. Biol. Evol. 2011, 28, 2443–2451. [Google Scholar] [CrossRef] [Green Version]
- Kirkpatrick, E.; Qiu, X.; Wilson, P.C.; Bahl, J.; Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Air, G.M. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc. Natl. Acad. Sci. USA 1981, 78, 7639–7643. [Google Scholar] [CrossRef] [Green Version]
- Nobusawa, E.; Aoyama, T.; Kato, H.; Suzuki, Y.; Tateno, Y.; Nakajima, K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 1991, 182, 475–485. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, C.; Zhang, H.; Liu, G.D.; Xue, C.; Cao, Y. Targeting hemagglutinin: Approaches for broad protection against the influenza a virus. Viruses 2019, 11, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benton, D.J.; Nans, A.; Calder, L.J.; Turner, J.; Neu, U.; Lin, Y.P.; Ketelaars, E.; Kallewaard, N.L.; Corti, D.; Lanzavecchia, A.; et al. Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. USA 2018, 115, 10112–10117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, D.-K.; Cheng, S.-F.; Kantchev, E.A.B.; Lin, C.-H.; Liu, Y.-T. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, A.L.; Freed, J.H. The Interaction between Influenza HA Fusion Peptide and Transmembrane Domain Affects Membrane Structure. Biophys. J. 2015, 109, 2523–2536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranaweera, A.; Ratnayake, P.U.; Ekanayaka, E.A.P.; Declercq, R.; Weliky, D.P. Hydrogen-Deuterium Exchange Supports Independent Membrane-Interfacial Fusion Peptide and Transmembrane Domains in Subunit 2 of Influenza Virus Hemagglutinin Protein, a Structured and Aqueous-Protected Connection between the Fusion Peptide and Soluble Ecto. Biochemistry 2019, 58, 2432–2446. [Google Scholar] [CrossRef]
- Schroth-Diez, B.; Ludwig, K.; Baljinnyam, B.; Kozerski, C.; Huang, Q.; Herrmann, A. The role of the transmembrane and of the intraviral domain of glycoproteins in membrane fusion of enveloped viruses. Biosci. Rep. 2000, 20, 571–595. [Google Scholar] [CrossRef]
- Ge, M.; Freed, J.H. Two conserved residues are important for inducing highly ordered membrane domains by the transmembrane domain of influenza hemagglutinin. Biophys. J. 2011, 100, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Scheiffele, P.; Roth, M.G.; Simons, K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 1997, 16, 5501–5508. [Google Scholar] [CrossRef]
- Takeda, M.; Leser, G.P.; Russell, C.J.; Lamb, R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 2003, 100, 14610–14617. [Google Scholar] [CrossRef] [Green Version]
- De Vries, M.; Herrmann, A.; Veit, M. A cholesterol consensus motif is required for efficient intracellular transport and raft association of a group 2 HA from influenza virus. Biochem. J. 2015, 465, 305–314. [Google Scholar] [CrossRef]
- Hu, B.; Höfer, C.T.; Thiele, C.; Veit, M. Cholesterol Binding to the Transmembrane Region of a Group 2 Hemagglutinin (HA) of Influenza Virus Is Essential for Virus Replication, Affecting both Virus Assembly and HA Fusion Activity. J. Virol. 2019, 93, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veit, M. Palmitoylation of virus proteins. Biol. Cell 2012, 104, 493–515. [Google Scholar] [CrossRef] [PubMed]
- Veit, M.; Serebryakova, M.V.; Kordyukova, L.V. Palmitoylation of influenza virus proteins. Biochem. Soc. Trans. 2013, 41, 50–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Zhou, J.; Liu, K.; Liu, Q.; Xue, C.; Li, X.; Zheng, J.; Luo, D.; Cao, Y. Mutations of two transmembrane cysteines of hemagglutinin (HA) from influenza A H3N2 virus affect HA thermal stability and fusion activity. Virus Genes 2013, 47, 20–26. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, S.; Ma, J.; Lei, W.; Liu, K.; Liu, Q.; Ren, Y.; Xue, C.; Cao, Y. Recombinant influenza A H3N2 viruses with mutations of HA transmembrane cysteines exhibited altered virological characteristics. Virus Genes 2014, 48, 273–282. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Y.; Liu, K.; Huang, M.; Li, R.; Wang, Y.; Liu, Q.; Zheng, J.; Xue, C.; Cao, Y. Recombinant influenza H9N2 virus with a substitution of H3 hemagglutinin transmembrane domain showed enhanced immunogenicity in mice and chicken. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, K.; Xue, C.; Zhou, J.; Li, X.; Luo, D.; Zheng, J.; Xu, S.; Liu, G.D.; Cao, Y. Recombinant influenza H1, H5 and H9 hemagglutinins containing replaced H3 hemagglutinin transmembrane domain showed enhanced heterosubtypic protection in mice. Vaccine 2014, 32, 3041–3049. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Xue, C.; Wu, Z.; Lin, Y.; Wei, Y.; Wei, X.; Qin, J.; Zhang, Y.; Wen, Z.; et al. A recombinant H7N9 influenza vaccine with the H7 hemagglutinin transmembrane domain replaced by the H3 domain induces increased cross-reactive antibodies and improved interclade protection in mice. Antivir. Res. 2017, 143, 97–105. [Google Scholar] [CrossRef]
- Liu, Q.; Xue, C.; Zheng, J.; Liu, K.; Wang, Y.; Wei, Y.; Liu, G.D.; Cao, Y. Influenza bivalent vaccine comprising recombinant H3 hemagglutinin (HA) and H1 HA containing replaced H3 hemagglutinin transmembrane domain exhibited improved heterosubtypic protection immunity in mice. Vaccine 2015, 33, 4035–4040. [Google Scholar] [CrossRef]
- Qin, J.; Zhang, Y.; Shen, X.; Gong, L.; Peng, O.; Liu, Y.; Xue, C.; Cao, Y. H7 virus-like particles assembled by hemagglutinin containing H3N2 transmembrane domain and M1 induce broad homologous and heterologous protection in mice. Vaccine 2018, 36, 5030–5036. [Google Scholar] [CrossRef]
- Senes, A.; Gerstein, M.; Engelman, D.M. Statistical analysis of amino acid patterns in transmembrane helices: The GxxxG motif occurs frequently and association with β-branched residues at neighboring positions. J. Mol. Biol. 2000, 296, 921–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unterreitmeier, S.; Fuchs, A.; Schäffler, T.; Heym, R.G.; Frishman, D.; Langosch, D. Phenylalanine Promotes Interaction of Transmembrane Domains via GxxxG Motifs. J. Mol. Biol. 2007, 374, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Worch, R. The helical hairpin structure of the influenza fusion peptide can be seen on a hydrophobic moment map. FEBS Lett. 2013, 587, 2980–2983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelman, D.M.; Steitz, T.A.; Goldman, A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 1986, 15, 321–353. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhang, F.; Davis, M.J.; Bodén, M.; Teasdale, R.D. Predicting the solvent accessibility of transmembrane residues from protein sequence. J. Proteome Res. 2006, 5, 1063–1070. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature 1982, 299, 371–374. [Google Scholar] [CrossRef]
- Influenza, Surveillance and Monitoring, Update 377. Available online: https://www.who.int/influenza/surveillance_monitoring/updates/latest_update_GIP_surveillance/en/ (accessed on 27 October 2020).
- Phoenix, D.A.; Harris, F. The hydrophobic moment and its use in the classification of amphiphilic structures (review). Mol. Membr. Biol. 2002, 19, 1–10. [Google Scholar] [CrossRef]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA 1984, 81, 140–144. [Google Scholar] [CrossRef] [Green Version]
- Dawson, J.P.; Weinger, J.S.; Engelman, D.M. Motifs of serine and threonine can drive association of transmembrane helices. J. Mol. Biol. 2002, 316, 799–805. [Google Scholar] [CrossRef]
- Worch, R.; Bökel, C.; Höfinger, S.; Schwille, P.; Weidemann, T. Focus on composition and interaction potential of single-pass transmembrane domains. Proteomics 2010, 10, 4196–4208. [Google Scholar] [CrossRef] [PubMed]
- Langosch, D.; Arkin, I.T. Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 2009, 18, 1343–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sal-Man, N.; Gerber, D.; Bloch, I.; Shai, Y. Specificity in transmembrane helix-helix interactions mediated by aromatic residues. J. Biol. Chem. 2007, 282, 19753–19761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, R.M.; Hecht, K.; Deber, C.M. Aromatic and cation-π interactions enhance helix-helix association in a membrane environment. Biochemistry 2007, 46, 9208–9214. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, L.V.; Serebryakova, M.V.; Polyansky, A.A.; Kropotkina, E.A.; Alexeevski, A.V.; Veit, M.; Efremov, R.G.; Filippova, I.Y.; Baratova, L.A. Linker and/or transmembrane regions of influenza A/Group-1, A/Group-2, and type B virus hemagglutinins are packed differently within trimers. Biochim. Biophys. Acta Biomembr. 2011, 1808, 1843–1854. [Google Scholar] [CrossRef] [Green Version]
- Lorent, J.H.; Diaz-Rohrer, B.; Lin, X.; Spring, K.; Gorfe, A.A.; Levental, K.R.; Levental, I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Turner, N.; Haga, K.L.; Hulbert, A.J.; Else, P.L. Relationship between body size, Na+-K+-ATPase activity, and membrane lipid composition in mammal and bird kidney. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Turner, N.; Else, P.L.; Hulbert, A.J. An allometric comparison of microsomal membrane lipid composition and sodium pump molecular activity in the brain of mammals and birds. J. Exp. Biol. 2005, 208, 371–381. [Google Scholar] [CrossRef] [Green Version]
- Levental, I.; Levental, K.R.; Heberle, F.A. Lipid Rafts: Controversies Resolved, Mysteries Remain. Trends Cell Biol. 2020, 30, 341–353. [Google Scholar] [CrossRef]
- Leser, G.P.; Lamb, R.A. Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 2005, 342, 215–227. [Google Scholar] [CrossRef] [Green Version]
- Hess, S.T.; Gould, T.J.; Gudheti, M.V.; Maas, S.A.; Mills, K.D.; Zimmerberg, J. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA 2007, 104, 17370–17375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerl, M.J.; Sampaio, J.L.; Urban, S.; Kalvodova, L.; Verbavatz, J.M.; Binnington, B.; Lindemann, D.; Lingwood, C.A.; Shevchenko, A.; Schroeder, C.; et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012, 196, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurcher, T.; Luo, G.; Palese, P. Mutations at palmitylation sites of the influenza virus hemagglutinin affect virus formation. J. Virol. 1994, 68, 5748–5754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, R.; Herwig, A.; Azzouz, N.; Klenk, H.D. Acylation-Mediated Membrane Anchoring of Avian Influenza Virus Hemagglutinin Is Essential for Fusion Pore Formation and Virus Infectivity. J. Virol. 2005, 79, 6449–6458. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.J.; Takeda, M.; Lamb, R.A. Influenza Virus Hemagglutinin (H3 Subtype) Requires Palmitoylation of Its Cytoplasmic Tail for Assembly: M1 Proteins of Two Subtypes Differ in Their Ability To Support Assembly. J. Virol. 2005, 79, 13673–13684. [Google Scholar] [CrossRef] [Green Version]
- Kordyukova, L.V.; Serebryakova, M.V.; Baratova, L.A.; Veit, M. S Acylation of the Hemagglutinin of Influenza Viruses: Mass Spectrometry Reveals Site-Specific Attachment of Stearic Acid to a Transmembrane Cysteine. J. Virol. 2008, 82, 9288–9292. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Gorfe, A.A.; Levental, I. Protein Partitioning into Ordered Membrane Domains: Insights from Simulations. Biophys. J. 2018, 114, 1936–1944. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, P.; Seo, A.Y.; Pasolli, H.A.; Song, Y.E.; Johnson, M.C.; Lippincott-Schwartz, J. A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 2019, 21, 452–461. [Google Scholar] [CrossRef]
- Hofmann, M.W.; Weise, K.; Ollesch, J.; Agrawal, P.; Stalz, H.; Stelzer, W.; Hulsbergen, F.; De Groot, H.; Gerwert, K.; Reed, J.; et al. De novo design of conformationally flexible transmembrane peptides driving membrane fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 14776–14781. [Google Scholar] [CrossRef] [Green Version]
- Coskun, Ü.; Simons, K. Cell membranes: The lipid perspective. Structure 2011, 19, 1543–1548. [Google Scholar] [CrossRef] [Green Version]
- Halbleib, K.; Pesek, K.; Covino, R.; Hofbauer, H.F.; Wunnicke, D.; Hänelt, I.; Hummer, G.; Ernst, R. Activation of the Unfolded Protein Response by Lipid Bilayer Stress. Mol. Cell 2017, 67, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-J.; Xu, L.; Kong, W.-P.; Shi, W.; Canis, K.; Stevens, J.; Yang, Z.-Y.; Dell, A.; Haslam, S.M.; Wilson, I.A.; et al. Comparative Efficacy of Neutralizing Antibodies Elicited by Recombinant Hemagglutinin Proteins from Avian H5N1 Influenza Virus. J. Virol. 2008, 82, 6200–6208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weldon, W.C.; Wang, B.Z.; Martin, M.P.; Koutsonanos, D.G.; Skountzou, I.; Compans, R.W. Enhanced immunogenicity of stabilized trimeric soluble influenza hemagglutinin. PLoS ONE 2010, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Wei, C.J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.R.; Rao, S.S.; Kong, W.P.; Wang, L.; Nabel, G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Kemble, G.W.; Bodian, D.L.; Rosé, J.; Wilson, I.A.; White, J.M. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J. Virol. 1992, 66, 4940–4950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemble, G.W.; Danieli, T.; White, J.M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 1994, 76, 383–391. [Google Scholar] [CrossRef]
- Xu, S.; Zhou, J.; Liu, Q.; Liu, K.; Xue, C.; Li, X.; Zheng, J.; Luo, D.; Cao, Y. Evidences for the existence of intermolecular disulfide-bonded oligomers in the H3 hemagglutinins expressed in insect cells. Virus Genes 2014, 48, 304–311. [Google Scholar] [CrossRef]


| Subtype | Host | Occurrence (%) | Sequence |
|---|---|---|---|
| H1 | Human | 63.1 | ILAIYSTVASSLVLVVSLGAI |
| H1 | Other mammalian | 38.9 | ILAIYSTVASSLVLLVSLGAI |
| H1 | Avian | 85.6 | ILAIYSTVASSLVLLVSLGAI |
| H3 | Human | 67.1 | -LWISFAISCFLLCVALLGFIM |
| H3 | Other mammalian | 55.0 | -LWISFAISCFLLCVVLLGFIM |
| H3 | Avian | 79.4 | -LWISFAISCFLLCVVLLGFIM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubiszewski-Jakubiak, S.; Worch, R. Influenza A H1 and H3 Transmembrane Domains Interact Differently with Each Other and with Surrounding Membrane Lipids. Viruses 2020, 12, 1461. https://doi.org/10.3390/v12121461
Kubiszewski-Jakubiak S, Worch R. Influenza A H1 and H3 Transmembrane Domains Interact Differently with Each Other and with Surrounding Membrane Lipids. Viruses. 2020; 12(12):1461. https://doi.org/10.3390/v12121461
Chicago/Turabian StyleKubiszewski-Jakubiak, Szymon, and Remigiusz Worch. 2020. "Influenza A H1 and H3 Transmembrane Domains Interact Differently with Each Other and with Surrounding Membrane Lipids" Viruses 12, no. 12: 1461. https://doi.org/10.3390/v12121461
APA StyleKubiszewski-Jakubiak, S., & Worch, R. (2020). Influenza A H1 and H3 Transmembrane Domains Interact Differently with Each Other and with Surrounding Membrane Lipids. Viruses, 12(12), 1461. https://doi.org/10.3390/v12121461






